ABSTRACT
We have investigated six COVID-19 recovered cases with two doses of Covishield vaccination followed by reinfection. The primary SARS-CoV-2 infection found to occur with B.1 and reinfection with Omicron BA.1 and BA.2 variants. The genomic characterization and duration between two infections confirms these cases as SARS-CoV-2 reinfection. The immune response determined at different time intervals demonstrated boost post two dose vaccination, decline in pre-reinfection sera post 7 months and rise post reinfection. In conclusion, it was observed that these cases got SARS-CoV-2 reinfection with declined hybrid immunity acquired from primary infection and two dose covishield vaccination. This findings suggests the need to protect the community through booster dose of vaccination and prevent further infections following personal hygiene and non-pharmaceutical interventions.
To the Editor,
We read the article by Hasan et al., describing a case of SARS-CoV-2 breakthrough infection post two-dose vaccination and reinfection with Delta variant. The study emphasized the need to understand the duration of vaccine-induced immunity and its effectiveness against newly emerged SARS-CoV-2 variants.Citation1 A recent study reported that the COVID-19 recovered cases are 16 times more likely to be reinfected with Omicron than Delta variant.Citation2 However, the understanding the characteristics of reinfection and correlation with immune response is of much significance.
Here, we investigated six individuals with primary SARS-CoV-2 infection during March 2020 to October 2021 (first wave). The cases were then vaccinated with two doses of Covishield vaccine post three months of primary infection. They were followed until the occurrence of reinfection during January 2022 to February 2022 (third wave) in India. The suspected reinfection cases were identified as a positive SARS-CoV-2 Real time RT-PCR (Ct value <30) ≥90 days after the initial positive test and at least one consecutive negative test result between two incidences.Citation3 The naso/oropharyngeal swab and blood specimens were collected at four different time points namely primary SARS-CoV-2 infection, post 60 days of second vaccine dose, pre-reinfection (post 7 months of second dose), reinfection (post 10 months of second dose). The SARS-CoV-2 confirmation and genomic characterization was done using E gene specific Real time RT-PCR and next-generation sequencing.Citation4,Citation5 The IgG immune response was determined using SARS-CoV-2 S1-RBD specific IgG ELISA.Citation6 We have also measured the neutralizing antibody (NAb) titers of sera against Delta, Omicron and ancestral SARS-CoV-2 B.1 variant using a plaque reduction neutralization test (PRNT).Citation7
All the six cases tested positive for SARS-CoV-2 using Real time RT-PCR specific for E gene (viral RNA copy number 8.9 × 103 to 1.6 × 106/ml). The next-generation sequencing of the six cases revealed the primary infection with SARS-CoV-2 prototype B.1 strain (). The mean IgG antibody titer of the cases at reinfection was found to be 300 (). Besides this, the NAb geometric mean titer (GMT) of the sera against B.1, Delta and Omicron was 643 (95% CI 295–1404), 43 (95% CI 6–291), 1.9 (95% CI 0.16–22), respectively ().
Figure 1. (a) Molecular phylogenetic tree designed based on a neighbor joining analysis of SARS-CoV-2 sequences retrieved from clinical specimens of cases during primary infection, reinfection and reference sequences with bootstrap of 1000 replicates. The sequence of primary infection (EI*) and reinfection (RI*) cases are highlighted with blue color and violet color, respectively. Reference sequences are marked with black color along with prototype Wuhan strain in red color. (b) Anti-SARS-CoV-2 IgG antibody response determined with S1-RBD ELISA during primary infection, post sixty days of second dose vaccination, pre-reinfection and reinfection. Neutralizing antibody titers determined with plaque reduction neutralization test (PRNT) during primary infection, post sixty days of second dose vaccination, pre-reinfection and reinfection against (c) B.1 variant, (d) Delta variant, (e) Omicron variant. The dotted line represents the limit of detection of the assay. Data are presented as mean values ± standard deviation (SD). The statistical significance was assessed using the Kruskal–Wallis test and Dunn’s multiple comparisons test. Two-tailed Mann–Whitney test was performed between the groups, if the p-value for the Kruskal–Wallis test was found to be significant; p-values less than .05 were considered to be statistically significant.
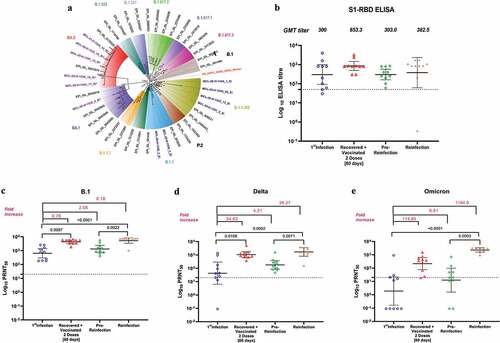
The serum samples collected after 2 months from second dose vaccination shown 2.8-fold rise in IgG and 6.8, 24.5, 475-fold rise in NAb titers against B.1, Delta and Omicron, respectively, than the primary infection. The pre-reinfection sera collected after 7 months of second vaccination demonstrated reduction in IgG antibodies titer (2.8-fold) and also in the NAb titers against B.1 (3.3-fold), Delta (5.9-fold) and Omicron (17.3-fold) ().
The SARS-CoV-2 reinfection amongst these COVID-19recovered two dose vaccinees found to occur after 10 months after second dose vaccination. The genomic characterization of six clinical specimens (viral RNA copy number 1.1 × 105 to 1.4 × 108/ml) revealed infecting SARS-CoV-2 variant to be Omicron sub lineage BA.1 (n = 1) and BA.2 (n = 5) (). The primary infection with B.1 and reinfection with BA.1 and duration between two infections confirms these cases as SARS-CoV-2 reinfection. The mutation analysis of the reinfection cases represented the specific mutations and deletions in BA.1 and common mutations/deletions of BA.1 and BA.2, of which common mutations found in BA.1 & BA.2 (K417N, S477N, T478K, E484A, Q493 R, Q498 R, N501Y, Y505 H) correlate with immune evasion potential of BA.1 and BA.2 sub lineages. The NAb GMT of reinfection cases sera against B.1, Delta and Omicron were found to be 5261 (95%CI: 3413–8111), 1685 (95%CI: 811–3501) and 2262 (95%CI: 1558–192.4), respectively. These titers represent significant boost in immune response post reinfection compared to pre-reinfection sera. As the reinfection occurred with Omicron sub lineages, about 1000-fold rise was observed in NAb titers in reinfection sera with Omicron than B.1 and Delta. Apparently, IgG titer showed only 1.3-fold increase at reinfection.
The findings of the study observed increasing immune response after two dose vaccination than at primary infection. Subsequently, this hybrid immunity found to decline to very low level at 7 month post second vaccination with which cases got reinfection. This suggests the need to protect the community through booster dose of vaccination and prevent further infections following personal hygiene and non-pharmaceutical interventions.
Acknowledgement
Authors gratefully acknowledge the staff of ICMR-NIV, Pune including Dr. Rajlaxmi Jain, Mrs. Triparna Majumdar, Mrs. Savita Patil, Mr. Annasaheb Suryawanshi, Ms. Pranita Gawande, Ms. Jyoti Yemul, Mr. Yash Joshi, Ms. Manisha Dudhmal, Ms. Aasha Salunkhe and Mr. Chetan Patil, for extending excellent technical support.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Hasan DA, Maulud SQ, Jalal PJ, Priyanka, Choudhary OP. SARS-CoV-2 vaccine breakthrough reinfection in a health-care worker of Iraq: a case report. Hum Vacc Immunother. 2022 Apr 14:1–3.
- UK Office for National Statistics. Coronavirus (COVID-19) infection survey, characteristics of people testing positive for COVID-19, UK: 19 January 2022 [Internet]; 2022 [accessed 2022 Jan 21]. https://www.ons.gov.uk/peoplepopulationandcommunity/healthandsocialcare/conditionsanddiseases/bulletins/coronaviruscovid19infectionsurveycharacteristicsofpeopletestingpositiveforcovid19uk/latest#reinfections-withcovid-19-uk.
- Yahav D, Yelin D, Eckerle I, Eberhardt CS, Wang J, Cao B, Kaiser L. Definitions for coronavirus disease 2019 reinfection, relapse and PCR re-positivity. Clin Microbiol Infect. 2021;27(3):315–18. doi:10.1016/j.cmi.2020.11.028.
- Choudhary ML, Vipat V, Jadhav S, Basu A, Cherian S, Abraham P, Potdar V. Development of in vitro transcribed RNA as positive control for laboratory diagnosis of SARS-CoV2 in India. Ind J Med Res. 2020;151(2):251. doi:10.4103/ijmr.IJMR_671_20.
- Yadav PD, Nyayanit DA, Shete AM, Jain S, Majumdar TP, Chaubal GY, Shil P, Kore PM, Mourya DT. Complete genome sequencing of Kaisodi virus isolated from ticks in India belonging to Phlebovirus genus, family Phenuiviridae. Ticks Tick Borne Dis. 2019;10(1):23–33. doi:10.1016/j.ttbdis.2018.08.012.
- Deshpande G, Kaduskar O, Deshpande K, Bhatt V, Yadav P, Gurav Y, Potdar V, Khutwad K, Vidhate S, Salunke A, et al. Longitudinal clinic-serological analysis of anti-nucleocapsid and anti-receptor binding domain of spike protein antibodies against SARS-CoV-2. Int J Infect Dis. 2021;112:103–10. doi:10.1016/j.ijid.2021.09.024.
- Deshpande GR, Sapkal GN, Tilekar BN, Yadav P, Gurav Y, Gaikwad S, Kaushal H, Deshpande K, Kaduskar O, Sarkale P, et al. Neutralizing antibody responses to SARS-CoV-2 in COVID-19 patients. Ind J Med Res. 2020 July;152(1):82. doi:10.4103/ijmr.IJMR_2382_20.
