ABSTRACT
Viral infection typically originates from a limited number of virions known as transmitted/founder (T/F) viruses. Studies of cross-species transmission, and intra-species transmission of antigenically variable viruses, indicates T/F variants may express distinct, transmissibility enhancing phenotypes. However, with evidence that transmissibility is associated with not only intrinsic virological features, such as virion composition, but also extrinsic factors, such as viral population structure, the challenge of resolving T/F signatures that can be targeted by rational vaccine or antiviral design is substantial. Nonetheless, failure to develop vaccines for antigenically variable viruses, such as HIV/HCV, and the ongoing risk of cross-species transmission with pandemic potential, recommends development of T/F targeting vaccines. In this commentary, the T/F phenomena is introduced, explored in both the classical (HIV) and non-canonical (coronaviruses) instances, and discussed in relation to rational and preemptive vaccine design.
Introduction
Viral transmission is characterized by a population bottleneck from which a minority of virions emerge to establish infection.Citation1 Recently it has been hypothesized that transmission may not be a simply stochastic process whereby a random sampling of the donor population establishes infection in the recipient.Citation2 Rather, it is believed that the population bottleneck itself may be selective, with only variants replicating in tissues sampled during transmission capable of establishing infection.Citation3 Further, only a subset of virions, termed transmitted/founder (T/F) viruses, within a viral population may express the phenotype required to infect and productively reproduce in a new host.Citation4 Importantly, though all viral transmission, like all genetic bottlenecks, implies a founder-effect, the role of T/F viruses is limited to two distinct phenomena: i) cross-species transmission of emergent viruses (SARS-CoV), and ii) intra-species transmission of variable viruses (HIV). This is because, unlike neutral founder-effects, which occur whenever a minority of members from a population emerge from a genetic bottleneck, the T/F phenomena requires that variance in transmissibility, either to a new species, or to a new host, exists within a viral population.Citation5 For the T/F phenomena to be sustained within a viral species, the fitness advantages of the transmissible phenotype must be in trade-off, or neutral, with reproducibility within hosts following transmission. If this criteria is not met, the T/F phenotype can become fixed, as observed in viruses emergent from cross-species transmission.Citation6 This balance differentiates common cross-species transmission events, which involve transmissible phenotypes that lack reproducibility in the new host setting (H5N1), from pandemics, wherein a variant both transmissible and reproducible in a new host-setting crosses species (COVID-19)Citation7,Citation8 (). Transmissibility is therefore not an intrinsic property of a virion, but emergent from the particular transmission setting, such that a flu virus which is highly cross-species transmissible may be non-transmissible in the new host-background – a caveat which likely spares mice and men from more frequent, emergent pandemics.
Figure 1. Transmission bottlenecks
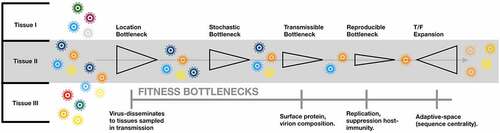
Deep sequencing of viral populations is resolving both cross-species and intra-species T/F dynamics.Citation9 However, many technical and conceptual challenges remain, including access to samples at early infection, availability of samples from transmission pairs, and the possibility that T/F phenotypes are not genetically defined, but emergent from epigenetic, structural, or tissue-localized features, the resolution of which exceeds the capacity of available technologies. Despite these challenges, continued investigation of the T/F phenomena may be critical to vaccine development for variable viruses, such as HCV and HIV, where problem-space limiting approaches to antigenic variability are urgently needed.Citation10
Founder effect
When a minority sub-population colonizes a new environment, the genetic composition of the colonizing population may change in response to novel selection pressures or genetic drift.Citation11 Loss of ancestral genetic variation, with subsequent enrichment of only some alleles in the new population upon expansion, is known as a founder effect.Citation12 These events can be stochastically determined, such as when, by random sampling, dominant alleles become most represented in the nascent population. Alternatively, if the founder population is nonrandomly selected, minority traits may become enriched.Citation13 Importantly, enrichment of minor traits is not definitive evidence of a selective bottleneck, as, in small founding populations, minority traits in the ancestral population may enrich simply by genetic drift.Citation13
Both selection and drift can be operative in viral founder events, and can be tracked by evaluating rates of synonymous (S) and nonsynonymous (NS) mutation.Citation14 For example, immune-meditated selective pressures favor nonsynomyous mutations in HCV during acute-infection, while S mutations (or NS that do not influence sensitivity to immune responses), present in the ancestral population, can persist solely by drift.Citation15 Because all viral transmissions are founder events, stochastic and selective forces must be differentiated when determining if the T/F phenomena is operative. One approach is to evaluate the ratio of S-to-NS mutations, another is determining if non-dominant variants in the ancestral population are consistently over-represented in the nascent population, and if these variants present unique phenotypes that enhance transmissibility.Citation16 The T/F phenomena is therefore distinct from a classical founder event, in that the colonizing population is related to the ancestral not only by the size of the bottleneck, but also it’s location, in terms of which tissues or stages of infection are sampled during transmission, and it’s selectively, in terms of which viral phenotypes are eligible for establishing infection in a new host ().
Cross-species
Cross-species transmission with subsequent speciation is the most significant T/F event. As discussed, because transmissibility and reproducibility are non-synonymous and emergent features, a majority of cross-species T/F events do not result in productive colonization of a new host.Citation17 Influenza provides a common case study, where cross-species transmission, though frequent, especially among farm workers, rarely results in the introduction of a transmissible, reproducible infection in humans.Citation18 Though this position is primarily supported by retrospective analysis of seroprevalence, functional evidence pertaining to differential representation of sialic acid residues across the respiratory track has also been obtained.Citation19,Citation20
For cross-species transmission to result in a broadened host range, the variants transmissible in the cross-species setting must also be reproducible in the new host, in terms of generating sufficiently high titers of mature, infectious virus, and replicate, or disseminate to, the appropriate tissues for subsequent transmission within the new host species.Citation20 Though these events are rare, they are even harder to detect, as host-reproducibility and subsequent transmission may directionally select minor phenotypes, ultimately resulting in speciation. Evidence of this has been obtained for SaRS-CoV, with positive selection on the spike protein during cross-species and early human-host evolution subsequently stabilizing late in the pandemic.Citation21 Adaptation to new host-species following cross-species transmission of murine hantavirus has also been observed, though speciation in this context seems largely driven by genetic drift.Citation6 Notwithstanding the technical challenges in contemporaneously detecting these events, their occurence implies pandemic potential. However, there are certain virological patterns which could help guide surveillance, with the potential for preemptive vaccine design. For example, cross-species T/F variants capable of sustaining high genetic diversity have a proportionally greater likelihood of generating variants balancing both transmissibility and reproducibility in the new host-setting.Citation22 This risk is magnified among pathogens capable of establishing chronic infection with more replication cycles. HIV’s cross-species transmission is one catastrophic example of a pathogen with both high variability and chronic potential.Citation23 However, even among species with lower variability, such as COVID-19, the emergence of a phenotype balancing transmissibility and reproducibility in a new host-setting may be anticipated by comparative analysis of other recent, coronavirus cross-species transmission events.Citation24 Indeed, groups had proposed this potential in identifying features common to cross-species T/F coronaviruses that could be targeted by vaccines or antivirals in the years preceding the global pandemic.Citation14
T/F variants in variable viruses
The T/F phenomenon was first described for intra-species transmission of highly variable pathogens.Citation25 In HIV transmission pairs, it was observed that founder variants differed from the dominant source quasispecies, implying the founding subpopulation is not a random sampling of the donor population, but is instead selected.Citation3 For example, if the quasi-species in the donor population did not differ in terms of transmissibility, then in accordance with a nonselective genetic bottleneck, recipient populations would tend to consist of the highest-frequency quasi-species in the donor.Citation26
These findings have recently been extended to HCV, with recipient quasispecies populations coalescing to minor or moderate, but not dominant, variants in the ancestral population.Citation2 These observations do not exclude the importance of variant frequency in transmission events, as very low frequency minority variants, even if manifesting the transmissible phenotype, may not be selected during transmission. The T/F phenomena for HIV/HCV is therefore consistent with some features of a genetic bottleneck, but with the additional caveat that variant frequency and variant transmissibility determine founder populations. Another caveat is that transmissibility may not represent a binary trait, but instead a spectrum, with the transmissibility of each variant relationally determined by its competing subpopulation.Citation2 If, rather than existing solely as an intrinsic property, the transmissibility of any given variant is described by its position within a viral subpopulation, the translational advantage of targeting T/F variants in vaccine design for variable pathogens may be limited.
Transmissible phenotypes
The identification of rare source variants in recipient populations prompted the search for T/F phenotypes, or intrinsic virological features associated with enhanced transmissibility.Citation3,Citation25,Citation26 Initially, these inquiries focused on surface glycoproteins, which mediate viral entry. In HIV-1, T/F variants were found to differ from chronic variants in terms of variable loop lengths and glycosylation.Citation27,Citation28 Mechanistically, these T/F signatures may increase affinity for mucosal receptors, and, with up to 50% of glycosylation sites on T/F Env variants variably glycosylated, this may translate into a substantial fitness advantage during transmission events.Citation29 An alternative explanation is that Env glycosylation results in greater sequestration of virus in the transmission fluid (potentially via interaction with host-lectins), increasing the relative transmissibility of variants with reduced glycosylation.Citation16 Cell-entry-related T/F traits can also be independent of sequence or post-translational modification, involving virion composition, with HIV-1 T/F variants presenting with higher surface Env density, potentially increasing interaction with cell-receptors.Citation30 Post-transmission pressures may then favor alternative phenotypes, such as higher glycosylation that reduces sensitivity to neutralizing Ab, illustrating how the T/F phenomena is favored in instances of i) high variability, ii) fitness trade-off between transmissibility and reproducibility, iii) long-infection duration (chronic).
Surface glycoprotein T/F signatures have also been identified among acute HCV variants, with 37% of residues in HVR1 (a highly variable epitope on HCV’s surface glycoprotein E2 implicated in receptor recognition) physiochemically differing, in terms of polarity and volume, from chronic stage HVR1.Citation31 However, in a separate study, though T/F variants were found to differ from the dominant donor quasispecies, no enhanced infectivity was observed in vitro, suggesting either that the complex fitness parameters affecting transmissibility may not be preserved across models, or that the observed T/F signatures in surface glycoproteins are only incidental.Citation32 Indeed, non-genetic features of transmissibility, such as lipidation, glycosylation, and virion composition, as observed for HIV-1, may be operative in HCV T/F events. To date, these issues have not been thoroughly explored, despite evidence that T/F signatures may not manifest as intrinsic haplotypes. For example, acute HCV variants were found to occupy more central positions in the sequence-space then chronic variants, implying that sequence-space position, rather than only sequence itself, may influence transmissibility.Citation2 Complicating analysis is the possibility that multiple, distinct T/F signatures may exist, with more than one fitness bottleneck providing an avenue for transmissibility. Given limited access to early-infection samples and transmission pairs, identifying multiple, non-overlapping T/F phenotypes is not a trivial technical challenge.
Sample selection may also influence T/F characterization if virions encounter tandem fitness bottlenecks in establishing infection. Tissue localization, transmission fluid/route, cell-entry, suppression of innate immunity, and intra-host infectivity may all impose distinct fitness bottlenecks on the donor population. Therefore, sampling in the first days after infection may yield different T/F signatures then after viral expansion, during which the relative reproducibility of quasispecies in the recipient population will determine dominance. Unfortunately, these challenges cannot be resolved theoretically or in silico, and require deep sequencing of the donor and recipient populations, at multiple timepoints following transmission, to identify which fitness bottleneck could be best targeted by vaccine design.
Targeting T/F variants in vaccine design
Evidence that productive infection in variable viruses (HIV/HCV) is established by a limited number of antigenically convergent variants has prompted efforts to direct vaccine efforts toward T/F variants.Citation33,Citation34 The advantage of T/F targeting vaccines is a dramatic reduction in the neutralizing breadth that must be elicited via vaccination.Citation34 Thus far T/F targeting vaccines remain mostly theoretical, despite findings that HIV-1 T/F variants elicit Ab with the greatest neutralization breadth in animal model.Citation35 To date, however, no group has shown greater-than-average cross-reactivity between T/F variants, either in HIV or HCV. The challenge is partly exacerbated by the lack of robust animal models that faithfully recapitulate transmission events, to which the advantage of T/F elicited Ab would presumably be restricted.Citation36,Citation37
Similarly, despite surveillance efforts, intra-family signatures of cross-species transmissibility have only been described for a limited number of pathogens, and have not manifested in preemptive vaccine design. Though a substantial undertaking, considering 52 human-transmissibility signatures were identified in avian Influenza A alone, targeted approaches based on population modeling, high-risk virus families, and pandemic potential of cross-species transmission events, could manageably reduce the problem space.Citation38
However, T/F targeting vaccines may simply introduce an additional fitness bottleneck which privileges distinct viral phenotypes, rather than blocking transmission. The feasibility and advantages of T/F targeting vaccines, though promising in theory, must be experimentally verified, beginning with unambiguous identification of transmissibility signatures.
Concluding remarks
Antigenic variability has been a major barrier to vaccine development. HIV and HCV exemplify this challenge, with highly heterogeneous populations preserving adaptability to host-specific immune pressures. Vaccine efforts have therefore focused on targeting conserved epitopes, under evolutionary constraint, thereby limiting the viruses adaptive space. However, so far these approaches have failed, with even conserved epitope targeting vaccines ultimately vulnerable to immune escape.Citation39 Another approach would be to focus on the transmitted viruses that establish infection, on the hypothesis that transmissible phenotypes are limited or antigenically convergent. The first step of T/F targeting vaccine design is elucidating signatures of transmissibility, which may be genetic, post-translational, or defined by virion composition. Though the individual signatures will be virus-specific, the approaches used to elucidate features of transmissibility can be more broadly applied across virus families, and include cross-species transmission, supporting surveillance efforts, and potentially guiding preemptive vaccine or antiviral design. Despite preliminary evidence that transmissibility signatures can be identified, both for intra-species and cross-species transmission, the advantages of using these features to guide vaccine design, for any virus, remains to be determined.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Abbreviations
| (T/F) | = | Transmitted/Founder |
| HIV | = | Human Immunodeficiency Virus |
| SARS-CoV | = | Severe Acute Respiratory Syndrome Coronavirus |
| COVID-19 | = | Coronavirus disease 2019 |
| HCV | = | Hepatitis C Virus |
| AB | = | Antibody |
| HVR1 | = | Hypervariable Region 1 |
| S | = | Synonymous Mutation |
| NS | = | Non-synonymous Mutation |
| HVR1 | = | Hypervariable Region I |
| Env | = | HIV Envelope Gene |
References
- McCrone JT, Lauring AS. Genetic bottlenecks in intraspecies virus transmission. Curr Opin Virol. 2018 Feb;28:20–25. PubMed Central. doi:10.1016/j.coviro.2017.10.008.
- Campo DS, Zhang J, Ramachandran S, Khudyakov Y. Transmissibility of intra-host hepatitis C virus variants. BMC Genomics. Dec 2017;18(Suppl 10). PubMed Central. doi:10.1186/s12864-017-4267-4.
- Boeras DI, Hraber PT, Hurlston M, Evans-Strickfaden T, Bhattacharya T, Giorgi EE, Mulenga J, Karita E, Korber BT, Allen S, et al. Role of donor genital tract HIV-1 diversity in the transmission bottleneck. Proc Natl Acad Sci U S A. 2011 Nov;108(46):E1156–1163. PubMed. doi:10.1073/pnas.1103764108.
- Gnanakaran S, Bhattacharya T, Daniels M, Keele BF, Hraber PT, Lapedes AS, Shen T, Gaschen B, Krishnamoorthy M, Li H, et al. Recurrent signature patterns in HIV-1 B clade envelope glycoproteins associated with either early or chronic infections. PLoS Pathog. Sept 2011;7 (9). PubMed Central. doi:10.1371/journal.ppat.1002209.
- Shaw GM, Hunter E. HIV transmission. Cold Spring Harb Perspect Med. Nov 2012;2(11). PubMed. doi:10.1101/cshperspect.a006965.
- Lin X-D, Wang W, Guo W-P, Zhang X-H, Xing J-G, Chen S-Z, Li M-H, Chen Y, Xu J, Plyusnin A, et al. Cross-species transmission in the speciation of the currently known murinae-associated hantaviruses. J Virol. 2012 Oct;86(20):11171–82. jvi.asm.org. doi:10.1128/JVI.00021-12.
- Weiss RA, Hamilton W, Weiss RA, Wain–Hobson S. The leeuwenhoek lecture 2001. Animal origins of human infectious disease. Philos Trans R Soc London Ser B. 2001 June;356(1410):957–77. PubMed Central. doi:10.1098/rstb.2001.0838.
- Klempner MS, Shapiro DS. Crossing the species barrier — one small step to man, one giant leap to mankind. N Engl J Med. 2004 Mar;350(12):1171–72. Taylor and Francis+NEJM. doi:10.1056/NEJMp048039.
- Borucki MK, Chen-Harris H, Lao V, Vanier G, Wadford DA, Messenger S, Allen JE. Ultra-deep sequencing of intra-host rabies virus populations during cross-species transmission. PLoS Negl Trop Dis. Nov 2013;7(11). PubMed Central. doi:10.1371/journal.pntd.0002555.
- Haynes BF, Shaw G, Korber B, Kelsoe G, Sodroski J, Hahn B, Borrow P, McMichael A. HIV-host interactions: implications for vaccine design. Cell Host Microbe. 2016 Mar;19(3):292–303. PubMed Central. doi:10.1016/j.chom.2016.02.002.
- Barton NH. Natural selection and random genetic drift as causes of evolution on islands. Philos Trans R Soc Lond B Biol Sci. 1996 June;351(1341):785–94. discussion 795. PubMed. doi:10.1098/rstb.1996.0073.
- Andersson S, Ellmer M, Jorgensen TH, Palmé A. Quantitative genetic effects of bottlenecks: experimental evidence from a wild plant species, nigella degenii. J Hered. 2010 June;101(3):298–307. PubMed. doi:10.1093/jhered/esp108.
- Matute DR. The role of founder effects on the evolution of reproductive isolation. J Evol Biol. 2013;26(11):2299–311. Wiley Online Library. doi:10.1111/jeb.12246.
- Forni D, Cagliani R, Clerici M, Sironi M. Molecular evolution of human coronavirus genomes. Trends Microbiol. 2017 Jan;25(1):35–48. ScienceDirect. doi:10.1016/j.tim.2016.09.001.
- Lapa D, Garbuglia A, Capobianchi M, Del Porto P. Hepatitis C virus genetic variability, human immune response, and genome polymorphisms: which is the interplay? Cells. Apr 2019;8(4). PubMed Central. doi:10.3390/cells8040305.
- Joseph SB, Swanstrom R, Kashuba AD, Cohen MS. Bottlenecks in HIV-1 transmission: insights from the study of founder viruses. Nat Rev Microbiol. 2015 July;13(7):414–25. PubMed Central. doi:10.1038/nrmicro3471.
- Nicholson KG, Wood JM, Zambon M. Influenza. Lancet (London, England). 2003 Nov;362(9397):1733–45. PubMed. doi:10.1016/S0140-6736(03)14854-4.
- Webster RG. Wet markets—a continuing source of severe acute respiratory syndrome and influenza? Lancet (London, England). 2004 Jan;363(9404):234–36. PubMed Central. doi:10.1016/S0140-6736(03)15329-9.
- Gomaa MR, Kandeil A, Kayed AS, Elabd MA, Zaki SA, Abu Zeid D, El Rifay AS, Mousa AA, Farag MM, McKenzie PP, et al. Serological evidence of human infection with avian influenza A H7virus in Egyptian poultry growers. Plos One. 2016 June;11(6):e0155294. PLoS Journals. doi:10.1371/journal.pone.0155294.
- Louz D, Bergmans HE, Loos BP, Hoeben RC. Cross‐species transfer of viruses: implications for the use of viral vectors in biomedical research, gene therapy and as live‐virus vaccines. J Gene Med. 2005 Oct;7(10):1263–74. PubMed Central. doi:10.1002/jgm.794.
- Tang X, Li G, Vasilakis N, Zhang Y, Shi Z, Zhong Y, Wang L-F, Zhang S. Differential stepwise evolution of SARS coronavirus functional proteins in different host species. BMC Evol Biol. Mar 2009; 9. PubMed Central. doi:10.1186/1471-2148-9-52.
- Bordería AV, Stapleford KA, Vignuzzi M. RNA virus population diversity: implications for inter-species transmission. Curr Opin Virol. 2011 Dec;1(6):643–48. ScienceDirect. doi:10.1016/j.coviro.2011.09.012.
- Ayouba A, Akoua-Koffi C, Calvignac-Spencer S, Esteban A, Locatelli S, Li H, Li Y, Hahn BH, Delaporte E, Leendertz FH, et al. Evidence for continuing cross-species transmission of SIVsmm to humans: characterization of a new HIV-2 lineage in rural côte d’ivoire. AIDS (London, England). Sept 2013;27 (15). PubMed Central. doi:10.1097/01.aids.0000432443.22684.50.
- Anthony SJ, Johnson CK, Greig DJ, Kramer S, Che X, Wells H, Hicks AL, Joly DO, Wolfe ND, Daszak P, et al. Global patterns in coronavirus diversity. Virus Evol. Jan 2017;3 (1). academic.oup.com. doi:10.1093/ve/vex012.
- Korber B, Myers G. Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res Hum Retroviruses. 1992 Sept;8(9):1549–60. PubMed. doi:10.1089/aid.1992.8.1549.
- Sagar M, Laeyendecker O, Lee S, Gamiel J, Wawer M, Gray R, Serwadda D, Sewankambo N, Shepherd J, Toma J, et al. Selection of HIV variants with signature genotypic characteristics during heterosexual transmission. J Infect Dis. 2009 Feb;199(4):580–89. PubMed Central. doi:10.1086/596557.
- Chohan B, Lang D, Sagar M, Korber B, Lavreys L, Richardson B, Overbaugh J. Selection for human immunodeficiency virus type 1 envelope glycosylation variants with shorter V1-V2 loop sequences occurs during transmission of certain genetic subtypes and may impact viral RNA levels. J Virol. 2005 May;79(10):6528–31. PubMed Central. doi:10.1128/JVI.79.10.6528-6531.2005.
- Ping L-H, Joseph SB, Anderson JA, Abrahams M-R, Salazar-Gonzalez JF, Kincer LP, Treurnicht FK, Arney L, Ojeda S, Zhang M, et al. Comparison of viral env proteins from acute and chronic infections with subtype C human immunodeficiency virus type 1 identifies differences in glycosylation and CCR5 utilization and suggests a new strategy for immunogen design. J Virol. 2013 July;87(13):7218–33. PubMed Central. doi:10.1128/JVI.03577-12.
- Rathore U, Saha P, Kesavardhana S, Kumar AA, Datta R, Devanarayanan S, Das R, Mascola JR, Varadarajan R. Glycosylation of the core of the HIV-1 envelope subunit protein Gp120 is not required for native trimer formation or viral infectivity. J Biol Chem. 2017 June;292(24):10197–219. PubMed Central. doi:10.1074/jbc.M117.788919.
- Parrish NF, Gao F, Li H, Giorgi EE, Barbian HJ, Parrish EH, Zajic L, Iyer SS, Decker JM, Kumar A, et al. Phenotypic properties of transmitted founder HIV-1. Proc Natl Acad Sci U S A. 2013 Apr;110(17):6626–33. PubMed Central. doi:10.1073/pnas.1304288110.
- Astrakhantseva I, Campo DS, Araujo A, Teo CG, Khudyakov Y, Kamili S. Variation in physicochemical properties of the hypervariable region 1 during acute and chronic stages of hepatitis C virus infection. In 2011 IEEE International Conference on Bioinformatics and Biomedicine Workshops (BIBMW), 2011. Semantic Scholar. doi:10.1109/BIBMW.2011.6112357
- D’Arienzo V, Moreau A, D’Alteroche L, Gissot V, Blanchard E, Gaudy-Graffin C, Roch E, Dubois F, Giraudeau B, Plantier JC, et al. Sequence and functional analysis of the envelope glycoproteins of hepatitis C virus variants selectively transmitted to a new host. J Virol. 2013 Dec;87(24):13609–18. jvi.asm.org. doi:10.1128/JVI.02119-13.
- Li, Hui, et al. “Molecular Identification of Transmitted/Founder Hepatitis C Viruses and Their Progeny by Single Genome Sequencing.” Methods in Molecular Biology (Clifton, N.J.), vol. 1911, 2019, pp. 139–55. PubMed, doi:10.1007/978-1-4939-8976-8_9
- Regenmortel V, Marc HV. Development of a preventive HIV vaccine requires solving inverse problems which is unattainable by rational vaccine design. Front Immunol. 2018;8. Frontiers. doi:10.3389/fimmu.2017.02009.
- Liao H-X, Tsao C-Y, Alam SM, Muldoon M, Vandergrift N, Ma B-J, Lu X, Sutherland LL, Scearce RM, Bowman C, et al. Antigenicity and immunogenicity of transmitted/ founder,consensus, and chronic envelope glycoproteins of human immunodeficiency virus type 1. J Virol. 2013 Apr;87(8):4185–201. jvi.asm.org. doi:10.1128/JVI.02297-12.
- Burm R, Collignon L, Mesalam AA, Meuleman P. Animal models to study hepatitis C virus infection. Front Immunol. May 2018; 9. PubMed Central. doi:10.3389/fimmu.2018.01032.
- Policicchio BB, Pandrea I, Apetrei C. Animal models for HIV cure research. Front Immunol. 2016;7. Frontiers. doi:10.3389/fimmu.2016.00012.
- Chen G-W, Chang S-C, Mok C-K, Lo Y-L, Kung Y-N, Huang J-H, Shih Y-H, Wang J-Y, Chiang C, Chen C-J, et al. Genomic signatures of human versus avian influenza A viruses. Emerg Infect Dis. 2006 Sept;12(7):1353–60. PubMed Central. doi:10.3201/eid1209.060276.
- Folgori A, Capone S, Ruggeri L, Meola A, Sporeno E, Ercole BB, Pezzanera M, Tafi R, Arcuri M, Fattori E, et al. A T-cell HCV vaccine eliciting effective immunity against heterologous virus challenge in chimpanzees. Nat Med. 2006 Feb;12(2):190–97. www.nature.com. doi:10.1038/nm1353.
