ABSTRACT
Current vaccines, which induce a B-cell-mediated antibody response against the spike protein of SARS-CoV-2, have markedly reduced infection rates. However, the emergence of new variants as a result of SARS-CoV-2 evolution requires the development of novel vaccines that are T-cell-based and that target mutant-specific spike proteins along with ORF1ab or nucleocapsid protein. This approach is more accommodative in inducing highly neutralizing antibodies, without the risk of antibody-dependent enhancement, as well as memory CD8+T-cell immunity.
To the Editor,
Immunity against severe acute respiratory syndrome coronavirus (SARS-CoV)-2 infection is an important topic of investigation in COVID-19 research. The analysis of immune correlates has helped in the design of many vaccines against SARS-CoV-2. To date, there are 108 candidate vaccines under clinical development and 184 vaccines in pre-clinical development. Three vaccines have been shown to have more than 90% efficacy in clinical trials. These include the mRNA-based vaccines BNT162b1Citation1 and mRNA-1273,Citation2 and the chimpanzee adenovirus vectored vaccine ChAdOx1 nCoV-19 (AZD1222).Citation3 The vaccine trial with BNT162b1, which is a lipid nanoparticle-formulated nucleoside-modified mRNA that encodes the receptor-binding domain (RBD) of the SARS-CoV-2 spike (S)-protein, reported efficacy in adults. In most participants, BNT162b1 elicited robust RBD-specific CD4+ type 1 T-helper (Th1)-biased responses and strong neutralizing antibody responses. The anti-RBD IgG levels were higher in individuals who received the vaccine compared to those who had a natural infection and were capable of neutralizing pseudoviruses with diverse SARS-CoV-2 S-protein variants.Citation1 Similarly, the vaccine trial with mRNA-1273, which is a lipid nanoparticle-encapsulated mRNA-based vaccine that encodes the prefusion stabilized full-length S-protein, elicited primary CD4+ Th1-biased responses and high levels of neutralizing antibodies.Citation2 The adenovirus-vectored vaccine AZD1222 was also shown to be successful in inducing anti-S-IgG responses.Citation3 Accordingly, these three vaccine types have been authorized by the World Health Organization (WHO) for emergency useCitation4,Citation5 and are mainly being administered in North America and Europe, among other countries. In addition, a number of other vaccines are being administered in various parts of the world; these include Ad26.COV2.S (viral vector; authorized for emergency use by WHO), BBIBP-CorV (inactivated virus; authorized for emergency use by WHO),Citation4,Citation5 CoviVac (viral vector), Gam-COVID-Vac (Sputnik V; viral vector), CoronaVac (inactivated virus), Covaxin (inactivated virus), QazCovid-in (inactivated virus), EpiVacCorona (protein subunit),Citation4 Sputnik Light (viral vector), Convidecia (viral vector), WIBP-CorV (inactivated virus), Minhai (inactivated virus), COVIran Barakat (inactivated virus), Zifivax (protein subunit), Abdala (protein subunit), Soberana 02 (protein subunit) and MVC-COV1901 (protein subunit). Moreover, early clinical data have shown great promise with the NVX-CoV2373 (Novavax) vaccine, which is a recombinant nanoparticle that contains the full-length S-glycoprotein of the prototype strain plus Matrix-M adjuvant.Citation6 Generally, the aforementioned vaccines confer between 50 and 95% protection against SARS-CoV-2 infection.
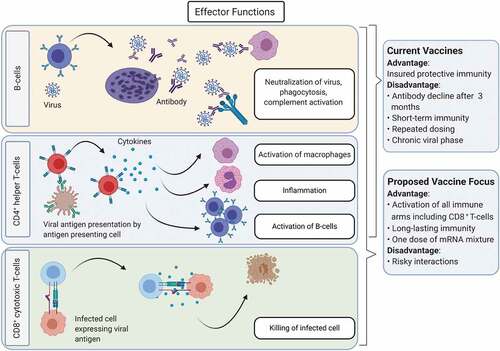
Most of the vaccine development efforts have focused on the B-cell-mediated antibody response against the S-protein of the virus, with a preference of inducing a Th1-biased CD4 T-cell response.Citation7 However, there are concerns regarding the sole focus on vaccines that target the S-protein. Although such vaccines induce a strong anti-S-IgG response and provide protective immunity against the original/current SARS-CoV-2 strains; these antibodies may fail to neutralize the virus or be less efficient in neutralizing variants in patients who become infected/re-infected with a mutated strain. As SARS-CoV-2 strains evolve, new mutations in the S-protein are being discovered. These include mutations in three main epitopes in the RBD, which affect binding by polyclonal antibodies. The impact of such mutations varies substantially among individuals and also over time;Citation8 however, mutations that mostly affect antibody binding are located at few sites in the RBD’s receptor-binding motif. Of particular concern was the E484 in the B.1.1.7 variant, which is the most critical site in the S-protein that may increase vaccine resistance, and where neutralization by some sera was reduced more than 10-fold.Citation8 However, with the emergence of new SARS-CoV-2 mutations/variants, the B.1.1.7 with E484 is replaced by more dominant variants. As of June 2021, 28 variants of concern/under investigation have been reported, with the B.1.617.2 variant (Delta Variant) becoming increasingly dominant.Citation9 Therefore, to combat such antigenic evolution by SARS-CoV-2, vaccines targeted against non-S-based protective epitopes/proteins in addition to S-epitopes/proteins must be considered.
Moreover, the pre-existence of anti-S-IgG (induced by vaccines targeting the S-protein or previous coronavirus infection(s)) that is sub-neutralizing, as a result of mutations in the S-protein, may accelerate infectivity through antibody-dependent enhancement (ADE) and lead to severe disease. Higher antibody titers have been reported in patients with severe COVID-19,Citation10 which may have been due to ADE as a result of prior coronavirus exposures/antigenic heterogeneity. Early in the pandemic, concerns were raised with regard to vaccine-induced enhancement of disease via ADE.Citation11 Thereafter, two different ADE risks for SARS-CoV-2 antibodies have been described. The first ADE risk is mediated by antibody-dependent infection of macrophages via Fc receptors. The second ADE risk is related to activation and degranulation of mast cells with Fc receptor-bound SARS-CoV-2 antibodies, which leads to increased histamine release;Citation12 this model is consistent with multisystem inflammatory syndrome in infants with maternally transferred antibodies to SARS-CoV-2.Citation12 These two ADE risks have critical implications for B-cell-based vaccines for subsets of the population; these are based on age, pregnancy, cross-reactive antibodies, and variabilities in antibody levels over time.Citation12
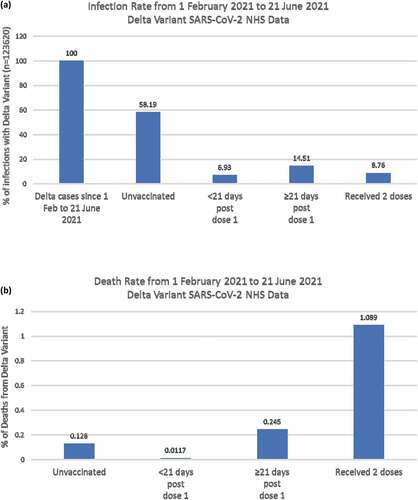
Analysis of data from the National Health Service (NHS) published by Public Health EnglandCitation9 has shown that the infection rate with the Delta variant post-vaccination (<21 days post-dose 1, ≥21 days post-dose 1, and after two doses) was drastically lower compared to unvaccinated cases (). However, the death rate as a result of infection with the Delta variant was 8-folds higher in cases that had received two doses of the vaccine when compared to unvaccinated cases ().Citation9 These data support the notion that pre-existing anti-S-IgG induced by the vaccine may be sub-neutralizing in a subset of individuals (hence becoming infected in spite of being vaccinated) and thus may accelerate infectivity via ADE and lead to higher death rates. Taken together with models presented by Ricke,Citation12 these data further emphasize the importance of developing novel vaccines that are T-cell-based and not dependent only upon antibodies.
Figure 1. Infection and death rates by vaccination status among Delta confirmed cases in England, 1 February 2021 to 21 June 2021. (a) This figure shows the infection rate in each of the specified groups compared to total Delta cases. The groups include unvaccinated cases, cases with <21 days post dose 1, cases with ≥21 days post dose 1, and cases that received 2 vaccine doses. (b) This figure shows the death rate in four groups of Delta cases, these include unvaccinated cases, cases with <21 days post dose 1, cases with ≥21 days post dose 1, and cases that received 2 vaccine doses. This figure was produced using NHS data published by Public Health England.Citation9
The involvement of ADE in other pathogenic coronavirus infections has been described.Citation13 In animals infected with SARS-CoV, anti-S-IgG altered inflammatory responses leading to severe acute lung injury. Of particular concern, animals vaccinated with SARS-CoV protein showed more pronounced acute lung injury compared to unvaccinated animals. Additionally, although the adoptive transfer of anti-S-IgG neutralizing antibodies reduced viral load upon subsequent challenge, these antibodies resulted in acute diffuse alveolar damage.Citation13 These findings suggest that pre-existing anti-S-IgG at the acute stage of SARS-CoV infection may have enhanced disease severity. Concerningly, the same may hold true for SARS-CoV-2 infection (post-vaccination or initial infection) whereby a certain subset of patients who produce early and/or sub-optimal antibodies, which fail to clear the virus but facilitate viral replication, succumb to severe disease. The early production of anti-S-IgG, prior to viral clearance, may lead to the formation of anti-S-IgG-Fc receptor (FcR) complexes on the surface of monocytes/macrophages. Such immune complexes may alter the polarization of alveolar macrophages leading to the accumulation of proinflammatory monocytes/macrophages and the production of inflammatory cytokines. This pathologic mechanism is supported by the finding that FcR blockade reduces inflammatory cytokine production.Citation13 To circumvent the potential risk of ADE as a result of SARS-CoV-2 evolution, new generation vaccines should encompass non-S-antigens that are not subject to mutational variation as well as the S-based antigens.
Critically, vaccines focused solely on eliciting neutralizing antibodies to the S-protein may fail to induce long-term immunity.Citation10 Indeed, anti-S-neutralizing antibodies decline three months post-infection with SARS-CoV-2. Therefore, vaccines that elicit both antibody and memory cellular immune responses are key for long-lasting immunity. Knowledge gained from SARS-CoV immunity suggests that cytotoxic CD8+T-cell responses are key for long-term immunity. In patients who recovered from SARS-CoV infection, memory CD8+T-cells persisted for 6–11 years, whereas memory B-cells and IgG antibodies were undetectable.Citation14,Citation15 Importantly, the vast majority of epitopes (26 of 29) that are recognized by memory CD8+ T-cells from patients convalescing from SARS-CoV-2 infection are located in the open reading frame (ORF)1ab, N, M, and ORF3a, rather than the S-protein (3 with only 1 in the RBD region).Citation16 Therefore, vaccines that target the S-protein solely may largely fail to induce memory CD8+T-cell responses, and as such, the vaccines’ efficacy may not be long-lasting. In addition, the ORF/N/M epitopes are located in regions of the virus that are not subject to significant mutational variation.Citation16 Accordingly, adding non-S-epitopes makes for a better vaccine candidate.
In conclusion, given the amenability of the mRNA-based vaccine platforms, designing mutant-specific S-proteins along with ORF1ab or nucleocapsid protein, may be more accommodative in inducing both highly neutralizing antibodies as well as memory CD8+T-cell immunity that truly mimic natural infection (). In addition, we believe that using the RBD domain as an antigen may not induce a robust immune response. As shown by Corbett et al., having 2 proline substitutions (2P) at the apex of the central helix and heptad repeat 1 stabilized the prefusion fusion protein in Middle East respiratory syndrome coronavirus, SARS-CoV, and the S-protein of SARS-CoV-2.Citation17 While prefusion-stabilized surface fusion proteins are necessary for inducing a robust immunogenic reaction, a similar design may be avoided for vaccines based on non-surface antigens.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Additional information
Funding
References
- Sahin U, Muik A, Derhovanessian E, Vogler I, Kranz LM, Vormehr M, Baum A, Pascal K, Quandt J, Maurus D, et al. COVID-19 vaccine BNT162b1 elicits human antibody and T H 1 T cell responses. Nature. 2020;586:594-599. doi:10.1038/s41586-020-2814-7
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384:403-416. doi:10.1056/NEJMoa2035389
- Voysey M, Clemens SAC, Madhi SA, Weckx LY, Folegatti PM, Aley PK, Angus B, Baillie VL, Barnabas SL, Bhorat QE, et al. Safety and efficacy of the ChAdOx1 nCoV-19 vaccine (AZD1222) against SARS-CoV-2: an interim analysis of four randomised controlled trials in Brazil, South Africa, and the UK. Lancet. 2021;397:99–111
- World Health Organization. COVID-19 vaccine tracker and landscape; 2021 Jul 16 [accessed 2021 July 24]. https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines.
- World Health Organization. Coronavirus disease (COVID-19) pandemic. [accessed 2021 July 24]. https://www.who.int/emergencies/diseases/novel-coronavirus-2019.
- Heath PT, Galiza EP, Baxter DN, Boffito M, Browne D, Burns F, Chadwick DR, Clark R, Cosgrove C, Galloway J, et al. Safety and efficacy of NVX-CoV2373 Covid-19 vaccine. N Engl J Med. 2021. doi:10.1056/NEJMoa2107659.
- Lambert PH, Ambrosino DM, Andersen SR, Baric RS, Black SB, Chen RT, Dekker CL, Didierlaurent AM, Graham BS, Martin SD, et al. Consensus summary report for CEPI/BC March 12- 13, 2020 meeting: assessment of risk of disease enhancement with COVID-19 vaccines. Vaccine. 2020;38(31):4783–91. doi:10.1016/j.vaccine.2020.05.064.
- Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. Greaney AJ, Loes AN, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. Comprehensive mapping of mutations in the SARS-CoV-2 receptor-binding domain that affect recognition by polyclonal human plasma antibodies. Cell Host & Microbe. 2021;29(3):463-476.e6. doi:10.1016/j.chom.2021.02.003.
- Public Health England. SARS-CoV-2 variants of concern and variants under investigation in England. Technical briefing 18. [online] Gov.uk; 2021 Jul 9 [accessed 2021 July 24]. https://assets.publishing.service.gov.uk/government/uploads/system/uploads/attachment_data/file/1001358/Variants_of_Concern_VOC_Technical_Briefing_18.pdf.
- Seow J, Graham C, Merrick B, Acors S, Pickering S, Steel KJA, Hemmings O, O’Byrne A, Kouphou N, Galao RP, et al. Longitudinal observation and decline of neutralizing antibody responses in the three months following SARS-CoV-2 infection in humans. Nat Microbiol. 2020. doi:10.1038/s41564-020-00813-8.
- Tetro JA. Is COVID-19 receiving ADE from other coronaviruses? Microbes Infect. 2020;22(2):72–73. doi:10.1016/j.micinf.2020.02.006.
- Ricke DO. Two different antibody-dependent enhancement (ADE) risks for SARS-CoV-2 antibodies. Front Immunol. 2021;12:640093. doi:10.3389/fimmu.2021.640093.
- Liu L, Wei Q, Lin Q, Fang J, Wang H, Kwok H, Tang H, Nishiura K, Peng J, Tan Z, et al. Anti-spike IgG causes severe acute lung injury by skewing macrophage responses during acute SARS-CoV infection. JCI Insight. 2019;4:4. doi:10.1172/jci.insight.123158.
- Tang F, Quan Y, Xin Z-T, Wrammert J, Ma M-J, Lv H, Wang T-B, Yang H, Richardus JH, Liu W, et al. Lack of peripheral memory B cell responses in recovered patients with severe acute respiratory syndrome: a six-year follow-up study. J Immunol. 2011;186(12):7264–68. doi:10.4049/jimmunol.0903490.
- Peng H, Yang LT, Wang LY, Li J, Huang J, Lu Z-Q, Koup RA, Bailer RT, Wu C-Y. Long-lived memory T lymphocyte responses against SARS coronavirus nucleocapsid protein in SARS-recovered patients. Virology. 2006;351(2):466–75. doi:10.1016/j.virol.2006.03.036.
- Ferretti AP, Kula T, Wang Y, Nguyen DMV, Weinheimer A, Dunlap GS, Xu Q, Nabilsi N, Perullo CR, Cristofaro AW, et al. Unbiased screens show CD8+ T cells of COVID-19 patients recognize shared epitopes in SARS-CoV-2 that largely reside outside the spike protein. Immunity. 2020;53(5):1095–107 e1093. doi:10.1016/j.immuni.2020.10.006.
- Corbett KS, Edwards DK, Leist SR, Abiona OM, Boyoglu-Barnum S, Gillespie RA, Himansu S, Schäfer A, Ziwawo CT, DiPiazza AT, et al. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586(7830):567–71. doi:10.1038/s41586-020-2622-0.