ABSTRACT
The coronavirus disease 2019 (COVID-19) pandemic has illustrated the critical need for effective prophylactic vaccination to prevent the spread of severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). Intranasal vaccination is an attractive approach for preventing COVID-19 as the nasal mucosa is the site of initial SARS-CoV-2 entry and viral replication prior to aspiration into the lungs. We previously demonstrated that a single intranasal administration of a candidate adenovirus type 5-vectored vaccine encoding the receptor-binding domain of the SARS-CoV-2 spike protein (AdCOVID) induced robust immunity in both the airway mucosa and periphery, and completely protected K18-hACE2 mice from lethal SARS-CoV-2 challenge. Here we show that a single intranasal administration of AdCOVID limits viral replication in the nasal cavity of K18-hACE2 mice. AdCOVID also induces sterilizing immunity in the lungs of mice as reflected by the absence of infectious virus. Finally, AdCOVID prevents SARS-CoV-2 induced pathological damage in the lungs of mice. These data show that AdCOVID not only limits viral replication in the respiratory tract, but it also prevents virus-induced inflammation and immunopathology following SARS-CoV-2 infection.
Introduction
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) emerged in late 2019 and has resulted in a global pandemic of an acute respiratory disease, named “coronavirus disease 2019” (COVID-19), which threatens human health and public safety. The infective dose for SARS-CoV-2 remains undetermined, but it is known that airborne transmission occurs via inhalation of respiratory droplets expelled from infected hosts.Citation1,Citation2 SARS-CoV-2 entry into host cells is dependent on engagement of the receptor-binding domain (RBD) of the spike protein to the viral entry receptor angiotensin-converting enzyme-2 (ACE2), initiating fusion of the virus with the cellular membrane.Citation3 ACE2 is expressed in the nasal goblet and ciliated epithelial cells lining the respiratory tract and the lungs, and it is hypothesized that the nasal cavity serves as the initial reservoir for seeding of SARS-CoV-2 to the lungs.Citation4,Citation5 High viral load in the nasopharyngeal space directly correlates with SARS-CoV-2 transmission and mortality.Citation6–10 The lung is the principle organ affected in COVID-19 patients with hypoxic respiratory failure being the primary cause of COVID-19 mortality.Citation11 The lungs of COVID-19 patients exhibit distinct histological characteristics including acute endothelial injury, pervasive capillary inflammation, fibrosis, thrombosis, and apoptotic or pyroptotic cell death.Citation12,Citation13 These data suggest that the optimal anti-SARS-CoV-2 immune response for preventing disease will contain a mucosal immunity component.
The urgent need for safe and effective SARS-CoV-2 countermeasures has resulted in eleven vaccines being marketed worldwide, with multiple vaccines being granted authorization by the U.S. Food and Drug Administration (USFDA) for adults, adolescents, and children.Citation14–17 The vast majority of SARS-CoV-2 vaccines are either inactivated whole virus vaccines or encode the full-length spike protein (S) as the antigen, and all are delivered by intramuscular (IM) injection.Citation18 IM injection elicits systemic immunity but cannot yield potent mucosal immunity.Citation19,Citation20 Alternatively, a noninvasive administration route such as intranasal delivery may be attractive.Citation21 Data suggest intranasal vaccination, acting in a manner analogous to viral infection, has the potential to confer sterilizing immunity in the respiratory tract, reducing the morbidity and transmission of COVID-19.Citation22–27 By removing the need for needles, intranasal delivery may also increase vaccine uptake, presenting a more efficient vaccine administration method.Citation28
We recently reported that a replication-deficient adenovirus serotype 5 (Ad5)-vectored vaccine candidate encoding only the SARS-CoV-2 RBD was highly immunogenic in mice when delivered by the intranasal route.Citation29 RBD-only vaccines may have multiple advantages over trimeric spike-encoding vaccines. Immunofocusing of the antibody repertoire may yield higher affinity antibodies and neutralizing antibodies directed against conserved subdominant epitopes, allowing for potent cross-variant neutralization.Citation30–33 Furthermore, the majority of neutralizing antibodies in human convalescent sera are directed against the RBD, highlighting the importance of anti-RBD responses in protective immunity.Citation34–36 Our vaccine, referred to as AdCOVID, elicited systemic neutralizing antibody responses in the sera of animals that persisted for at least six months.Citation29 AdCOVID administration also resulted in a robust population of anti-RBD T cells and anti-spike IgA antibodies in the lungs of all vaccinated mice. Importantly, AdCOVID was completely protective against lethal SARS-CoV-2 challenge in K18-hACE2 mice, with no animals exhibiting overt morbidity or mortality.
Here, we further investigated the protective efficacy of AdCOVID in K18-hACE2 mice. We report that a single intranasal administration of AdCOVID significantly decreased the copies of viral RNA in SARS-CoV-2 challenged mice compared to vehicle controls. AdCOVID vaccination also prevented the seeding of infectious virus in the lungs of challenged mice. Finally, AdCOVID vaccination significantly reduced SARS-CoV-2-induced immunopathology in challenged mice.
Methods
Ethics statement and mice
K18-hACE2 breeding pairs were originally purchased from Jackson Laboratory (Strain #034860) and subsequent generations were bred at the University of Alabama at Birmingham (UAB). Animal procedures performed at UAB were conducted in accordance with Public Health Service Policy on the Humane Care and Use of Laboratory Animals and Guide for the Care and Use of Laboratory Animals. Studies were performed in the Southeastern Biosafety Lab (SEBLAB) ABSL3 located at UAB. Studies were performed under UAB’s Institutional Animal Care and Use Committee (IACUC: D16–00162 A3255–01) Protocol 22065 approved on 27 April 2020.
Vaccine candidate
The vaccine candidate evaluated in this study was based on a replication-deficient, E1- and E3-deleted Ad5 vector platform and expresses a human codon-optimized gene for the RBD domain (residues 302 to 543) of SARS-CoV-2 spike protein (accession number QHD43416.1).Citation37 The Ad5-vectored RBD transgene includes a human tissue plasminogen activator leader sequence and is expressed under the control of the cytomegalovirus immediate early promoter/enhancer.Citation29 An initial seed stock was obtained from transfection of recombinant vector plasmid into E1-complementing PER.C6 cells as previously described.Citation29 Vaccine titers were measured in HEK 293 cells as previously described.Citation29
Propagation of SARS-CoV-2
The original SARS-CoV-2 isolate USA-WA1/2020 was obtained from BEI resources (#NR-52281) and propagated in Vero E6 cells (ATCC, #CRL-1586). Confluent cells in a 25 CM2 flask were inoculated with virus (0.015 MOI) in serum free Eagle’s MEM (Life Technologies, #11430–030) with added supplements, and incubated with rocking at 35°C (5% CO2) for 2 hours. The inoculum was then removed, cells were overlaid with 2% FBS in media, and virus growth continued until obvious cytopathic effect (CPE) was observed in 2 to 3 days post infection. A large-scale working stock was obtained through 2–3 more passages of virus in cells in 150 CM2 flasks until significant viral titer was achieved.
K18-hACE2 vaccination and challenge
Mixed sex K18-hACE2 mice of at least 6 weeks of age were randomly allocated into vaccination groups for immunogenicity experiments. 2.2E + 09 infectious units (ifu) of replication-deficient Ad5 vector encoding the RBD (AdCOVID) from the SARS-CoV-2 spike protein were administered intranasally at the described doses in a volume of 50 µL (25 µL/nostril). Briefly, mice were sedated with isoflurane and placed in the supine position. AdCOVID was delivered into the nasal mucosa by pipette during normal inhalation, and mice were allowed to recover naturally. The control group received 50 µL of vehicle alone by intranasal administration. On study day 32, all mice were challenged by the intranasal route with LD50 doses of either 2.0E + 04 or 0.5E + 04 plaque forming units (PFU) SARS-CoV-2 strain USA/WA-1/2020. Mice were euthanized either 3- or 4-days post-infection (DPI) as indicated.
Serum collection
Blood samples were collected from the submandibular vein of vaccinated mice into BD Microtainer blood collection tubes (BD Biosciences, #365963). The samples were centrifuged at 13,000 rpm at RT for 10 minutes and the serum was collected, aliquoted and frozen at −80ºC until analyzed.
Recombinant SARS-CoV-2 protein production
To produce recombinant SARS-CoV-2 spike ectodomain protein, two human codon-optimized constructs were generated with linear sequence order encoding: a human IgG leader sequence, the SARS-CoV-2 spike ectodomain (amino acids 14–1211), a GGSG linker, T4 fibritin foldon sequence, a GS linker, and finally an AviTag (construct 1) or 6X-HisTag (construct 2). Each construct was engineered with two sets of mutations to stabilize the protein in a pre-fusion conformation as previously described.Citation29 IgG standards were generated as previously described.Citation29
SARS-CoV-2 spike cytometric bead array
To generate the spike cytometric bead array (CBA), recombinant SARS-CoV-2 ectodomain trimers were passively absorbed onto streptavidin functionalized fluorescent microparticles (3.6 µm, Spherotech, # CPAK-3567-4K). 500 µg of biotinylated SARS2-CoV-2 was incubated with 2E + 07 streptavidin functionalized fluorescent microparticles in 400 µL of 1% BSA in PBS. Following coupling, the SARS-CoV-2 spike conjugated beads were washed twice in 1 mL of 1% BSA, PBS, 0.05% NaN3, resuspended at 1E + 08 beads/mL and stored at 4°C. The loading of recombinant SARS2-CoV-2 spike onto the beads was evaluated by staining 1E + 05 beads with dilutions ranging from 1 µg/mL to 2 ng/mL of the recombinant anti-SARS spike antibody CR3022 (IDT Biologika) and visualized with an anti-human IgG secondary antibody.
CBA measurement of spike-specific IgG responses
The quantification of SARS-CoV-2 spike IgG was performed in serum samples obtained from all animals using the spike CBA described above. 5 µL of a suspension containing 5E + 05 of each SARS-CoV-2 spike and anti-IgG beads was added to the diluted samples and CBA measurement was conducted as previously described.Citation29
Nasal swipe collection
Nasal swipes were collected immediately prior to intranasal challenge to establish baseline viral load, and then 1-hour post-infection. Nasal swipes were then collected daily through study termination (DPI +4). Awake mice were scruffed and the exterior surface of their nose was swiped for 5–10 seconds with a polyester-tipped swab (ThermoFisher, #22-029-574) that had been dipped in a screw-cap tube containing 300 µL Viral Transport Medium (VTM; HBSS (+Ca2+ +Mg2+) containing 2% FBS, 100 µg/mL Gentamicin and 0.5 µg/mL AmphotericinB). The tip of the swab was cut and dropped inside the VTM-containing tube. Once all swab samples were collected, the tubes were capped and vortexed, and then the swab tips were removed and discarded.
Nasal wash collection
The upper respiratory tract was washed postmortem with 400 µL VTM instilled into the trachea with a blunt 19-gauge needle and flushed out of the nose. 200–300 µL of the nasal wash collected was used for quantification of viral RNA by qRT-PCR. The left-over sample volumes were used for antibody titer quantification.
Tissue processing and single cell isolation
The lungs were excised at euthanasia (without pre-collection of the bronchoalveolar lavage (BAL) fluid) and placed in a 2 mL Lysing Matrix M tube (MP Biomedicals, #116923050-CF) containing 1 mL VTM. The lungs were homogenized using a FastPrep-24 classic bead beating grinder lysis system (MP Biomedicals, #116004500). The total lung homogenate was centrifuged at 10,000 rpm for 10 minutes at room temperature. 300 µL of the homogenate supernatant was used for quantification of viral load by qRT-PCR. The remaining homogenate supernatants were used for quantification of viral load by plaque assay. Brains were excised and placed in a 2-mL Lysing Matrix M tube containing 800 µL VTM and homogenized using a FastPrep-24 classic bead beating grinder lysis system. The homogenate was centrifuged at 10,000 rpm for 10 minutes at room temperature. 300 µL of the homogenate supernatant was used for quantification of viral load by qRT-PCR. The remaining homogenate supernatants were used for quantification of viral load by plaque assay.
qRT-PCR
The quantification of SARS-CoV-2 viral RNA was performed from nasal swipe (300 µL), nasal wash (300 µL), total lung (tissue + BAL) homogenate (300 µL), and brain homogenates (300 µL) isolated from individual mice. Each sample was mixed 1:1 with lysis buffer containing 10% proteinase K from the Maxwell RSC Viral Total Nucleic Acid Purification kit (Promega, #AS1330). The samples were mixed and incubated at 56ºC for 10 minutes. RNA was extracted utilizing the Maxwell RSC 48 Instrument (Promega, Madison, WI) and qRT-PCR performed utilizing QuantStudio™ 5 Real-Time PCR instrument (ThermoFisher, Waltham, MA). The genomic RNA PCR primers (forward 5’-GAC CCC AAA ATC AGC GAA AT-3’ and reverse 5’-TCT GGT TAC TAC TGC CAG TTG AAT CTG-3’) and probe (/SFAM/ACC CCG CAT TAC GTT TGG TGG ACC/BHQ_1) targeted the nucleocapsid (N) gene and were purchased from Integrated DNA Technologies, Inc. (IDT, Coralville, Iowa). AccuPlex™ SARS-CoV-2 Reference Material (SeraCare Life Sciences, Inc, #0505–0126) was extracted and amplified in parallel to generate a standard curve enabling viral quantitation. The lowest limit of quantification for this assay is 25 viral RNA copies/mL. The subgenomic RNA PCR assay utilized a forward primer targeting the leader sequence (5’-CGA TCT CTT GTA GAT CTG TTC TC-3’) and the same reverse primer targeting the N gene with RT-PCR analysis utilizing the Power SYBR™ Green RNA-to-CT™ 1-Step Kit (ThermoFisher, #4391178).
Plaque assay
Six-well plates were seeded with Vero E6 cells (4E + 05/well) 16 hours before the start of the assay. Samples were thawed at 37ºC and centrifuged at 10,000 rpm for 5 minutes to separate the supernatant from any residual tissue debris. The sample supernatants were serially diluted ten-fold in 1x PBS containing 1% FBS. To obtain the lower limit of detection, a 1:2 dilution was also included. The plated Vero E6 cells were infected with each virus dilution and incubated for 1 hour at 37ºC with shaking every 10 minutes. Post-incubation, the virus solution was aspirated, and the cells were overlayed with 0.6% Avicel solution supplemented with 3% FBS and 1x MEM. The plates were incubated in a CO2 incubator for 72 hours. The Avicel overlay was aspirated, and the cell monolayer was fixed with 10% neutral-buffered formalin for 1 hour. The cells were stained with 1% crystal violet solution and plaques were counted for each dilution. The virus titers were determined by multiplying the number of plaques by the dilution factor. The lowest limit of quantification for this assay was determined by the dilution of the lung tissue plated.
Histology and lung pathology assessment
Lung tissues were collected and fixed for seven days in 10% neutral-buffered formalin and then embedded dorsal side down in paraffin using standard procedures. Samples were sectioned at 5 µm, and resulting slides were stained with hematoxylin and eosin (H&E). All tissue slides were evaluated by light microscopy by a board-certified veterinary pathologist blinded to study group allocations. Representative photo images were collected using a Nikon Eclipse Ci microscope (Nikon Inc., Melville, NY) and analyzed with NIS-Elements software (Nikon Inc., Melville, NY). To assess lung histopathologic lesions in mice, we used an algorithm developed at UC Davis.Citation38 Additional scoring of airway, alveolar, and vascular pathology in affected areas was scored as previously indicated.Citation39–41 Briefly, to quantify total airway score, the extent of airway involvement, including the (i) extent of inflammation, (ii) bronchiolar epithelial hyperplasia and (iii) necrosis were individually scored (scale 0–4) and summed. To generate a total alveolar score, (i) alveolar inflammation, (ii) alveolar damage and (iii) type II pneumocyte hyperplasia were individually assessed (scored 0–4) and summed. To determine the vascular score, (i) vascular involvement and inflammation were assessed (scale 0–4) with the presence of perivascular hemorrhage/edema, fibrosis, and fibrin individually measured (absent = 0, present = 1). The airway, alveolar, and vascular scores were added together to generate a total lung pathology score.
Quantification and statistical analysis
Statistical significance was assigned when P values were <0.05 using Prism Version 9.3.1 (GraphPad Software, San Diego, CA). Data was converted to a log scale prior to graphing and statistical analysis. All tests and values are indicated in the relevant figure legends.
Results
Intranasal vaccination with AdCOVID elicits SARS-CoV-2 spike-specific antibody responses
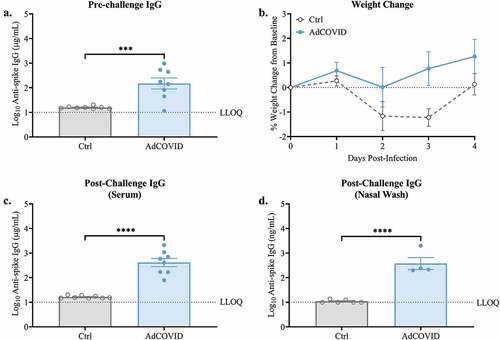
AdCOVID is a replication-deficient, E1- and E3-deleted Ad5 vector platform expressing a human codon-optimized gene for the RBD (residues 302 to 543) from the spike antigen of the Wuhan-1 strain of SARS-CoV-2 (accession number QHD43416).Citation29,Citation37 We previously showed that AdCOVID, administered intranasally as a single dose, was highly immunogenic in both inbred C57BL/6J and outbred CD-1 mice through the induction of mucosal IgA, serum neutralizing antibodies, and CD4+ and CD8+ T cells.Citation29 Here, we vaccinated K18-hACE2 mice with a single dose of AdCOVID (2.2E + 09 ifu) via the intranasal route. At 21 days post-vaccination, anti-SARS-CoV-2 spike IgG antibodies were measured in sera samples using a spike cytometric bead array (CBA). All but 1 mouse (7/8) receiving AdCOVID seroconverted, generating SARS-CoV-2-specific IgG (). Vehicle control mice did not develop anti-spike IgG antibodies. On day 32, all mice were intranasally challenged with 1 x LD50 dose of 2.0E + 4 PFU of SARS-CoV-2 strain USA/WA-1/2020. Following challenge, mice were weighed daily until day post-infection (DPI) +4. As expected, AdCOVID vaccinated animals did not lose weight, and weight loss in the control group was minimal over the first four days of infection (). On DPI +4, all mice were euthanized, and post-challenge spike-specific antibodies were quantified by CBA. All AdCOVID vaccinated mice had anti-spike serum IgG antibodies at DPI +4, while vehicle control mice failed to generate antigen-specific antibodies at this timepoint (). The presence of anti-spike IgG in all AdCOVID vaccinated mice, and the absence of anti-spike IgG antibodies in all vehicle control mice on DPI +4, suggests that the single AdCOVID mouse without detectable antibody levels prior to challenge had been successfully immunized. We next investigated the ability of intranasal AdCOVID to elicit immunity within the upper respiratory tract. Nasal washes were collected from a subset of mice at euthanasia (n = 4 or 6/group) and anti-spike antibodies were quantified by CBA. Vehicle control mice did not have quantifiable levels of anti-spike IgG in nasal washes. Conversely, AdCOVID vaccinated mice had high levels of anti-spike IgG present in the nasal cavity (). To confirm that viral challenge causes morbidity in our model, we performed a separate study wherein K18-hACE2 mice were intranasally challenged with an infectious dose of 0.5E + 4 PFU of SARS-CoV-2 strain USA/WA-1/2020. Following challenge, mice were weighed daily until DPI +13. As was seen in the above experiment, AdCOVID vaccinated animals did not lose weight while the non-vaccinated control group had weight loss starting at DPI +3 and peaking between DPI +7–9 (Figure S1).
Figure 1. Spike-specific serum IgG responses following single dose intranasal vaccination with AdCOVID. K18-hACE2 mice (8 mice/group) were intranasally vaccinated with 2.2E + 09 ifu AdCOVID or vehicle control on day 0. All mice were challenged on day 32 with 2.0E + 04 PFU of SARS-CoV-2 strain USA/WA-1/2020 by the intranasal route. (a) Sera were collected on day 21 and analyzed individually for quantification of spike-specific IgG by CBA. (b) Body weight was measured daily. Four days post-challenge (DPI +4), spike-specific IgG in the (c) serum and (d) nasal wash was quantified by CBA. IgG results are expressed in either µg/ml or ng/ml as indicated. Bars represent mean ± SEM. Body weights are presented as the mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. ***, P < .001; ****, P < .0001.
Intranasal vaccination with AdCOVID limits SARS-CoV-2 replication in the upper respiratory tract
Intranasal vaccination has the potential to reduce viral shedding within the nasal cavity of individuals experiencing breakthrough infection. We therefore measured the viral load in the nasal cavity of the AdCOVID-vaccinated and vehicle control K18-hACE2 mice challenged with 2.0E + 04 ifu SARS-CoV-2. Nasal swipes were taken daily through DPI +4. Both vehicle control and AdCOVID vaccinated mice had detectable levels of viral RNA () and subgenomic nucleocapsid (N) RNA indicative of replicated virus () after twenty-four hours of infection. Importantly, AdCOVID mice had a significant reduction in viral RNA on DPI +3. Similarly, AdCOVID vaccinated mice had subgenomic N RNA values that trended lower than those measured in vehicle control mice throughout the course of the study. At study termination on DPI +4, viral load in the nasal cavity was measured by collecting nasal wash from a subset of animals from each vaccination group. AdCOVID vaccinated mice exhibited an 2-fold decrease in viral RNA () and an 2-fold decrease in replicated subgenomic RNA () compared to vehicle control mice.
Figure 2. AdCOVID limits viral load in the nasal cavity following intranasal SARS-CoV-2 challenge. K18-hACE2 mice were intranasally vaccinated with 2.2E + 09 ifu AdCOVID or vehicle control on day 0, challenged on day 32 with (a-d) 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 and analyzed between 0–4 days post-infection or (E-H) challenged on day 32 with 0.5E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 and analyzed on DPI +3. (a-d) Nasal swipes (n = 8 mice/group) were taken daily for qRT-PCR quantification of (A) total viral RNA and (b) subgenomic N RNA. On DPI +4, animals (n = 4–6 mice/group) were euthanized, and nasal wash was collected for qRT-PCR quantification of (c) total viral N RNA and (d) subgenomic N RNA. (e-h) Spike-specific IgG antibodies measured in serum (panel E, n = 11 mice/group) on day 21 post-vaccination. Spike-specific IgA antibodies measured in BAL and nasal wash on DPI +3 (panel F, n = 4–5 mice/group). qRT-PCR quantification of (g) total viral N RNA and (h) subgenomic N RNA in nasal wash on DPI +3 (n = 4–5 mice/group). Viral quantitation presented as RNA copies. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. **, P < .01; ***, P < .001; ****, P < .0001.
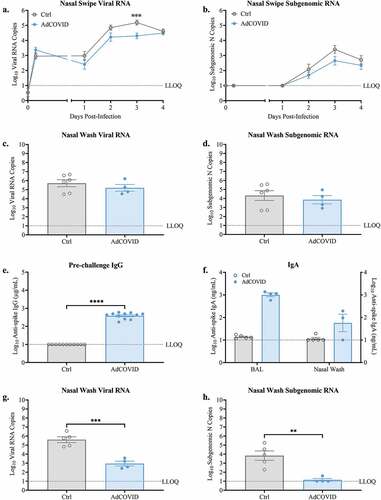
Since the upper respiratory tract represents the first site of infection with SARS-CoV-2 and the nasal swab data indicated that the peak viral shedding from the nasal passage of unvaccinated animals was at DPI +3, we analyzed a separate cohort of AdCOVID and vehicle control mice (). As before, we observed that the AdCOVID-vaccinated animals exhibited high titers of anti-spike IgG antibodies in serum 21 days post-vaccination (). All mice were challenged on day 32 (0.5E + 04 PFU) and euthanized on DPI +3. On DPI +3, we detected spike specific IgA antibodies in both the nasal wash and bronchial alveolar lavage (BAL) derived from the AdCOVID vaccinated mice (), which was not detected in the non-vaccinated animals. Consistent with our prior experiment showing decreased viral RNA in the nasal secretions of AdCOVID vaccinated mice on DPI +3, we observed a highly significant decrease in both total viral RNA () and replicating virus subgenomic RNA () in the nasal wash of the DPI +3 AdCOVID vaccinated mice. Indeed, subgenomic viral RNA was only detected in the nasal wash from 1 of 4 of the vaccinated animals compared to 5/5 of the vehicle control mice. These data show that intranasal vaccination with AdCOVID promotes humoral immune responses in the mucosa and can significantly decrease replicating viral load in the nasal passage at early timepoints following exposure to SARS-CoV-2.
Intranasal vaccination with AdCOVID limits SARS-CoV-2 replication in the lower respiratory tract
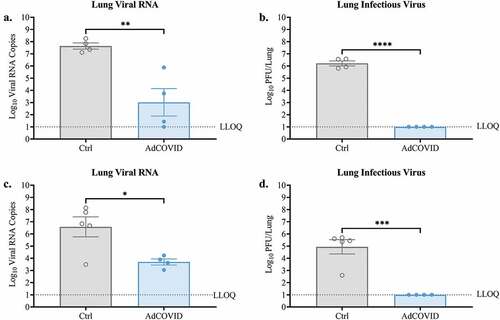
Since early replication of SARS-CoV-2 was diminished in the upper respiratory tract of the AdCOVID-vaccinated animals, we next tested the ability of AdCOVID to decrease SARS-CoV-2 replication in the lungs. On DPI +4 we observed that the lung tissue of vehicle control mice had high levels of SARS-CoV-2 RNA (). Consistent with this result, viral plaque assay using lung tissue from non-vaccinated mice revealed high levels of infectious virus (). Conversely, AdCOVID vaccinated mice had significantly reduced viral RNA levels (375-fold decrease) in the lungs, with only two mice rising above the lowest limit of quantification (LLOQ) (). Importantly, we were unable to recover infectious virus from the lungs of AdCOVID vaccinated mice on DPI +4 (). The same outcome was observed in an independent experiment when we measured total viral RNA () and infectious virus () in the lungs of AdCOVID-vaccinated and non-vaccinated animals on DPI +3. Therefore, intranasal AdCOVID vaccination decreased infectious virus in the lungs to undetectable levels within 3 days of exposure to SARS-CoV-2.
Figure 3. AdCOVID limits viral replication in the lungs following intranasal SARS-CoV-2 challenge. AdCOVID-vaccinated and control K18-hACE2 mice were challenged on day 32 with either (a, b) 2.0E + 04 PFU or (c, d) 0.5E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020. On (a-b) DPI +4 or (c, d) DPI +3, animals were euthanized, and both lung lobes were harvested. (A, C) Total viral N RNA was quantified by qRT-PCR. Results are expressed as RNA copies. (b, d) Infectious virus was quantified in the lungs by plaque assay. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests. *, P < .05; **, P < .01; ***, P < .001; ****, P < .0001.
Intranasal vaccination with AdCOVID limits lung inflammation following SARS-CoV-2 exposure
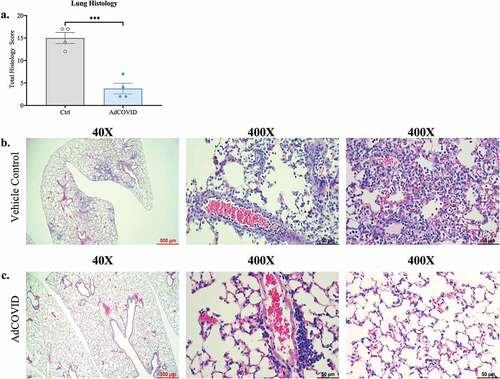
As AdCOVID vaccinated mice exhibited significantly reduced viral load in the lungs on DPI +3 and DPI +4, we used histology to assess inflammation and damage within the lung airways, the alveoli, and the lung vasculature (Table S1), and determined a total lung pathology score that encompassed each of these measurements. Consistent with our findings showing lower viral load in the lungs of AdCOVID-vaccinated mice, we observed a 4-fold reduction in total lung pathology scores in AdCOVID-vaccinated mice compared to vehicle control mice on DPI +4 (). Notably, mice receiving vehicle control showed evidence of viral pneumonia following SARS-CoV-2 challenge (, left panel) as characterized by immune (polymorphonuclear and mononuclear) cell accumulation in perivascular, intimal (, middle panel), and alveolar interstitium (, right panel) as well as evidence of pneumocyte and bronchial epithelial hyperplasia (Table S1). In contrast, AdCOVID-vaccinated mice showed a marked reduction in alveolar inflammation (, left panel) with the exception of the accumulation of mononuclear cells around medium to large caliber pulmonary vessels (, middle panel), and no evidence of alveolar interstitial inflammation (, right panel). Thus, intranasal vaccination with AdCOVID greatly reduces early virus-elicited airway, alveolar, and vascular inflammation, possibly preventing alveolar damage and hyperplasia of Type II pneumocytes and bronchial epithelial cells.
Figure 4. AdCOVID protects mice from SARS-CoV-2 induced lung inflammation. AdCOVID and control vaccinated K18-hACE2 mice were challenged with 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 on day 32. Mice were euthanized on DPI +4 (n = 4/group) and evaluated for lung inflammation and damage. (a) Total lung pathology score representing airway, alveolar, and vascular changes associated with inflammation, cellular hyperplasia and damage. Individual points represent the total pathology score/mouse (See Table S1 for individual scores per mouse/measurement). (b, c) Representative lung sections stained with hematoxylin and eosin and imaged at 40X (left; scale bar, 500 µm) and 400X (middle & right; scale bar, 50 µm). Bars in panel a represent mean ± SEM. Statistical analysis (n = 4 mice/group) was performed with an unpaired Student’s t test. ***, P < .001.
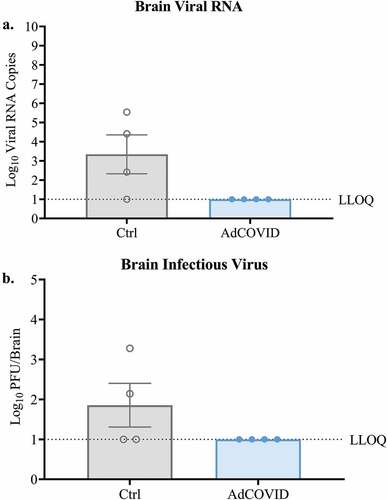
Intranasal AdCOVID vaccination prevents SARS-CoV-2 replication in the periphery of K18-hACE2 mice
It was previously reported that intranasal SARS-CoV-2 infection can cause encephalitis and, in some cases, death in a fraction of infected K18-hACE2 mice.Citation42 To determine whether AdCOVID vaccination can prevent neuroinvasion in the K18-hACE2 animals, we quantified viral RNA and infectious virus in the brain of the SARS-CoV-2 challenged mice. Consistent with other reports,Citation43 3/4 SARS-CoV-2 infected control mice had detectable viral RNA in the brain on DPI +4 () and infectious virus was recovered by plaque assay from the brains of two of these animals (). In contrast, neither viral RNA () nor infectious virus () was recovered from the brains of any of the AdCOVID vaccinated-SARS-CoV-2 infected animals. Collectively, these data suggest that a single vaccination with AdCOVID was sufficient to decrease viral load in the nasal epithelium and lungs of SARS-CoV-2 challenged mice at early timepoints post-infection and was able to minimize the risk of viral spread to the brain.
Figure 5. AdCOVID prevents neuroinvasion following intranasal SARS-CoV-2 challenge. K18-hACE2 mice were challenged with 2.0E + 04 PFU SARS-CoV-2 strain USA/WA-1/2020 on day 32. On DPI +4, mice (n = 4/group) were euthanized, and the brains were harvested. (a) Total viral N RNA was quantified by qRT-PCR. Results are expressed as RNA copies. (b) Infectious virus was quantified in the whole brain by plaque assay. Bars represent mean ± SEM. Lowest limit of quantification (LLOQ). Statistical analyses were performed with unpaired Student’s t tests.
Discussion
Preclinical and clinical studies imply that intranasal vaccination has the potential to be efficacious against respiratory viruses.Citation44–47 Intranasal administration bypasses preexisting adenovector immunity and elicits comparable immunity to intramuscular injection.Citation22,Citation46 Consistent with these reports, we previously showed that AdCOVID, an intranasal Ad5-vectored vaccine encoding the RBD of the SARS-CoV-2 spike protein, elicited long-lived, protective systemic and mucosal immunity following a single administration in mice.Citation29 Here, we further expanded upon these findings. A single administration of AdCOVID limited viral replication in the upper and lower respiratory tracts following intranasal SARS-CoV-2 challenge. AdCOVID vaccinated K18-hACE2 mice had significantly reduced levels of viral RNA in both the nasal wash and nasal swipes. AdCOVID vaccination also markedly impaired viral replication in the lungs of challenged mice, as evidenced by the reduction in subgenomic viral RNA and the absence of infectious virus. Importantly, AdCOVID vaccinated mice exhibited little evidence of viral pneumonia, with a near absence of alveolar inflammation.
It is becoming increasingly apparent that the nasal cavity serves as the principal point of viral entry.Citation48 The nasal epithelium has a high concentration of ACE2, and ciliated cells in the nasal cavity are the primary targets for SARS-CoV-2 replication during the early stages of infection.Citation4,Citation49 Studies utilizing the ferret model of influenza indicate that viral seeding of lower airways is heavily dependent on viral replication and shedding in the nasal cavity.Citation50 Our findings suggest that intranasal vaccination has the potential to substantially limit viral replication within the nasal cavity, thereby preventing productive infection of the lower respiratory tract. AdCOVID vaccinated mice had significant reductions in the level of viral RNA within the upper respiratory tract. Reduced viral seeding, combined with AdCOVID-elicited anti-RBD IgA and anti-RBD T cell populations in the lungs, most likely prevented productive SARS-CoV-2 infection of alveolar Type II cells and histopathological damage.Citation29,Citation51 Additionally, the lack of lung inflammation implies protection from immunopathology,Citation52,Citation53 and may reduce the possibility of long-term COVID disease. Similar pathology results have been reported using other intranasal vaccine modalities.Citation27,Citation54
Of further importance, it is thought that intranasal vaccines have the potential to block transmission between vaccinated hosts and antigenically naïve bystanders.Citation55,Citation56 It was recently reported that intranasal vaccination with a parainfluenza virus type 5 (PIV5)-based SARS-CoV-2 vaccine prevented viral transmission from vaccinated ferrets to co-housed, naïve ferrets.Citation57 AdCOVID’s ability to block SARS-CoV-2 transmission remains to be confirmed, but the reduced viral burden in the respiratory tract suggests that such a beneficial effect is feasible.
While effective at preventing severe COVID-19 disease, evidence suggests that IM vaccination does not elicit long-lasting, sterilizing immunity within the respiratory tract.Citation20 IM vaccination of rhesus macaques with an adenoviral vectored vaccine candidate prevented viral pneumonia, but viral replication in the upper respiratory tract was unaffected.Citation58 It has also been reported that IM vaccination with CoronaVac, an inactivated virus vaccine, failed to produce antibody responses in the nasal epithelial lining fluid (NELF) of human vaccinees.Citation19 This same study showed that the Comirnaty mRNA vaccine elicited low levels of neutralizing antibodies in the NELF which waned within two months of vaccination.Citation19 Pre-clinical studies suggest that intranasal SARS-CoV-2 vaccination is capable of inducing sustained mucosal immunity,Citation29 but it remains to be seen if intranasal vaccines are sufficiently immunogenic to yield protective immunity in humans without preexisting immunity.Citation59 The dearth of marketed intranasal vaccines suggests that the path to licensure as a standalone vaccine may be difficult.
Despite ever-increasing numbers of infected persons and extensive deployment of SARS-CoV-2 vaccines, intranasal vaccines may still offer advantages for the control of the COVID-19 pandemic by serving as a priming regimen or a booster intervention. Vaccination regimens consisting of IM priming and intranasal boosting have been well studied and it appears to be an effective approach. Intramuscular prime-Ad5 intranasal boosting resulted in improved peripheral and local humoral immunity in SARS-CoV-2 vaccinated mice compared to two doses delivered by either route alone.Citation60 Antibodies in the lungs had broad potency and were capable of neutralizing multiple SARS-CoV-2 viral variants. Heterologous prime-boost also generated significantly higher numbers of lung-resident T cells compared to IM vaccination alone. Furthermore, a heterologous prime-boost strategy completely protected Syrian hamsters from COVID-19 disease and immunopathology.Citation61 Indeed, similar results were reported in a recent intranasal vaccine Phase 1 trial where heterologous delivery routes yielded improved neutralizing antibody titers while maintaining T cell immunity.Citation59 These data suggest that heterologous delivery vaccination regimens may result in balanced systemic and mucosal immunity capable of protecting against divergent SARS-CoV-2 strains.
In summary, there remains a need for SARS-CoV-2 vaccines capable of eliciting mucosal immunity. The reduced viral shedding measured in the upper and lower respiratory tracts of pre-clinical challenge models establishes the ability of intranasal vaccines to activate local immune cell populations. How best to deploy intranasal SARS-CoV-2 vaccine regimens remains unclear, but they have the potential to be key weapons in the fight against COVID-19.
Author contributions
Conceptualization: J.J.S., R.G.K., T.D.R, B.G., F.E.L., and M.S.R.; Methodology: M.D.S., J.J.S., D.B., A.S-S., R.G.K, C.D.M., J.B.F., K.S.H., S.M.L., T.D.R., M.S.R., F.E.L. and B.G.; Investigation: R.G.K., T.D.R., and F.E.L.; Reagent Development and Data Collection: M.D.S., D.B., A.S-S.,R.G.K., T.W.D., C.D.M. J.B.F., F.Z., J.L.T.; Vaccine candidate design/production: J.Z., B.G., and M.S.R; Writing-Original Draft Preparation: J.J.S., B.G., and M.S.R.; Writing-Review and Editing: All authors; All authors had full access to all the data in the study and take responsibility for integrity of the data and the accuracy of the data analysis.
Supplemental Material
Download MS Word (206 KB)Acknowledgments
The authors would like to thank Tsungwei Feng, Young Lee, and Bethlehem Shiberu (Altimmune) for their contributions to the AdCOVID program; Uma Mudunuru, Thomas ‘Scott’ Simpler and Rebecca Burnham (UAB) for ABSL2 animal husbandry; Donovan J. Murphy (UAB) for ABSL3 animal husbandry; Erik D. Dohm, Justin Roth and Moustafa A. Awaden (UAB) for BSL3 training and access to the SEBLAB; Derek B. Moates (UAB Fungal Reference Lab) for coordinating sample drop-off and data delivery; Emily E. Helmen and Sherri Coffman (UAB Comparative Pathology Lab) for histology; and Andrea J. Osborne (UAB Animal Resources Program) for veterinary care of animals.
Disclosure statement
Investigators at The University of Alabama at Birmingham (UAB: F.E.L. as project lead) were funded by a sponsored research agreement from Altimmune to perform these studies. F.E.L., R.G.K., and T.D.R. serve as paid consultants for Altimmune. J.J.S., M.S.R., and B.G. are employees of Altimmune Inc. and may have received stock options and compensation as part of their employment. Altimmune Inc. has filed the following patent applications related to AdCOVID: PCT/US21/17920; US 17/175,144 and US 17/175,131. All other authors declare no potential conflicts of interest.
Supplemental material
Supplemental data for this article can be accessed on the publisher’s website at https://doi.org/10.1080/21645515.2022.2127292.
Additional information
Funding
References
- Chan JF, Zhang AJ, Yuan S, Poon VK, Chan CC, Lee AC, Chan WM, Fan Z, Tsoi HW, Wen L, et al. Simulation of the clinical and pathological manifestations of coronavirus disease 2019 (COVID-19) in a golden Syrian Hamster Model: implications for disease pathogenesis and transmissibility. Clin Infect Dis. 2020;71:1–11.
- Prather KA, Marr LC, Schooley RT, McDiarmid MA, Wilson ME, Milton DK. Airborne transmission of SARS-CoV-2. Science. 2020;370(6514):303–04. doi:10.1126/science.abf0521.
- Letko M, Marzi A, Munster V. Functional assessment of cell entry and receptor usage for SARS-CoV-2 and other lineage B betacoronaviruses. Nature Microbiol. 2020;5(4):562–69. doi:10.1038/s41564-020-0688-y.
- Sungnak W, Huang N, Becavin C, Berg M, Queen R, Litvinukova M, Talavera-López C, Maatz H, Reichart D, Sampaziotis F, et al. SARS-CoV-2 entry factors are highly expressed in nasal epithelial cells together with innate immune genes. Nat Med. 2020;26:681–87.
- Hou YJ, Okuda K, Edwards CE, Martinez DR, Asakura T, Dinnon KH, 3rd, Kato T, Lee RE, Yount BL, Mascenik TM, et al. SARS-CoV-2 reverse genetics reveals a variable infection gradient in the respiratory tract. Cell. 2020;182(2):429–46 e14. doi:10.1016/j.cell.2020.05.042.
- Kawasuji H, Takegoshi Y, Kaneda M, Ueno A, Miyajima Y, Kawago K, Fukui Y, Yoshida Y, Kimura M, Yamada H, et al. Transmissibility of COVID-19 depends on the viral load around onset in adult and symptomatic patients. PloS One. 2020;15:e0243597.
- Pujadas E, Chaudhry F, McBride R, Richter F, Zhao S, Wajnberg A, Nadkarni G, Glicksberg BS, Houldsworth J, Cordon-Cardo C. SARS-CoV-2 viral load predicts COVID-19 mortality. Lancet Respir Med. 2020;8(9):e70. doi:10.1016/S2213-2600(20)30354-4.
- Fajnzylber J, Regan J, Coxen K, Corry H, Wong C, Rosenthal A, Worrall D, Giguel F, Piechocka-Trocha A, Atyeo C, et al. SARS-CoV-2 viral load is associated with increased disease severity and mortality. Nat Commun. 2020;11(1):5493. doi:10.1038/s41467-020-19057-5.
- Kim JY, Ko JH, Kim Y, Kim YJ, Kim JM, Chung YS, Kim HM, Han MG, Kim SY, Chin BS. Viral load kinetics of SARS-CoV-2 infection in first two patients in Korea. J Korean Med Sci. 2020;35:e86.
- Zou L, Ruan F, Huang M, Liang L, Huang H, Hong Z, Yu J, Kang M, Song Y, Xia J, et al. SARS-CoV-2 viral load in upper respiratory specimens of infected patients. N Engl J Med. 2020;382(12):1177–79. doi:10.1056/NEJMc2001737.
- Siddiqi HK, Mehra MR. COVID-19 illness in native and immunosuppressed states: a clinical–therapeutic staging proposal. J Heart Lung Transplant. 2020;39(5):405–07. doi:10.1016/j.healun.2020.03.012.
- Yap JKY, Moriyama M, Iwasaki A. Inflammasomes and pyroptosis as therapeutic targets for COVID-19. J Immunol. 2020;205(2):307–12. doi:10.4049/jimmunol.2000513.
- Pons S, Fodil S, Azoulay E, Zafrani L. The vascular endothelium: the cornerstone of organ dysfunction in severe SARS-CoV-2 infection. Crit Care. 2020;24(1):353. doi:10.1186/s13054-020-03062-7.
- Polack FP, Thomas SJ, Kitchin N, Absalon J, Gurtman A, Lockhart S, Perez JL, Marc GP, Moreira ED, Zerbini C, et al. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N Engl J Med. 2020;383:2603–15.
- Baden LR, El Sahly HM, Essink B, Kotloff K, Frey S, Novak R, Diemert D, Spector SA, Rouphael N, Creech CB, et al. Efficacy and Safety of the mRNA-1273 SARS-CoV-2 Vaccine. N Engl J Med. 2021;384(5):403–16. doi:10.1056/NEJMoa2035389.
- Sadoff J, Gray G, Vandebosch A, Cardenas V, Shukarev G, Grinsztejn B, Goepfert PA, Truyers C, Fennema H, Spiessens B, et al. Safety and efficacy of single-dose Ad26.COV2.S vaccine against Covid-19. N Engl J Med. 2021;384:2187–201.
- Administration USFaD. COVID-19 vaccines. U.S. Food and Drug Administration; 2021.
- Khan WH, Hashmi Z, Goel A, Ahmad R, Gupta K, Khan N, Alam I, Ahmed F, Ansari MA. COVID-19 pandemic and vaccines update on challenges and resolutions. Front Cell Infect Microbiol. 2021;11:690621. doi:10.3389/fcimb.2021.690621.
- Chan RWY, Liu S, Cheung JY, Tsun JGS, Chan KC, Chan KYY, Fung GPG, Li AM, Lam HS. The mucosal and serological immune responses to the novel coronavirus (SARS-CoV-2) Vaccines. Front Immunol. 2021;12:744887. doi:10.3389/fimmu.2021.744887.
- Corbett KS, Flynn B, Foulds KE, Francica JR, Boyoglu-Barnum S, Werner AP, Flach B, O’Connell S, Bock KW, Minai M, et al. Evaluation of the mRNA-1273 Vaccine against SARS-CoV-2 in nonhuman primates. N Engl J Med. 2020;383:1544–55.
- Taddio A, Ipp M, Thivakaran S, Jamal A, Parikh C, Smart S, Sovran J, Stephens D, Katz J. Survey of the prevalence of immunization non-compliance due to needle fears in children and adults. Vaccine. 2012;30:4807–12. doi:10.1016/j.vaccine.2012.05.011.
- Croyle MA, Patel A, Tran KN, Gray M, Zhang Y, Strong JE, Feldmann H, Kobinger GP. Nasal delivery of an adenovirus-based vaccine bypasses pre-existing immunity to the vaccine carrier and improves the immune response in mice. PloS One. 2008;3:e3548. doi:10.1371/journal.pone.0003548.
- Zhang JTE, Feng T, Shi Z, Van Kampen KR, Tang DC. Adenovirus-vectored drug-vaccine duo as a rapid-response tool for conferring seamless protection against influenza. PloS One. 2011;6:e22605.
- Krishnan V, Andersen BH, Shoemaker C, Sivko GS, Tordoff KP, Stark GV, Zhang J, Feng T, Duchars M, Roberts MS, et al. Efficacy and immunogenicity of single-dose AdVAV intranasal anthrax vaccine compared to anthrax vaccine absorbed in an aerosolized spore rabbit challenge model. Clin Vaccine Immunol. 2015;22:430–39. doi:10.1128/CVI.00690-14.
- Wold WS, Toth K. Adenovirus vectors for gene therapy, vaccination and cancer gene therapy. Curr Gene Ther. 2013;13:421–33.
- Zhang C, Zhou D. Adenoviral vector-based strategies against infectious disease and cancer. Hum Vaccin Immunother. 2016;12:2064–74.
- Hassan AO, Kafai NM, Dmitriev IP, Fox JM, Smith BK, Harvey IB, Chen RE, Winkler ES, Wessel AW, Case JB, et al. A single-dose intranasal ChAd vaccine protects upper and lower respiratory tracts against SARS-CoV-2. Cell. 2020;183:169–84 e13. doi:10.1016/j.cell.2020.08.026.
- Yusuf H, Kett V. Current prospects and future challenges for nasal vaccine delivery. Hum Vaccin Immunother. 2017;13:34–45.
- King RG, Silva-Sanchez A, Peel JN, Botta D, Dickson AM, Pinto AK, Meza-Perez S, Allie SR, Schultz MD, Liu M, et al. Single-dose intranasal administration of AdCOVID elicits systemic and mucosal immunity against SARS-CoV-2 and fully protects mice from lethal challenge. Vaccines (Basel). 2021;9. doi:10.3390/vaccines9080881.
- Ma C, Wang L, Tao X, Zhang N, Yang Y, Tseng CK, Li F, Zhou Y, Jiang S, Du L. Searching for an ideal vaccine candidate among different MERS coronavirus receptor-binding fragments—the importance of immunofocusing in subunit vaccine design. Vaccine. 2014;32(46):6170–76. doi:10.1016/j.vaccine.2014.08.086.
- Piccoli L, Park YJ, Tortorici MA, Czudnochowski N, Walls AC, Beltramello M, Silacci-Fregni C, Pinto D, Rosen LE, Bowen JE, et al. Mapping neutralizing and immunodominant sites on the SARS-CoV-2 spike receptor-binding domain by structure-guided high-resolution serology. Cell. 2020;183:1024–42.
- Starr TN, Czudnochowski N, Liu Z, Zatta F, Park YJ, Addetia A, Pinto D, Beltramello M, Hernandez P, Greaney AJ, et al. SARS-CoV-2 RBD antibodies that maximize breadth and resistance to escape. Nature. 2021;597:97–102.
- Greaney AJ, Loes AN, Gentles LE, Crawford KHD, Starr TN, Malone KD, Chu HY, Bloom JD. Antibodies elicited by mRNA-1273 vaccination bind more broadly to the receptor binding domain than do those from SARS-CoV-2 infection. Sci Transl Med. 2021;13:eabi9915.
- Zost SJ, Gilchuk P, Case JB, Binshtein E, Chen RE, Nkolola JP, Schäfer A, Reidy JX, Trivette A, Nargi RS, et al. Potently neutralizing and protective human antibodies against SARS-CoV-2. Nature. 2020;584:443–49. doi:10.1038/s41586-020-2548-6.
- Dan JM, Mateus J, Kato Y, Hastie KM, Yu ED, Faliti CE, Grifoni A, Ramirez SI, Haupt S, Frazier A, et al. Immunological memory to SARS-CoV-2 assessed for up to 8 months after infection. Science. 2021;371:eabf4063.
- Seydoux E, Homad LJ, MacCamy AJ, Parks KR, Hurlburt NK, Jennewein MF, Akins NR, Stuart AB, Wan YH, Feng J, et al. Analysis of a SARS-CoV-2-infected individual reveals development of potent neutralizing antibodies with limited somatic mutation. Immunity. 2020;53:98–105.
- Tang DC, Zhang J, Toro H, Shi Z, Van Kampen KR. Adenovirus as a carrier for the development of influenza virus-free avian influenza vaccines. Expert Rev Vaccines. 2009;8:469–81.
- Carroll T, Fox D, van Doremalen N, Ball E, Morris MK, Sotomayor-Gonzalez A, Servellita V, Rustagi A, Yinda CK, Fritts L, et al. The B.1.427/1.429 (epsilon) SARS-CoV-2 variants are more virulent than ancestral B.1 (614G) in Syrian hamsters. PLoS Pathog. 2022;18:e1009914. doi:10.1371/journal.ppat.1009914.
- Bewley KR, Coombes NS, Gagnon L, McInroy L, Baker N, Shaik I, St-Jean JR, St-Amant N, Buttigieg KR, Humphries HE, et al. Quantification of SARS-CoV-2 neutralizing antibody by wild-type plaque reduction neutralization, microneutralization and pseudotyped virus neutralization assays. Nat Protoc. 2021;16:3114–40. doi:10.1038/s41596-021-00536-y.
- Gruber AD, Osterrieder N, Bertzbach LD, Vladimirova D, Greuel S, Ihlow J, Horst D, Trimpert J, Dietert K. Standardization of reporting criteria for lung pathology in SARS-CoV-2-infected Hamsters: what matters? Am J Respir Cell Mol Biol. 2020;63:856–59.
- Li Q, Vijaykumar K, Philips SE, Hussain SS, Huynh VN, Fernandez-Petty CM, Lever JE, Foote JB, Ren J, Campos-Gómez J, et al. Mucociliary transport deficiency and disease progression in Syrian Hamsters with SARS-CoV-2 infection. bioRxiv. 2022.
- Kumari P, Rothan HA, Natekar JP, Stone S, Pathak H, Strate PG, Arora K, Brinton MA, Kumar M. Neuroinvasion and encephalitis following intranasal inoculation of SARS-CoV-2 in K18-hACE2 mice. Viruses. 2021;13:132. doi:10.3390/v13010132.
- Song E, Zhang C, Israelow B, Lu-Culligan A, Prado AV, Skriabine S, Lu P, Weizman O-E, Liu F, Dai Y, et al. Neuroinvasion of SARS-CoV-2 in human and mouse brain. J Exp Med. 2021;218. doi:10.1084/jem.20202135.
- Gould VMW, Francis JN, Anderson KJ, Georges B, Cope AV, Tregoning JS. Nasal IgA provides protection against human influenza challenge in volunteers with low serum influenza antibody titre. Front Microbiol. 2017;8:900.
- Belshe RB, Gruber WC, Mendelman PM, Mehta HB, Mahmood K, Reisinger K, Treanor J, Zangwill K, Hayden F, Bernstein D, et al. Correlates of immune protection induced by live, attenuated, cold-adapted, trivalent, intranasal influenza virus vaccine. J Infect Dis. 2000;181:1133–37. doi:10.1086/315323.
- Tasker S, Wight O’Rourke A, Suyundikov A, Jackson Booth P-G, Bart S, Krishnan V, Zhang J, Anderson KJ, Georges B, Roberts MS. Safety and immunogenicity of a novel intranasal influenza vaccine (NasoVAX): a phase 2 randomized, controlled trial. Vaccines. 2021;9:224.
- Kim E, Okada K, Beeler JA, Crim RL, Piedra PA, Gilbert BE, Gambotto A. Development of an adenovirus-based respiratory syncytial virus vaccine: preclinical evaluation of efficacy, immunogenicity, and enhanced disease in a cotton rat model. J Virol. 2014;88:5100–08. doi:10.1128/JVI.03194-13.
- Li H, Wang Y, Ji M, Pei F, Zhao Q, Zhou Y, Hong Y, Han S, Wang J, Wang Q, et al. Transmission routes analysis of SARS-CoV-2: a systematic review and case report. Front Cell Dev Biol. 2020;8:618. doi:10.3389/fcell.2020.00618.
- Ahn JH, Kim J, Hong SP, Choi SY, Yang MJ, Ju YS, Kim YT, Kim HM, Rahman MDT, Chung MK, et al. Nasal ciliated cells are primary targets for SARS-CoV-2 replication in the early stage of COVID-19. J Clin Invest. 2021;131:131. doi:10.1172/JCI148517.
- Richard M, van den Brand JMA, Bestebroer TM, Lexmond P, de Meulder D, Fouchier RAM, Lowen AC, Herfst S. Influenza A viruses are transmitted via the air from the nasal respiratory epithelium of ferrets. Nat Commun. 2020;11:766.
- Mulay A, Konda B, Garcia G, Jr., Yao C, Beil S, Villalba JM, Koziol C, Sen C, Purkayastha A, Kolls JK, et al. SARS-CoV-2 infection of primary human lung epithelium for COVID-19 modeling and drug discovery. Cell Rep. 2021;35:109055. doi:10.1016/j.celrep.2021.109055.
- Tian S, Xiong Y, Liu H, Niu L, Guo J, Liao M, Xiao S-Y. Pathological study of the 2019 novel coronavirus disease (COVID-19) through postmortem core biopsies. Mod Pathol. 2020;33:1007–14. doi:10.1038/s41379-020-0536-x.
- Bourgonje AR, Abdulle AE, Timens W, Hillebrands JL, Navis GJ, Gordijn SJ, Bolling MC, Dijkstra G, Voors AA, Osterhaus AD, et al. Angiotensin-Converting enzyme 2 (ACE2), SARS-CoV -2 and the pathophysiology of coronavirus disease 2019 (COVID -19). J Pathol. 2020;251:228–48. doi:10.1002/path.5471.
- Ku MW, Bourgine M, Authie P, Lopez J, Nemirov K, Moncoq F, Noirat A, Vesin B, Nevo F, Blanc C, et al. Intranasal vaccination with a lentiviral vector protects against SARS-CoV-2 in preclinical animal models. Cell Host Microbe. 2021;29:236–49 e6. doi:10.1016/j.chom.2020.12.010.
- Lowen AC, Steel J, Mubareka S, Carnero E, Garcia-Sastre A, Palese P. Blocking interhost transmission of influenza virus by vaccination in the guinea pig model. J Virol. 2009;83:2803–18.
- Price GE, Lo CY, Misplon JA, Epstein SL, García-Sastre A. Mucosal immunization with a candidate universal influenza vaccine reduces virus transmission in a mouse model. J Virol. 2014;88:6019–30. doi:10.1128/JVI.03101-13.
- An D, Li K, Rowe DK, Diaz MCH, Griffin EF, Beavis AC, Johnson SK, Padykula I, Jones CA, Briggs K, et al. Protection of K18-hACE2 mice and ferrets against SARS-CoV-2 challenge by a single-dose mucosal immunization with a parainfluenza virus 5–based COVID-19 vaccine. Sci Adv. 2021;7. doi:10.1126/sciadv.abi5246.
- van Doremalen N, Lambe T, Spencer A, Belij-Rammerstorfer S, Purushotham JN, Port JR, van Doremalen N, Avanzato VA, Bushmaker T, Flaxman A, et al. ChAdox1 nCov-19 vaccine prevents SARS-CoV-2 pneumonia in rhesus macaques. Nature. 2020;586:578–82. doi:10.1038/s41586-020-2608-y.
- Wu S, Huang J, Zhang Z, Wu J, Zhang J, Hu H, Zhu T, Zhang J, Luo L, Fan P, et al. Safety, tolerability, and immunogenicity of an aerosolised adenovirus type-5 vector-based COVID-19 vaccine (Ad5-nCov) in adults: preliminary report of an open-label and randomised phase 1 clinical trial. Lancet Infect Dis. 2021;21:1654–64. doi:10.1016/S1473-3099(21)00396-0.
- Lapuente D, Fuchs J, Willar J, Vieira Antao A, Eberlein V, Uhlig N, Issmail L, Schmidt A, Oltmanns F, Peter AS, et al. Protective mucosal immunity against SARS-CoV-2 after heterologous systemic prime-mucosal boost immunization. Nat Commun. 2021;12:6871. doi:10.1038/s41467-021-27063-4.
- Bosnjak B, Odak I, Barros-Martins J, Sandrock I, Hammerschmidt SI, Permanyer M, Patzer GE, Greorgiev H, Jauregui RG, Tscherne A, et al. Intranasal delivery of MVA vector vaccine induces effective pulmonary immunity against SARS-CoV-2 in rodents. Front Immunol. 2021;12:772240.
