ABSTRACT
Transcriptomics studies the set of RNA transcripts produced by the genome using high-throughput sequencing and bioinformatics. This growing field has revolutionized our understanding of host-pathogen interactions, revealing new insights into the host response to influenza infection and vaccination. Studies using transcriptomics have identified a unique immunosignature for influenza discernable from other bacterial and viral pathogens, key transcriptional factors that discriminate early from late, mild versus severe, and symptomatic versus asymptomatic infection. Recent studies evaluating the host response to influenza vaccines have revealed key differences in live versus inactivated influenza vaccines, identified early transcriptional signatures that predict hemagglutinin antibody production following vaccination, increased our understanding of how adjuvants enhance the immune response to influenza vaccine antigens, and demonstrate biologic variability in the response to vaccination due to host factors. These studies demonstrate the potential for influenza transcriptomics to be applied to clinical care, understanding the mechanisms of infection, and informing vaccine development.
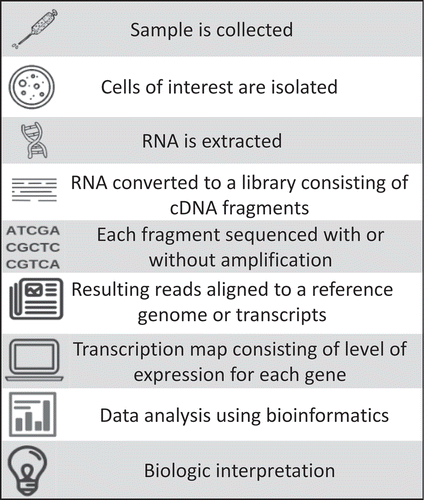
Transcriptomics is the study of the complete set of RNA transcripts that are produced by the genome (under specific circumstances or in a specific cell or group of cells) using high-throughput sequencing. Comparing transcriptomes within groups enables the identification of genes that are differentially expressed in specific cell populations or in response to different treatments. Technologies used to conduct transcriptomics include microarrays and RNA sequencing. Microarrays are a fixed-probe technology, while RNA sequencing is more dynamic, and measures both known as well as new transcripts in a given sample. Microarrays measure the relative amount of mRNA activity of target genes with existing sequences. RNA sequencing uses high-throughput sequencing to record all transcripts and provides information regarding the gene sequence in addition to the expression level. Such tools generate a transcriptional profile (gene expression signature), representing a snapshot of genes expressed at a specific point in time. The steps involved in transcriptomics utilizing RNA sequencing are outlined in . Briefly, after sample collection, cells of interest are isolated, RNA is extracted, and converted to a library consisting of cDNA fragments. Each molecule is then sequenced with or without amplification. The resulting reads are then aligned to a reference genome or transcripts (or assembled de novo) to produce a genome-scale transcription map that consists of the level of expression for each gene. RNA sequencing data can produce over 109 short DNA sequences, which must then be analyzed using bioinformatics.
Transcriptomics has revolutionized our understanding of how genes are expressed, by providing a comprehensive, unbiased and integrated analysis of the complexities of cellular activity. However, transcriptomic analyses require significant computation and proper experimental design to produce meaningful data, and this technology assumes that mRNA transcription is a proxy for protein products of a cell, which is not always the case. Not only is RNA unstable, but post-transcriptional modifications further modulate protein synthesis, such that mRNA and protein abundance do not always correlate. Further, gene expression is highly tissue-specific, and caution is needed in interpretation of gene expression patterns from a mix of cell populations.
Transcriptomics is emerging as an important tool in immunological and infectious diseases research. The transcriptomic study of peripheral blood mononuclear cells – including B cells, T cells, monocytes, dendritic cells, and natural killer cells – can provide a comprehensive summary of the immune response to infection. More recently, newer platforms enable whole blood analysis and single cell sorting to allow for a more comprehensive analysis, and immune cells found in respiratory secretions are being used to study the local host response to influenza infection.
Immunopathology of influenza infection
Complex co-ordinated immune responses are triggered in the host following an acute influenza infection, involving both innate and adaptive immunologic processes.
Innate immune response
The first mechanisms of defense against influenza infection come into play at the portal of entry in the respiratory tract. The virus must cross the mucous layer that covers the respiratory epithelium in order to attach to the cell membrane and invade the cell. Infected epithelial cells, tissue macrophages and plasmacytoid dendritic cells (pDC) identify the viral RNA as a foreign element through pattern recognition receptors (PRRs), which activate an innate immune cascade resulting in the downstream secretion of type I interferons, other inflammatory cytokines, and chemokines.1-Citation3 The type I interferons stimulate hundreds of genes collectively known as interferon-stimulated genes (ISGs) in the surrounding cells, establishing potent innate local antiviral activity.Citation4 The inflammatory mediators released at this stage of infection may result in systemic symptoms including fever and malaise. They also instruct the adaptive immune response. The chemokines released by epithelial cells and local immune cells attract natural killer cells (NKs), monocytes and neutrophils at the site of infection. These recognize the virally infected cells and eliminate them through NK-mediated cytotoxicity followed by monocyte and neutrophil phagocytosis of the dead cells.Citation5 This process is generally sufficient to eliminate the virus in most immune-competent individuals.Citation6,Citation7
Adaptive immunity – T cell response
If the influenza virus overcomes the innate protective immune responses and establishes a successful infection in the respiratory tract, the ultimate clearance of the virus requires the participation of the adaptive immune system. The adaptive immune response is generated in the lymphoid tissue. Typically, conventional dendritic cells (cDC) carry antigen to the draining lymph node where both T cells and B cells become primed. Primed CD4+ and CD8+ T cells differentiate into T helper 1 (Th1), T follicular helper (Tfh) and cytotoxic T cells (CTL), all of which contribute to the clearance of influenza infection.Citation8 Regulatory T cells (Treg) and other types of T helper cells may also be generated, but they contribute to the overall outcome of the viral clearance process mostly through their interactions with the influenza-specific Th1, CTL, Tfh and B cells.Citation9,Citation10 T cells migrate to the site of infection where they continue to proliferate and differentiate, activate themselves and local NKs and phagocytes until the virus is eliminated. The process of viral clearance is accompanied by tissue destruction and systemic inflammation. Once the foreign antigenic stimulus disappears, T helper cell proliferation ceases, effector T cells die or differentiate into memory cells and the tissue repair process predominates. Influenza-specific tissue-resident memory T cells (Trm) have been recently recognized.Citation11 While these cells lack the receptors necessary to leave the lung, they have a relatively long life and high replicative capacity like memory T cells, but also have the ability to accelerate cytokine production after encountering cognate antigens, similar to effector T cells. These T cells will quickly generate an adaptive immune response upon re-exposure to influenza viruses. It is important to note that T cells recognize small epitopes that are shared not only by homotypic, but also by heterosubtypic influenza viruses, thus enabling cross-protection against different influenza subtypes. Treg are another important component of the antiviral process. They are essential in quenching the effector immune response and redirecting the Th1 cell differentiation from effector to memory. They also play an important role in tissue repair that has only recently been recognized.Citation12 The T cell response must strike a fine balance between tissue destruction and viral elimination.
Adaptive immunity – antibody response
Tfh and Th1 cells cross into the germinal center of the lymph node where they provide support for the influenza-specific plasmablasts to differentiate into memory B cells and long-lived plasma cells, which are the main producers of high-affinity antibodies against influenza. Antibodies against the major glycoproteins of the viral envelope, including the hemagglutinin (HA), neuraminidase (NA) and matrix protein 2 (M2), mediate several adaptive immune mechanisms. Antibodies against the HA block the influenza virus attachment to the target cell thus neutralizing the virus. Most antibodies generated against the HA recognize the variable portion of the HA. These antibodies confer protection against influenza A and B homotypic viruses but very limited protection against heterosubtypic viruses. There are conserved areas in the HA of influenza A viruses that have been extensively studied because they generate broadly neutralizing antibodies.Citation13,Citation14 However, broadly neutralizing antibodies are rarely synthesized by the host during infection. The NA has a critical role in the release of newly formed viruses from the infected cells that controls the spread of the infection. The NA contains a higher proportion of conserved amino acid sequences compared with the HA and; therefore, anti-NA antibodies can neutralize a larger spectrum of viruses.Citation15,Citation16 Anti-M2 antibodies do not induce neutralizing antibodies, but are expressed in abundance on the surface of the infected cells and may be an important target for viral clearance through phagocytosis or antibody-mediated cellular cytotoxicity through the recruitment of NK cells, monocytes, macrophages, and phagocytic DC.Citation17-Citation19
What have we learned about influenza pathogenesis through transcriptomics?
Each stage of the immune response against influenza infection may be characterized through its gene expression, cytokine release, and cell activation signatures or patterns, which can be studied using transcriptomics.Citation20 Transcriptomics has helped elucidate immune pathways specific to influenza infection, and has delineated immune profiles that differentiate asymptomatic from symptomatic infection, mild from severe disease, and early versus late infection. Further, transcriptional profiling enables the detection of infection prior to the onset of peak respiratory symptoms and has identified strain-specific differences in the host response to infection. A summary of the most recent findings from studies using transcriptomics to study influenza infection and vaccination and their potential clinical and research applications are provided in . A summary of study methodology and main findings from studies evaluating the host response to influenza infection are provided in .
Table 1. Transcription profiles of host responses to influenza infection and vaccination with potential clinical and research applications.
Table 2. Summary of findings from studies evaluating the host response to influenza infection.
Immune profiling of host PBMCs can differentiate bacterial versus viral infections in febrile adults with 89% sensitivity and 94% specificity.Citation21 Moreover, transcriptional profiling can also identify signatures unique to influenza infection,Citation22-Citation24 which can be distinguished from uninfected individuals with 94% accuracy. Compared with other viral pathogens, infection with influenza was associated with a higher magnitude and longer duration of the illness biosignature, which reflected upregulation of interferon pathway and innate immunity genes. Gene expression patterns 21 days post-infection were identical to baseline gene expression.Citation23 A unique molecular signature consisting of eight gene clusters was shown to correlate with symptomatic disease. This included genes coding for innate viral RNA sensors (TLR7, RNA helicases, and interferon induced with helicase C domain 1), which were transcribed 36 h before the peak symptoms. The expression of suppressor of cytokine signaling genes (SOC) 1 and 3 (which are important inhibitory modulators in limiting the inflammatory effect of interferon signaling during viral infection) declines early among asymptomatic individuals but strongly increases among symptomatic individuals. Further, ribosomal protein gene transcription was upregulated in asymptomatic compared with symptomatic individuals.Citation25
Several key immune pathways have been found to discriminate early from late phases of infection. For example, a large increase in components of the type-1 interferon antiviral response and innate immunity were upregulated, whereas the expression of genes involved in translational elongation and protein synthesis were decreased during acute infection in one study.Citation23 Four days later, there was a characteristic recovery phase, with the upregulation of genes involving antigen binding and antibody secretion, and genes regulating cell morphogenesis.
Several transcriptional patterns associated with the outcome of infection have also been described. Signatures characterized by decreased type I interferon and ubiquitination gene transcription were associated with a more severe outcome of influenza A infection.Citation26 In contrast, transcription of interferon-induced transmembrane protein genes were associated with less severe disease.
Transcriptomics studies have the additional advantage of identifying infection based on immune signatures prior to the onset of symptoms. In a study of adults experimentally infected with influenza A H1N1 or H3N2, there was a specific genomic signature for infection that was present as early as 53-h post viral exposure, over 24 h before the onset of symptoms. Predictive genes included interferon response elements, the myxovirus-resistance gene MX1, and cytokine response pathways.Citation27 When applied to a population visiting the Emergency Department, this signature differentiated H1N1 pdm-infected from other patients with 92% accuracy.
There is evidence of strain-specific variability in gene signatures involved in the host response to influenza infections. Seasonal influenza A H1N1 and H3N2 infections result in gene expression profiles that share 44 out of the top 50 expressed genes but significantly differ from the transcriptional profile of the avian H5N1. Infection with H7N9 induces a gene transcription profile that was more similar to seasonal than avian influenza infection indicating better adaptation of H7N9 to human hosts compared with other avian viruses.Citation28
Immunity conferred by influenza vaccines
Vaccination is the primary strategy for protection against influenza infections. Although there are multiple influenza vaccine preparations, they can be largely grouped into live attenuated (LAIV) and inactivated vaccines (IIV). In addition, selective IIV are adjuvanted but currently are not licensed for use in children. LAIV and IIV differ in their immune mechanisms and correlates of protection. Influenza vaccines are licensed based on the hemagglutination inhibition antibody (HAI) titers that they generate. This is based on an older study in adults that showed that HAI titers ≥1:40 after vaccination were associated with 50% decrease in the incidence of symptomatic influenza infections.Citation29 Recent studies have challenged this dogma, but this continues to be a licensure criterion of influenza vaccines and is also considered the main mechanism of protection of IIV. IIV also generate T cell responses and IgA antibodies that are not measured by the HAI assay but may contribute to protection against influenza infection. Inactivated vaccines generate demonstrable, but limited heterosubtypic protection against influenza A and B viruses. The most obvious reasons for the decreased cross-protection potential of IIV are the low T cell responses and the emphasis on the antibodies against the highly variable HA at the expense of the more conserved NA or M2.Citation30 The addition of adjuvants increases the antibody production and/or decreases the amount of antigen that is needed to generate HAI titers ≥1:40 in response to vaccination. Less is known about CMI responses to adjuvanted IIV but they arguably confer higher cross protection than non-adjuvanted IIV, suggesting that they may also generate higher CMI responses. It is important to understand that HAI antibodies are not necessary nor sufficient for protection against influenza infection.Citation31 This was well demonstrated by studies in children and older individuals in whom CMI or nasal IgA correlated with protection against influenza and HAI titers did not.Citation32-Citation35 Conversely, hematopoietic stem cell transplant recipients have regular infusions of HAI-containing IVIG for the first 6 months after transplantation but continue to be at high risk of severe influenza infection. LAIV exemplifies best this paradigm. HAI titers generated by LAIV are low and do not correlate with protection against influenza infection. Conversely, LAIV generates stronger CMI responses and broader cross-protection compared with IIV. LAIV is more efficacious in children than in adults.
Insights into the immunobiology of influenza vaccines using transcriptomics
Recent studies have described gene expression signatures associated with influenza vaccination. Advanced bioinformatics analytical tools allowed classifying the gene expression patterns in modules characteristic for activation and proliferation of different immune cells, such as B cells, T cells, NK cells, and dendritic cells.Citation36 Differences have been observed in the immune profiles of IIV compared with LAIV recipients.Citation37-Citation39 IIV has generally been associated with increased transcription of B cells, plasmablasts, plasma cells, and conventional DC modules.Citation40,Citation41 Furthermore, the early transcriptional signatures (involved in interferon signaling, antigen processing, and presentation and IL-6 regulation) of IIV recipients predicted HAI production after vaccination.Citation40 The molecular signatures associated with antibody responses to IIV have been reproduced in different populations and seasons, utilizing different techniques, underscoring the solidity of the data.Citation37,Citation42
The addition of adjuvants to IIV may create signature patterns specific for each adjuvant in addition to the antigen. This is suggested by initial studies of MF59- and ASO3-adjuvanted vaccines.Citation43-Citation45 ASO3 induces NK cell division activity and interferon signaling and antigen processing and presentation several days after vaccination.Citation45 However, the relationship between the adjuvant signature and protection conferred by the vaccines still needs to be elucidated.
The transcriptional signature of LAIV was characterized by increased activity of plasmacytoid DC, T cell and NK cell modules. Interferon-signaling pathways were induced 7 days after vaccination.Citation38 Transcriptional signatures in LAIV recipients differed from those of IIV recipients and were not associated with the magnitude of the HAI responses to the vaccine.Citation38,Citation39 The LAIV signature revealed five upregulated genes that represented an interferon-stimulated gene response. It is interesting to note that the transcriptional signature of LAIV more closely resembled that of yellow fever vaccine, which is also a live attenuated virus vaccine, than the IIV signature. The transcriptional signature of yellow fever, however, is predictive of the antibody response to this vaccine which also correlates with protection against yellow fever viral infection.
Finally, transcriptional signatures following influenza vaccination differ among children, young adults and older adults,Citation43,Citation46 racial groupsCitation47 and between men and women.Citation48,Citation49 Further research is required to understand key differences in the immune response among our more vulnerable populations.
Conclusion
Transcriptomics is a rapidly evolving discipline that provides an unbiased, accurate and sensitive method to study host-pathogen interactions. This approach requires significant computation, and thoughtful experimental design and data interpretation are required to ensure that meaningful conclusions can be reached. The studies presented in this review demonstrate the potential for transcriptomics to provide valuable applications for research and clinical use. This technology has provided novel insights into the host response to influenza natural infection and vaccination, which can help guide the development and selection of future influenza vaccines and therapeutics. Further, combining pathogen detection with the host immune response can improve interpretation of pathogens identified in a biological sample and enhance the classification of disease states. Currently, transcriptomics techniques are labor intensive and require high-level bioinformatics support, limiting their clinical application. However, further advancements in this technology may allow for faster acquisition of transcriptomic profiles in the clinical setting, in order to enhance diagnostics, monitor response to therapy, and identify markers of severity.
Disclosure of potential conflicts of interest
Adriana Weinberg receives grant support from Medimmune, Merck and GSK. Suchitra Rao receives grant support from GSK and Biofire. Edwin Asturias receives grant support from GSK. All other authors have indicated that there are no conflicts of interest relevant to this article to disclose.
References
- Tamura S, Kurata T. Defense mechanisms against influenza virus infection in the respiratory tract mucosa. Jpn J Infect Dis. 2004;57:236–47.
- Okamoto M, Tsukamoto H, Kouwaki T, Seya T, Oshiumi H. Recognition of Viral RNA by Pattern Recognition Receptors in the Induction of Innate Immunity and Excessive Inflammation During Respiratory Viral Infections. Viral Immunol. 2017;30(6):408–20. doi:10.1089/vim.2016.0178.
- Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–38. doi:10.1038/35099560.
- Cao P, Yan AW, Heffernan JM, Petrie S, Moss RG, Carolan LA, Guarnaccia TA, Kelso A, Barr IG, McVernon J, et al. Innate immunity and the inter-exposure interval determine the dynamics of secondary influenza virus infection and explain observed viral hierarchies. PLoS Comput Biol. 2015;11(8):e1004334. doi:10.1371/journal.pcbi.1004334.
- Schultz-Cherry S. Role of NK cells in influenza infection. Curr Top Microbiol Immunol. 2015;386:109–20. doi:10.1007/82_2014_403.
- Iwasaki A, Pillai PS. Innate immunity to influenza virus infection. Nat Rev Immunol. 2014;14(5):315–28. doi:10.1038/nri3665.
- Oshansky CM, Gartland AJ, Wong SS, Jeevan T, Wang D, Roddam PL, Caniza MA, Hertz T, Devincenzo JP, Webby RJ, et al. Mucosal immune responses predict clinical outcomes during influenza infection independently of age and viral load. Am J Respir Crit Care Med. 2014;189(4):449–62. doi:10.1164/rccm.201309-1616OC.
- Brown DM, Lampe AT, Workman AM. The differentiation and protective function of cytolytic CD4 T cells in influenza infection. Front Immunol. 2016;7:93. doi:10.3389/fimmu.2016.00093.
- Braciale TJ, Sun J, Kim TS. Regulating the adaptive immune response to respiratory virus infection. Nat Rev Immunol. 2012;12:295. doi:10.1038/nri3166.
- Medzhitov R, Schneider DS, Soares MP. Disease tolerance as a defense strategy. Science (New York, NY). 2012;335(6071):936–41. doi:10.1126/science.1214935.
- Pizzolla A, Nguyen THO, Sant S, Jaffar J, Loudovaris T, Mannering SI, Thomas PG, Westall GP, Kedzierska K, Wakim LM. Influenza-specific lung-resident memory T cells are proliferative and polyfunctional and maintain diverse TCR profiles. J Clin Invest. 2018;128(2):721–33. doi:10.1172/JCI96957.
- Zhao H, Liao X, Kang Y. Tregs: where we are and what comes next? Front Immunol. 2017;8:1578. doi:10.3389/fimmu.2017.01578.
- Avnir Y, Tallarico AS, Zhu Q, Bennett AS, Connelly G, Sheehan J, Sui J, Fahmy A, Huang C-Y, Cadwell G, et al. Molecular signatures of hemagglutinin stem-directed heterosubtypic human neutralizing antibodies against influenza A viruses. PLoS Pathog. 2014;10(5):e1004103. doi:10.1371/journal.ppat.1004103.
- Fonville JM, Fraaij PL, de Mutsert G, Wilks SH, van Beek R, Fouchier RAM, Rimmelzwaan GF. Antigenic maps of influenza A(H3N2) produced with human antisera obtained after primary infection. J Infect Dis. 2016;213(1):31–38. doi:10.1093/infdis/jiv367.
- Monto AS, Petrie JG, Cross RT, Johnson E, Liu M, Zhong W, Levine M, Katz JM, Ohmit SE. Antibody to influenza virus neuraminidase: an independent correlate of protection. J Infect Dis. 2015;212(8):1191–99. doi:10.1093/infdis/jiv195.
- Halbherr SJ, Ludersdorfer TH, Ricklin M, Locher S, Berger Rentsch M, Summerfield A, Zimmer G. Biological and protective properties of immune sera directed to the influenza virus neuraminidase. J Virol. 2015;89(3):1550–63. doi:10.1128/JVI.02949-14.
- Ana-Sosa-Batiz F, Vanderven H, Jegaskanda S, Johnston A, Rockman S, Laurie K, Barr I, Reading P, Lichtfuss M, Kent SJ, et al. Influenza-specific antibody-dependent phagocytosis. PLoS One. 2016;11(4):e0154461. doi:10.1371/journal.pone.0154461.
- Jegaskanda S, Luke C, Hickman HD, Sangster MY, Wieland-Alter WF, McBride JM, Yewdell JW, Wright PF, Treanor J, Rosenberger CM, et al. Generation and protective ability of influenza virus-specific antibody-dependent cellular cytotoxicity in humans elicited by vaccination, natural infection, and experimental challenge. J Infect Dis. 2016;214(6):945–52. doi:10.1093/infdis/jiw262.
- He W, Tan GS, Mullarkey CE, Lee AJ, Lam MMW, Krammer F, Henry C, Wilson PC, Ashkar AA, Palese P, et al. Epitope specificity plays a critical role in regulating antibody-dependent cell-mediated cytotoxicity against influenza A virus. Proc Natl Acad Sci U S A. 2016;113(42):11931–36. doi:10.1073/pnas.1609316113.
- Bonduelle O, Carrat F, Luyt CE, Leport C, Mosnier A, Benhabiles N, Krivine A, Rozenberg F, Yahia N, Samri A, et al. Characterization of pandemic influenza immune memory signature after vaccination or infection. J Clin Invest. 2014;124(7):3129–36. doi:10.1172/JCI74565.
- Mejias A, Dimo B, Suarez NM, Garcia C, Suarez-Arrabal MC, Jartti T, Blankenship D, Jordan-Villegas A, Ardura MI, Xu Z, et al. Whole blood gene expression profiles to assess pathogenesis and disease severity in infants with respiratory syncytial virus infection. PLoS Med. 2013;10(11):e1001549. doi:10.1371/journal.pmed.1001549.
- Parnell GP, McLean AS, Booth DR, Armstrong NJ, Nalos M, Huang SJ, Manak J, Tang W, Tam O-Y, Chan S, et al. A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care. 2012;16(4):R157. doi:10.1186/cc11477.
- Zhai Y, Franco LM, Atmar RL, Quarles JM, Arden N, Bucasas KL, Wells JM, Niño D, Wang X, Zapata GE, et al. Host transcriptional response to influenza and other acute respiratory viral infections–A prospective cohort study. PLoS Pathog. 2015;11(6):e1004869. doi:10.1371/journal.ppat.1004869.
- Andres-Terre M, McGuire HM, Pouliot Y, Bongen E, Sweeney TE, Tato CM, Khatri P. Integrated, multi-cohort analysis identifies conserved transcriptional signatures across multiple respiratory viruses. Immunity. 2015;43(6):1199–211. doi:10.1016/j.immuni.2015.11.003.
- Yongsheng Huang, Aimee K. Zaas, Arvind Rao, Nicolas Dobigeon, Peter J. Woolf, Timothy Veldman, N. Christine Øien, Micah T. McClain, Jay B. Varkey, Bradley Nicholson, et al. Temporal dynamics of host molecular responses differentiate symptomatic and asymptomatic influenza a infection. PLoS Genet. 2011;7(8):e1002234. doi:10.1371/journal.pgen.1002138.
- Hoang LT, Tolfvenstam T, Ooi EE, Khor CC, Naim ANM, Ho EXP, Ong SH, Wertheim HF, Fox A, Van Vinh Nguyen C, et al. Patient-based transcriptome-wide analysis identify interferon and ubiquination pathways as potential predictors of influenza A disease severity. PLoS One. 2014;9(11):e111640. doi:10.1371/journal.pone.0111640.
- Josset L, Zeng H, Kelly SM, Tumpey TM, Katze MG. Transcriptomic characterization of the novel avian-origin influenza A (H7N9) virus: specific host response and responses intermediate between avian (H5N1 and H7N7) and human (H3N2) viruses and implications for treatment options. MBio. 2014;5(1):e01102–01113. doi:10.1128/mBio.01102-13.
- Nakaya HI, Wrammert J, Lee EK, Racioppi L, Marie-Kunze S, Haining WN, Means AR, Kasturi SP, Khan N, Li G-M, et al. Systems biology of vaccination for seasonal influenza in humans. Nat Immunol. 2011;12(8):786–95. doi:10.1038/ni.2067.
- Cao RG, Suarez NM, Obermoser G, Lopez SMC, Flano E, Mertz SE, Albrecht RA, García-Sastre A, Mejias A, Xu H, et al. Differences in antibody responses between trivalent inactivated influenza vaccine and live attenuated influenza vaccine correlate with the kinetics and magnitude of interferon signaling in children. J Infect Dis. 2014;210(2):224–33. doi:10.1093/infdis/jiu079.
- Cole KS, Martin JM, Horne WT, Lin CJ, Nowalk MP, Alcorn JF, Zimmerman RK. Differential gene expression elicited by children in response to the 2015–16 live attenuated versus inactivated influenza vaccine. Vaccine. 2017;35(49 Pt B):6893–97. doi:10.1016/j.vaccine.2017.09.019.
- Nakaya HI, Clutterbuck E, Kazmin D, Wang L, Cortese M, Bosinger SE, Patel NB, Zak DE, Aderem A, Dong T, et al. Systems biology of immunity to MF59-adjuvanted versus nonadjuvanted trivalent seasonal influenza vaccines in early childhood. Proc Natl Acad Sci U S A. 2016;113(7):1853–58. doi:10.1073/pnas.1519690113.
- Kurupati R, Kossenkov A, Haut L, Kannan S, Xiang Z, Li Y, Doyle S, Liu Q, Schmader K, Showe L, et al. Race-related differences in antibody responses to the inactivated influenza vaccine are linked to distinct pre-vaccination gene expression profiles in blood. Oncotarget. 2016;7(39):62898–911. doi:10.18632/oncotarget.11704.
- Tan Y, Tamayo P, Nakaya H, Pulendran B, Mesirov JP, Haining WN. Gene signatures related to B-cell proliferation predict influenza vaccine-induced antibody response. Eur J Immunol. 2014;44(1):285–95. doi:10.1002/eji.201343657.
- Bucasas KL, Franco LM, Shaw CA, Bray MS, Wells JM, Niño D, Arden N, Quarles JM, Couch RB, Belmont JW. Early patterns of gene expression correlate with the humoral immune response to influenza vaccination in humans. J Infect Dis. 2011;203(7):921–29. doi:10.1093/infdis/jiq156.
- Woods CW, McClain MT, Chen M, Zaas AK, Nicholson BP, Varkey J, Veldman T, Kingsmore SF, Huang Y, Lambkin-Williams R, et al. A host transcriptional signature for presymptomatic detection of infection in humans exposed to influenza H1N1 or H3N2. PLoS One. 2013;8(1):e52198. doi:10.1371/journal.pone.0052198.
- Hobson D, Curry RL, Beare AS, Ward-Gardner A. The role of serum haemagglutination-inhibiting antibody in protection against challenge infection with influenza A2 and B viruses. J Hyg (Lond). 1972;70:767–77.
- Zhong W, Reed C, Blair PJ, Katz JM, Hancock K. Influenza Serology Working G. Serum antibody response to matrix protein 2 following natural infection with 2009 pandemic influenza A(H1N1) virus in humans. J Infect Dis. 2014;209(7):986–94. doi:10.1093/infdis/jit811.
- Jegaskanda S, Reading PC, Kent SJ. Influenza-specific antibody-dependent cellular cytotoxicity: toward a universal influenza vaccine. J Immunol. 2014;193(2):469–75. doi:10.4049/jimmunol.1400432.
- Gould VMW, Francis JN, Anderson KJ, Georges B, Cope AV, Tregoning JS. Nasal IgA provides protection against human influenza challenge in volunteers with low serum influenza antibody titre. Front Microbiol. 2017;8:900. doi:10.3389/fmicb.2017.00900.
- Asahi-Ozaki Y, Yoshikawa T, Iwakura Y, Suzuki Y, Tamura S-I, Kurata T, Sata T. Secretory IgA antibodies provide cross-protection against infection with different strains of influenza B virus. J Med Virol. 2004;74(2):328–35. doi:10.1002/jmv.20173.
- Treanor J, Wright PF. Immune correlates of protection against influenza in the human challenge model. Dev Biol (Basel). 2003;115:97–104.
- Forrest BD, Pride MW, Dunning AJ, Capeding MRZ, Chotpitayasunondh T, Tam JS, Rappaport R, Eldridge JH, Gruber WC. Correlation of cellular immune responses with protection against culture-confirmed influenza virus in young children. Clin Vaccine Immunol. 2008;15(7):1042–53. doi:10.1128/CVI.00397-07.
- Avey S, Mohanty S, Wilson J, Zapata H, Joshi SR, Siconolfi B, Tsang S, Shaw AC, Kleinstein SH. Multiple network-constrained regressions expand insights into influenza vaccination responses. Bioinformatics. 2017;33(14):i208–i216. doi:10.1093/bioinformatics/btx260.
- Nakaya HI, Hagan T, Duraisingham SS, Lee EK, Kwissa M, Rouphael N, Frasca D, Gersten M, Mehta AK, Gaujoux R, et al. Systems analysis of immunity to influenza vaccination across multiple years and in diverse populations reveals shared molecular signatures. Immunity. 2015;43(6):1186–98. doi:10.1016/j.immuni.2015.11.012.
- Olafsdottir T, Lindqvist M, Harandi AM. Molecular signatures of vaccine adjuvants. Vaccine. 2015;33(40):5302–07. doi:10.1016/j.vaccine.2015.04.099.
- Howard LM, Hoek KL, Goll JB, Samir P, Galassie A, Allos TM, Niu X, Gordy LE, Creech CB, Prasad N, et al. Cell-based systems biology analysis of human AS03-adjuvanted H5N1 avian influenza vaccine responses: a phase I randomized controlled trial. PLoS One. 2017;12(1):e0167488. doi:10.1371/journal.pone.0167488.
- Lambert ND, Ovsyannikova IG, Pankratz VS, Jacobson RM, Poland GA. Understanding the immune response to seasonal influenza vaccination in older adults: a systems biology approach. Expert Rev Vaccines. 2012;11(8):985–94. doi:10.1586/erv.12.61.
- Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiébaut R, Tibshirani RJ, Davis MM. Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination. Proc Natl Acad Sci USA. 2014;111(2):869–74. doi:10.1073/pnas.1321060111.
- Shen-Orr SS, Furman D. Variability in the immune system: of vaccine responses and immune states. Curr Opin Immunol. 2013;25(4):542–47. doi:10.1016/j.coi.2013.07.009.