ABSTRACT
Gossypium arboreum (Desi Cotton) holds a special place in cotton industry because of its inherent ability to withstand drought, salinity, and remarkable resistance to sucking pests and cotton leaf curl virus. However, it suffers yield losses due to weeds and bollworm infestation. Genetic modification of G. arboreum variety FBD-1 was attempted in the current study to combat insect and weedicide resistance by incorporating cry1Ac, cry2A and cp4-EPSPS genes under control of 35S promoter in two different cassettes using kanamycin and GUS as markers through Agrobacterium-mediated shoot apex cut method of cotton transformation. The efficiency of transformation was found to be 1.57%. Amplification of 1700 bp for cry1Ac, 167 bp for cry2A and 111 bp for cp4-EPSPS confirmed the presence of transgenes in cotton plants. The maximum mRNA expression of cry1Ac and cp4-EPSPS was observed in transgenic cotton line L3 while minimum in transgenic cotton line L1. The maximum protein concentrations of Cry1Ac, Cry2A and Cp4-EPSPS of 3.534 µg g−1, 2.534 µg g−1 and 3.58 µg-g−1 respectively were observed for transgenic cotton line L3 as compared to control cotton line. On leaf-feed-based insect bioassay, almost 99% mortality was observed for Helicoverpa armigera on the transgenic cotton plant (L3). It completely survived the 1900 ml hectare−1 glyphosate spray assay as compared to non-transgenic cotton plants. The necrotic spots appeared on the third day, leading to the complete death of control plants on the fifth day of assay. The successful multiple gene-stacking in G. arboreum FBD-1 variety could be further used for qualitative improvement of cotton fiber through plant breeding techniques.
1. Introduction
Gossypium arboreum, also called the Asiatic cotton (desi cotton), is one of the four cultivable cotton species around the world. It is a diploid cotton species (2 n), having absorbable, short and coarse fiber, highly favorable for the surgical industry. Gossypium arboreum is known to possess drought/salinity tolerance, Cotton Leaf Curl Virus (CLCuV) resistance,Citation1 nematode and root rot resistance.Citation2 However, G. arboreum is severely affected by bollworm, especially Helicoverpa armigera.Citation3 Due to the increasing attack of CLCuV on imported varieties of Gossypium hirsutum, the need for the hour is to put efforts toward G. arboreum to improve its fiber characteristics and resistance against chewing insects and herbicides by genetic manipulation of its genome.Citation4,Citation5
The most important pests are Helicoverpa punctigera, H. armigera (spotted bollworm), and Tetranychus urticae (spider mite).Citation6 Conventionally, insects are controlled by spraying insecticides, but this is very expensive.Citation7 In 2018–19, about 21,175 metric tons of pesticides were consumed in Pakistan’s agriculture sector as per Economic Survey of Pakistan (2018–2019).Citation8 To cope with such challenges through biotechnology more than 100 Bacillus thuringiensis (Bt) crystal genes have been sequenced so far, which encode for Cry and Cyt proteins.Citation9 Such crystal proteins are toxic to the larvae of Lepidoptera, Diptera and Coleoptera. Several reports revealed the resistance against lepidopteron and dipterans insects in G. hirsutum when different cry genes (cry1Ab, cry1Ac, cry1A, cry2A, cry1F etc.) were transformed.Citation10 Cotton cultivars expressing both Cry2A and Cry1Ac endotoxins were more toxic to Spodoptra frugiperda, H. zea and S. exiguaCitation11 and H. armigera as compared to those expressing single toxin.Citation12,Citation13 Stacking different Cry proteins (Cry1Ac and Cry2A) can delay the development of resistance in Lepidopterans.Citation13 The Cry1Ac and Cry2A proteins do not share the high-affinity binding sites on brush border membrane vesicles in the insect midgutCitation12 and therefore stacking of cry1Ac and cry2A genes in the same crop has a potential advantage for resistance management.
Gossypium arboreum is highly vulnerable to weeds competition and 37% yield losses occur due to weeds.Citation14 Weeds can serve as hosts for diseases and pests, reduce the nutritional quality of soil and adversely affect cotton harvesting or its lint quality.Citation7 Weed removal through chemical herbicide sprays provided an alternative to manual weed management but chemical herbicides result in plant damage, especially the cotton field.Citation15 The glyphosate spray is broad-spectrum broad-leaf weedicide and inhibits the shikimate pathway enzyme.Citation16 Glyphosate (N-phosphonomethyl glycine) kills the plant by blocking an enzyme 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) – shikimate biosynthetic pathway enzyme. EPSPS converts phosphoenolpyruvate and shikimate-3-phosphate (S3P) to inorganic phosphate and 5-enolpyruvylshikimate-3-phosphate. This pathway is involved in the production of amino acids, folic acids, phytoalexins, lignins, plastoquinones and many other secondary substances.Citation16 Blockage of EPSPS enzyme results in accumulating shikimic acid and hydroxybenzoic acid in leaves or nodes, leading to the plant’s death.Citation16 EPSPS-encoding bacterial genes are being introduced to crop plants by stable genetic transformation to develop resistance from glyphosate herbicides in transgenic crops. A Calotropis procera EPSPSII gene transformation in upland cotton have been reported to successfully induce glyphosate tolerance.Citation11 In the current study design, we aim to transform the cry genes along with Glyphosate Tolerant Gene (cp4-EPSPS) or GTG in G. arboreum with a proposed resistance development in transgenic variety against pest attack and glyphosate, respectively.Citation14
2. Materials and Methods
2.1. Field Setup and Sampling
This study was carried out at Center of Excellence in Molecular Biology, University of the Punjab, Lahore, during the years 2015–2016 (31°33ʹN, 74°19ʹE) in which a completely randomized block design was set out in two consecutive growing seasons (2015–2016) of cotton in the same field.
2.2. Plasmid Construction and Transformation into Agrobacterium
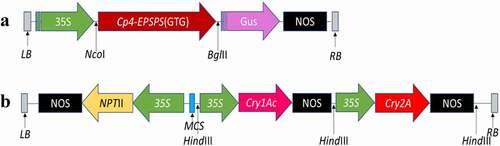
Two Bt genes cry1Ac+cry2A of 1400bp were sub-cloned in pKHG4 at the HindIII site to produce recombinant pKHG4-cry1Ac+ cry2A vector under the expression of 35S promoter, ( A) hereafter referred to as cry construct. Likewise, a 1700bp cp4-EPSPS expressed under 35S promoter was cloned at NcoI and BglII sites of pCAMBIA1301 ( B). The two recombinant plasmids were separately transformed into Agrobacterium tumefaciens LBA4404 by electroporation and confirmed through colony PCR by using gene-specific primers.
2.3. Plant Material and Transformation Procedure
About 5599 delinted seeds of G. arboreum were used for transformation experiments. Seeds were sterilized with a working concentration of 1% SDS and 1% HgCl2 for 5 min and rinsed completely afterward with autoclaved water. These sterilized seeds were incubated in dark at 30°C overnight. Germination index of G. arboreum (Desi Cotton) was determined by soaking thirty seeds in Petri-plate at 30°C for 48 hours and determined by using the following formula:
Germination Index (%age) = (Germinated Seeds/Total Seeds) x 100.
Overnight grown cultures of Agrobacterium harboring cry and cp4-EPSPS constructs were mixed and centrifuged at 3000 rpm for 15 min at 4°C. The supernatant was decanted, and the pellets were dissolved in 10 ml of MS broth. Shoot apices of the 48 h germinated embryos were transformed according to Rao et al. (2011).Citation17 The co-cultivated embryos were embedded in MS plates (hypocotyl region downward in the MS phytagel and epicotyl facing upward above the MS phytagel) supplemented with B5 vitamins and cefotaxime (200 mg L−1) and incubated at 28°C with a 16/8 h photoperiod (90 µmolm−2s−1). After 3–5 days of co-cultivation, plantlets with healthy shoots and roots were transferred to a fresh MS tube containing 50 mg L−1 kanamycin, 200 mg L−1 of cefotaxime and B5 vitamins.
After 8–10 weeks of growth in MS tubes, six-inch-long plantlets were transplanted to pots having sterilized soil mixture (equal proportion of clay, sand and peat moss). The plants were covered with plastic bags, for first 15 days, to maintain proper humidity at a temperature of 25 ± 2°C with a photoperiod of 16:8 hours of light and dark cycle. The plants were acclimatized according to the method of Gul et al., (2020).Citation18 About 48 putative transgenic cotton plants were transplanted to cotton tunnels under controlled environmental conditions. Transgenic cotton lines were successfully self-pollinated to produce T1-generation and similarly for the T2-generation.
2.4. Detection of Transgenes through Conventional PCR
Genomic DNA from putative transgenic cotton plants in T0 generation was isolated using G-spinII plant genomic DNA extraction kit (Cat# 17,271). Putative positive plants carrying cp4-EPSPS, cry1Ac and cry2A genes were confirmed by PCR from genomic DNA using gene-specific primers (). The PCR reaction consisted of; 2 µl genomic DNA, 2 µl of 10x pfu buffer (MgSO4 added) (Fermentas cat# B34), 2 µl of 2 mM dNTPs, 2 µl of 10 pM forward and reverse primers each, 0.5 µl of 500 U Taq polymerase (Fermentas cat# EP0402) and nuclease-free water upto 20 µl. The PCR for all three genes was performed in three steps; the first step of one cycle of initial denaturation at 95°C for 5 min, second step of 40 cycles of each denaturation at 95°C for 45s, annealing at 60°C for 45s and amplification at 72°C for 1–2 min, and third step of final amplification at 72°C for 5 min and hold at 4°C.
Table 1. Gene sequences of the primer pairs used for PCR
2.5. Quantification of Proteins through Immunostrip Test Assay and ELISA
Immunostrip assayCitation19 and Enzyme-Linked Immunosorbent Assay (ELISA) were performed to detect and quantify proteins, respectively, in T3 generation. For immunostrip assay, total crude protein was isolated from the fresh plant leaves. Immunostrip test assay was performed by placing strips about 1/4th of an inch of the strip in the diluted protein samples for 30 minutes. Strips were observed for desired bands after 30 min. For ELISA, Envirologix Qualiplate ELISA kit for Cry1Ac (cat# AP003), Cp4-EPSPS (cat# AP010), and Quantiplate ELISA kit (cat# AP005) for Cry2A were used. The ELISA was performed as per manufacturer’s instructions. The protein was quantified by measuring OD on microtiter ELISA plate reader at 450 nm absorbance.
2.6. Quantitative Real-Time Expression of Transgenic Cotton Plants
Young leaves of transgenic and non-transgenic control cotton plants in T1-generation were used to extract RNA according to the procedure performed as described by.Citation20 The cDNA was prepared as follows: total RNA (1pg- 5 µg), oligo (dT)18 primers 1 µl; dNTPs mix(10 mM) 1 µl, nuclease-free water up to 14.5 µl, 5x RT buffer 4 µl, RiboLock RNase Inhibitor 0.5 µl, Maxima reverse transcriptase 1 µl and then incubated at 50°C for 30 min and the reaction was terminated at 85°C for 5 min. The cDNA was stored at −80 °C. The quantitative RT-PCR was carried out using gene-specific primers (). The reaction mix contained: Maxima SYBR Green/ROX qPCR Master Mix (2X) 12.5 µl, forward primer 0.5 µl, reverse primer 0.5 µl, cDNA 2 µl and nuclease-free water to 25 µl. The reaction mix was prepared in the dark to prevent light exposure of SYBR green. The cycling protocol of all qPCR reactions was performed with the following conditions: one cycle at 94◦C for 10 min, 40 cycles at 94◦C for 45s, 52◦C for 45s and 72◦C for 30s. The endogenous expression of glyceraldehyde 3 phosphate dehydrogenase (GAPDH) was used as the internal control for the normalization of expression data.
2.7 Glyphosate Spray Assay
Herbicide resistance status of transgenic cotton plants was evaluated by spraying glyphosate on field plants in T1, T2 and T3 generation following method of.Citation14 Cotton was planted in the field and manual weeding was restrained. All the wild weeds could grow uncontrollably until the glyphosate solution (1900 ml hectare−1) was sprayed on transgenic and control cotton lines. Vegetative lesions and complete killing of the plants were observed daily for 5–7 days.
2.8 Insect Mortality Assay
The mature leaves of the cotton plants in T3 generation were used for insect mortality assay. Detached leaves from the cotton plants were put on the moist filter paper in a petri plate. The age synchronized population of insects reared on non-transgenic plants was used in this study. The H. armigera larvae were released onto transgenic G. arboreum plants’ detached leaves. Five leaves from five biological replicates of each transgenic plants were selected and to each biological replicate, five larvae were released. The petri plates were kept at 28°C ± 2°C in 16 hours light and 8 hours dark. The mortality data was recorded for 3–5 days of assay.
3. RESULTS
3.1. Transformation of cry1Ac+cry2A and cp4-EPSPS Genes in G. arboreum
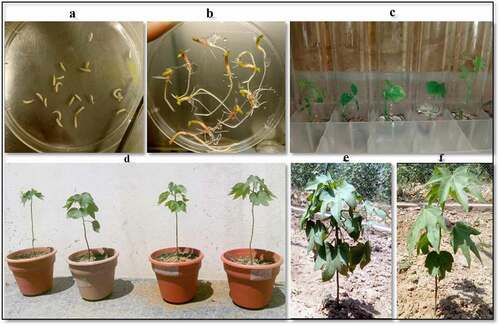
The germination index was calculated to be 73%. The seeds germinated at 28°C after 2 days were transformed through shoot apex explant by Agrobacterium-mediated transformation (). The Agrobacterium containing both cry and cp4-EPSPS constructs were combined as 1:1 ratio for cotton transformation. Healthy seedlings (3055), which developed roots and shoots (), were transferred to MS medium culture tubes for selection with kanamycin. Only 262 plantlets () survived the selection pressure of antibiotics and transplanted to soiled pots. (). The fungal contamination and acclimatization of the plants further reduced the number of surviving plants to 48, finally transplanted to the tunnel (). The transformation efficiency as calculated from the data () was found to be 1.57%. The frequency of transplantation at each developmental stage in the T1-generation seriously affected the plant survival regardless of the transformation efficiency.
Table 2. Numerical data of transformation experiments and transformation efficiency
Figure 2. Process of cotton transformation. (a) Embryos isolated from germinated seeds of cotton. (b) Transplantation of five days old cotton plantlets into culture tubes. (c) Twenty days old plantlets in glass tubes supplemented with kanamycin (50 mg L−1), cefotaxime (200 mg L−1), IBA and kinetin (100 mg L−1). (d) Shifting of putative transgenic cotton plants to plastic pots and tunnel. (e) Two months old putative transgenic cotton plants in the tunnel. (f) Two months old non-transgenic cotton plant in the tunnel
Transformation Efficiency = (No. of positive plants shifted to tunnel/No. of embryos subjected to selection) x 100
3.2. Molecular Analysis of Putative Transgenic Cotton Plants
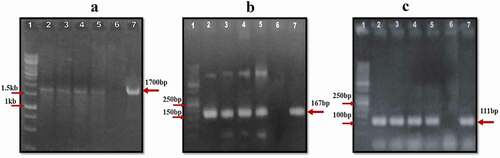
Putative transgenic cotton plants harboring cry1Ac, cry2A and cp4-EPSPS genes were analyzed through high throughput molecular techniques. These include PCR, ELISA, DIPSTICK test and Real-Time (RT) PCR. PCR amplification from genomic DNA of transgenic cotton plants with gene-specific primers revealed a 1700bp fragment of cry1Ac ( A) and 167bp of cry2A ( B) while 111bp for cp4-EPSPS ( C).
Figure 3. PCR detection of transgenic cotton plants. (a) PCR amplification of cry1Ac. Lane 1: 1kb DNA Ladder; Lane 2–5: putative transgenic cotton plants showing amplification of 1700bp PCR product of cry1Ac; Lane 6: Negative control; Lane 7: Positive control. (b) PCR amplification of cry2A. Lane 1: 50bp DNA Ladder; Lane 2–5: putative transgenic cotton plants showing amplification of 167bp PCR product of cry2A; Lane 6: Negative control; Lane 7: Positive control. (c) PCR amplification of cp4-EPSPS. Lane 1: 50bp DNA Ladder; Lane 2–5: putative transgenic cotton plants showing amplification of 111bp PCR product of cp4-EPSPS; Lane 6: Negative control; Lane 7: Positive control
3.3 Gene Expression Analysis of cp4-EPSPS through Quantitative Real-Time PCR
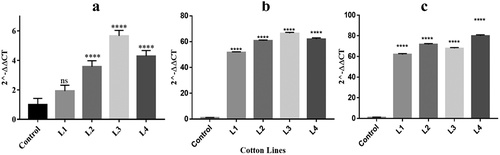
Quantitative real-time mRNA expression analysis was conducted to determine the relative expression of transgene cp4-EPSPS in cotton plants. The expression determination of three genes revealed a several-fold increase of mRNA in different transgenic cotton lines (represented by L1, L2, L3, L4 throughout the rest of the analysis). The highest mRNA expression (5.6-fold) of cry1Ac was found to be in L3 ( A), while cry2A expression was highest in transgenic line L4 up to 79.9-fold ( B). The maximum mRNA expression of cp4-EPSPS in transgenic plant L3 was found to be 66.47 ( C).
Figure 4. Relative quantification of mRNA expression through quantitative real time PCR. Non-transgenic wild type plant is represented by Control, and transgenic cotton plants as L1 to L4. (a) mRNA expression of cry1Ac gene. (b) mRNA expression of cry2A gene. (c) mRNA expression of cp4-EPSPS. Significant difference at p < .0001 is represented by **** as calculated by Dunnett’s multiple comparison test and one-way ANOVA
3.4 Quantification of Cry1Ac, Cry2A and Cp4-EPSPS Proteins through ELISA
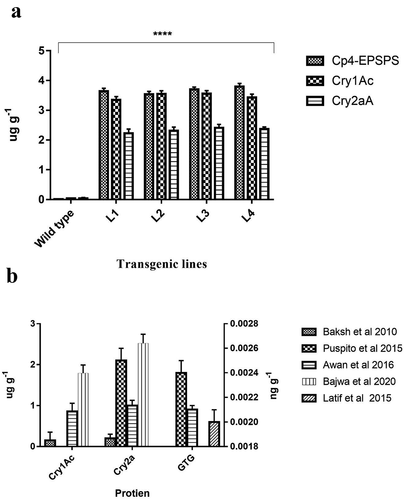
Crude protein was freshly extracted from fresh leaves of cotton plants for dipstick assay. Protein detection through dipstick assay revealed red bands for Cry1Ac and Cry2A in all four transgenic cotton plants. The appearance of a band in standard control confirmed the presence of respective proteins ( A & B). For ELISA, the sample was taken from three different parts of the plant i.e., upper, middle and lower canopy. ELISA assay was conducted to quantify the protein concentration in transgenic cotton plants. The assay results showed that maximum concentration of Cry1Ac and Cry2A was found to be 3.534 µg g−1 and 2.534 µg g−1 respectively, in fresh leaves of transgenic cotton plants. The Cry1Ac protein concentration in G. arboretum (diploid) is very much improved () than the similar protein’s expression in the G. hirsutum (tetraploid) as reported by previous studies ().Citation11,Citation14 The quantification of Cp4-EPSPS protein revealed maximum concentration to be 3.870 µg g−1 in transgenic cotton line L4 ()) which is also much higher than tetraploid version cotton reported in previous studies ().Citation11,Citation14
Figure 5. Immunostrip or dipstick assay. (a)Detection of Cry1Ac protein. The upper dark red line is the control line; Lower faint line shows a positive test line. (b) Detection of Cry2A protein. The upper dark red line is the control line; Lower faint line shows a positive test line
Figure 6. Comparison of transgene protein expression in diploid and tetraploid cotton. (a) shows the obtained concentration of GTG (Cp4-EPSPS) in diploid transgenic lines (Gossypium arboretum). (b) depicts the obtained concentration of Cp4-EPSPS, Cry1Ac and Cry2a in tetraploid cotton (Gossypium hirsutum) as determined in different studies.Citation11,Citation14,Citation21–23
3.5 Insect Resistance and Herbicide-Tolerance Status of Transgenic Cotton
The leaves of cotton plants were fed to H. armigera larvae. The efficacy of Bt proteins on larvae was evaluated by amount of leaf consumed, weight gain of larvae, and larvae’s death. After 3 days of the assay, complete mortality was observed in L3 and L4 transgenic cotton lines while the amount of plant leaf consumed in L3 transgenic cotton line was more than L4. In the non-transgenic control cotton plants, 90% larvae survived, while in and L1 and L2 transgenic cotton lines, 60% and 90% survival were observed, respectively, along with maximum consumption of the plant leaf as shown in .
Figure 7. Resistance evaluation of transgenic cotton lines (cp4-EPSPS). (a) Insect mortality assay on detached leaves of representative cotton lines. (b) Glyphosate spray assay conducted in field to determine the herbicide resistance status of transgenic cotton plants

Resistance to cp4-EPSPS gene was evaluated by spraying glyphosate on cotton plants. All the transgenic cotton lines survived when observed after five and ten days of spray while all type of weeds were dried up after glyphosate spray in all transgenic cotton lines ().
4. Discussion
Gossypium arboreum is an Asiatic cotton or “Old World” cotton, originating from Indian Sub-continent. This cotton is cultivated in Pakistan due to its disease and insect resistance,Citation24 early maturity, drought and stress toleranceCitation25 and fiber characteristics.Citation26 G. arboreum is naturally resistant to Bemisia tabaci which is the biological vector of CLCuV transmission. This results in a loss of millions of dollars on CLCuV management.Citation18 Because of the natural resistance of G. arboreum to B. tabaci and CLCuV disease,Citation27 Bt genes integration could further enhance broad-spectrum resistance. Glyphosate – commercially known as “Roundup” – is a general weed killer and capable of suppressing the growth of broad leaved weeds along with crop plants.Citation28 We wanted to indirectly improve the yield and quality of fiber from G. arboreum by stacking herbicide resistance genes and insect resistance genes. The current study was proposed to control chewing insect pests and weeds through genetic modification of G. arboreum with a codon-optimized cry1Ac and cry2A gene construct for resistance against chewing insects and the cp4-EPSPS gene for tolerance against glyphosate herbicide.
Shoot apex transformation method has already been proved successful for recalcitrant crops like cotton. The transformation efficiency for G. arboreum was found to be 1.59%, though promising but less than other reports,Citation3,Citation18,Citation29 and still holds the potential for improvement. PCR amplification confirmed the successful integration of all genes, cry1Ac, cry2A and cp4-EPSPS genes in G. arboreum. The transcript accumulation analysis by qRT-PCR revealed 51- (minimum) to 66.47-fold (maximum) cp4-EPSPS transcripts in transgenic event L1 and L3, respectively. Increase in expression in transgenic cotton leaves demonstrates the successful constitutive expression of CaMV35S promoter in G. arboreum. This study’s result is consistent with the study byCitation30 who obtained upto 12-fold expression. For cry1Ac, about 5.66-fold increase in L3 and 4.29-fold increase in L4 was observed compared to the non-transgenic control line. Similarly, cry2A transcript level ranged between 61-fold to 79-fold in L1 and L3 transgenic cotton lines, respectively. Qualitative analysis with dipstick immunostrip assay confirmed Cry proteins’ presence (Cry1Ac and Cry2A) in transgenic cotton plant leaves in accordance with the study by.Citation31,Citation32 The quantitative analysis of Cry1Ac, Cry2A and Cp4-EPSPS protein with ELISA from fresh leaves of transgenic cotton plants showed 3.534 µg g−1 and 2.534 µgg−1 and 3.8 µg g−1 proteins, respectively. Comparison with other studies revealed that G. arboreum has more expression than G. hirsutum.Citation11,Citation21 In this study, Cry1Ac protein concentration is well above (3.534µg g−1) the LD95 concentration of 2.20 µg g−1 as determined by.Citation33 and 159 µg g−1 as determined by.Citation34 For a toxin to be effective, a higher LD95 value is a prerequisite for effective control of insects. The evolving reports of H. armigera resistance suggest that Bt-protein’s sub-lethal expression level is one of the key factors in resistance development in Pakistan among many other management-related issues.Citation33 There was also maximum concentration of Cp4-EPSPS protein in the transgenic line L4 (3.84ug g−1) as compared to control cotton plant which is consistent with the study by.Citation30 Also higher Cp4-EPSPS protein concentration was evident in G. arboreum as compared to G. hirsutum previously reported byCitation11,Citation14,Citation21 which may be due to the fact that diploid cotton can have higher expression of traits which make it tolerant to CLCuV and other insects pests as well. Uncontrolled weed growth, alongside crop plants, compete for soil nutrition and harbor insect pests and viruses.Citation35 This escalates the investment cost for farmer and poses economic burden. Stacking insect-resistant genes and herbicide-tolerance genes in the same transgenic plants can substantially enhance yields, relief from insect infestation as well as less consumption of pesticides.Citation11,Citation14
The transgenic cotton plants’ insect bioassay revealed 100% mortality of H. armigera larvae in transgenic cotton lines L3 and L4 (). The insect bioassay result was consistent with the study by.Citation11 andCitation21 Pyramiding the cotton plant with cry genes and cp4-EPSPS gene in G. hirsutum also showed success by killing 100% insects and tolerance from glyphosateCitation11,Citation21 this study for G. arboreum. However, this study’s uniqueness is that the data for insect mortality and glyphosate tolerance was analyzed in the T3 generation of G. arboreum transgenic plants, depicting stable gene expression in highly selective transgenic lines.
The transgenic cotton plants’ efficacy against glyphosate challenge assay demonstrated the survival of the transgenic cotton plants in the field and successful removal of all type of weeds (). There have already been reports about the introgression of cp4-EPSPS in G. hirsutum from our labCitation11,Citation14,Citation21 and other labs.Citation30 Still, to our knowledge, this is the first report of in-planta transformation of these genes into G. arboreum with a higher transformation efficiency and stable gene expression. Improvement of G. arboreum fiber yield, which is already resistant for CLCuV,Citation36 is a great asset for cotton germplasm and cost-efficient cotton production by farmers. The insect-resistant and herbicide-resistant G. arboreum variety can serve as a resource for fiber and yield improvement in the future.
5. Conclusion
Cotton farmers in Pakistan suffer huge economic burden due to insects, weeds, fiber quality and droughts/floods. In this study, the Gene pyramiding approach demonstrates the successful transformation and expression of Cry1Ac and Cry2A toxins and Cp4-EPSPS protein in a local variety of cotton (FBD-1), which is already resistant to the devastating insect B. tabaci. This approach would help farmers increase cotton yields, reduce insecticidal spray and manual weeding of cotton and decrease soil quality through excessive weedicides and insecticides. This improved variety of G. arboreum will provide opportunity to utilize this material further to improve fiber traits and its yield through breeding in the future.
Author’s Contribution
AQR came up with the idea and approval of this research project. MST carried out all the research work. AL supervised the plant transformation experiments. NS supervised the protein quantification assays. SB and MAUK supervised the insect bioassay. AG prepared the initial manuscript and final editing and formatting. AAS and TH evaluated the material for field experiments. All authors reviewed and approved the article for submission.
Acknowledgments
We are thankful to Higher Education Commission for providing resources to complete this research project.
Disclosure statement
The authors declare that they have no competing interests.
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
References
- Mushtaq R, Shahzad K, Shah ZH, Alsamadany H, Alzahrani HA, Alzahrani Y, Mujtaba T, Ahmed Z, Mansoor S, Bashir A. Isolation of biotic stress resistance genes from cotton (gossypium arboreum) and their analysis in model plant tobacco (nicotiana tabacum) for resistance against cotton leaf curl disease complex. Virol Methods. 2020;276:113760. doi:https://doi.org/10.1016/j.jviromet.2019.113760.
- Blaise D, Kranthi KR, Ravindran CD, Thalal K. High plant density can improve the productivity of rainfed asiatic cotton (gossypium arboreum L.). Arch Agron Soil Sci. 2020:1–13. doi:https://doi.org/10.1080/03650340.2020.1741553.
- Nandeshwar S, Moghe S, Chakrabarty P, Deshattiwar M, Kranthi K, Anandkumar P, Mayee C, Khadi B. Agrobacterium-mediated transformation of cry1Ac gene into shoot-tip meristem of diploid cotton gossypium arboreum cv. RG8 and regeneration of transgenic plants. Plant Mol Biol Rep. 2009;27(4):549–57. doi:https://doi.org/10.1007/s11105-009-0102-7.
- Akhtar K, Haidar S, Khan M, Ahmad M, Sarwar N, Murtaza M, Aslam M. Evaluation of gossypium species for resistance to cotton leaf curl burewala virus. Ann Appl Biol. 2010;157(1):135–47. doi:https://doi.org/10.1111/j.1744-7348.2010.00416.x.
- Ali MA, Khan IA, Awan SI, Ali S, Niaz S. Genetics of fibre quality traits in cotton (Gossypium hirsutum L.). Aust J Crop Sci. 2008;2:10–17.
- Farrell T, Johnson A. Cotton pest management guide 2005/06. NSW Department of Primary Industries:99. 2005.
- Charles G, Roberts G. Managing weeds in cotton. Australian Cotton Cooperative Research Centre, ed WEED Pak Cotton Research & Development Corporation Narrabri, NSW. 2002.
- Pakistan Economic Survey 2018-2019. (trans: economic Advisor’s Wing FD, Government of Pakistan, Islamabad.). Finance Division of Pakistan. 2019.
- Schnepf E, Crickmore N, Van Rie J, Lereclus D, Baum J, Feitelson J, Zeigler D, Dean D. Bacillus thuringiensis and its pesticidal crystal proteins. Microbiol Mol Bio Rev. 1998;62:775–806.
- Rocha-Munive MG, Soberón M, Castañeda S, Niaves E, Scheinvar E, Eguiarte LE, Mota-Sánchez D, Rosales-Robles E, Nava-Camberos U, Martínez-Carrillo JL. Evaluation of the impact of genetically modified cotton after 20 years of cultivation in Mexico. Front Bioeng Biotech. 2018;6:82. doi:https://doi.org/10.3389/fbioe.2018.00082.
- Puspito AN, Rao AQ, Hafeez MN, Iqbal MS, Bajwa KS, Ali Q, Rashid B, Abbas MA, Latif A, Shahid AA. Transformation and evaluation of Cry1Ac+ Cry2A and GTGene in gossypium hirsutum L. Front Plant Sci. 2015;6:943. doi:https://doi.org/10.3389/fpls.2015.00943.
- Hernández-Rodríguez CS, Van Vliet A, Bautsoens N, Van Rie J, Ferré J. Specific binding of bacillus thuringiensis cry2A insecticidal proteins to a common site in the midgut of Helicoverpa species. Appl Environ Microbiol. 2008;74(24):7654–59. doi:https://doi.org/10.1128/AEM.01373-08.
- Luo S, Wu K, Tian Y, Liang G, Feng X, Zhang J, Guo Y. Cross-resistance studies of cry1Ac-resistant strains of helicoverpa armigera (lepidoptera: noctuidae) to cry2Ab. Econ Entomol. 2007;100(3):909–15. doi:https://doi.org/10.1603/0022-0493(2007)100[909:CSOCSO]2.0.CO;2.
- Latif A, Rao AQ, Khan MA, Shahid N, Bajwa KS, Ashraf MA, Abbas MA, Azam M, Shahid AA, Nasir IA. Herbicide-resistant cotton (gossypium hirsutum) plants: an alternative way of manual weed removal. BMC Res Notes. 2015;8(1):453. doi:https://doi.org/10.1186/s13104-015-1397-0.
- Suarez L, Apan A, Werth J. Hyperspectral sensing to detect the impact of herbicide drift on cotton growth and yield. ISPRS J Photogramm Remote Sens. 2016;120:65–76. doi:https://doi.org/10.1016/j.isprsjprs.2016.08.004.
- Santos-Sánchez NF, Salas-Coronado R, Hernández-Carlos B, Villanueva-Cañongo C. Shikimic acid pathway in biosynthesis of phenolic compounds. Plant Physiological Aspects of Phenolic Compounds, Vol. 1. IntechOpen. 2019. doi:https://doi.org/10.5772/intechopen.83815.
- Rao AQ, Irfan M, Saleem Z, Nasir IA, Riazuddin S, Husnain T. Overexpression of the phytochrome B gene from Arabidopsis thaliana increases plant growth and yield of cotton (gossypium hirsutum). Zhejiang Univ Sci B. 2011;12(4):326–34. doi:https://doi.org/10.1631/jzus.B1000168.
- Gul A, Hussain G, Iqbal A, Rao AQ, Ud Din S, Yasmeen A, Shahid N, Ahad A, Latif A, Azam S. Constitutive expression of asparaginase in gossypium hirsutum triggers insecticidal activity against bemisia tabaci. Sci Rep. 2020;10(1):1–11. doi:https://doi.org/10.1038/s41598-020-65249-w.
- Kamo K, Jordan R, Guaragna MA, H-t H, Ueng P. Resistance to cucumber mosaic virus in gladiolus plants transformed with either a defective replicase or coat protein subgroup II gene from cucumber mosaic virus. Plant Cell Rep. 2010;29(7):695–704. doi:https://doi.org/10.1007/s00299-010-0855-3.
- Zhao L, Ding Q, Zeng J, FR W, Zhang J, SJ F, XQ H. An improved CTAB–ammonium acetate method for total RNA isolation from cotton. Phytochem Anal. 2012;23(6):647–50. doi:https://doi.org/10.1002/pca.2368.
- Awan M, Ali A, Muzaffar A, Abbas M, Rao AQ, Qamar Z, Butt S, Khan G, Rashid B, Nasir IA, et al. Transgenic cotton: harboring board term resistance against insect and weeds through Incorporation of CEMB double Bt and cp4EPSPS genes. Pak J Agri Sci. 2016;53(3):501–05. doi:https://doi.org/10.21162/PAKJAS/16.2007.
- Bajwa KS, Shahid AA, RaoAQ, BashirA, Aftab A, HusnainT. Stable transformation and expression of GhEXPA8 fiber expansin gene to improve fiber length and micronaire value in cotton. Front Plant Sci. 2015;6:838. doi:https://doi.org/10.3389/fpls.2015.00838.
- Bakhsh A, Rao AQ, Shahid AA, Husnain T, Riazuddin S. CaMV 35S is a developmental promoter being temporal and spatial in expression pattern of insecticidal genes (Cry1ac & Cry2a) in cotton. Aust J Basic & Appl Sci. 2010;4:37–44.
- Gong Q, Yang Z, Chen E, Sun G, He S, Butt HI, Zhang C, Zhang X, Yang Z, Du X. A phi-class glutathione S-transferase gene for Verticillium wilt resistance in gossypium arboreum identified in a genome-wide association study. Plant Cell Physiol. 2018;59(2):275–89. doi:https://doi.org/10.1093/pcp/pcx180.
- Lu P, Magwanga RO, Guo X, Kirungu JN, Lu H, Cai X, Zhou Z, Wei Y, Wang X, Zhang Z. Genome-wide analysis of multidrug and toxic compound extrusion (MATE) family in gossypium raimondii and gossypium arboreum and its expression analysis under salt, cadmium, and drought stress. G3: Genes, Genomes, Genetics. 2018;8(7):2483–500. doi:https://doi.org/10.1534/g3.118.200232.
- Guo WZ, Zhou BL, Yang LM, Wang W, Zhang TZ. Genetic diversity of landraces in gossypium arboreum L. race sinense assessed with simple sequence repeat markers. Integr Plant Biol. 2006;48(9):1008–17. doi:https://doi.org/10.1111/j.1744-7909.2006.00316.x.
- Khan MI, Haq HA, Ullah K, Arshad M, Majid A. Genetic diversity and correlation studies for cotton leaf curl disease (CLCuD), fiber & yield related attributes in exotic lines of gossypium arboreum L. Am J Plant Sci. 2017;8(3):615–24. doi:https://doi.org/10.4236/ajps.2017.83042.
- Schmid J, Amrhein N. Molecular organization of the shikimate pathway in higher plants. Phytochemistry. 1995;39(4):737–49. doi:https://doi.org/10.1016/0031-9422(94)00962-S.
- Keshamma E, Rohini S, Rao K, Madhusudhan B, Kumar MU. Tissue culture-independent in planta transformation strategy: an agrobacterium tumefaciens-mediated gene transfer method to overcome recalcitrance in cotton (Gossypium hirsutum L.). J Cotton Sci. 2008;12:264–72.
- Karthik K, Nandiganti M, Thangaraj A, Singh S, Mishra P, Rathinam M, Sharma M, Singh NK, Dash PK, Sreevathsa R. Transgenic cotton (gossypium hirsutum L.) to combat weed vagaries: utility of an apical meristem-targeted in planta transformation strategy to introgress a modified CP4-EPSPS gene for glyphosate tolerance. Front Plant Sci. 2020;11(768). doi:https://doi.org/10.3389/fpls.2020.00768.
- Bakhsh A, Rao AQ, Shahid AA, Husnain T. Spatio temporal expression pattern of an insecticidal gene (cry2A) in transgenic cotton lines. Not Sci Biol. 2012;4(4):115. doi:https://doi.org/10.15835/nsb448217.
- Shahid AA, Bano S, Khalid S, Samiullah TR, Bajwa KS, Ali MA. Biosafety assessment of transgenic Bt cotton on model animals. Adv Life Sci. 2016;3(3):97–108. doi:https://doi.org/10.1631/jzus.B1000168.
- Jamil S, Shahzad R, Rahman SU, Iqbal MZ, Yaseen M, Ahmad S, Fatima R. The level of cry1Ac endotoxin and its efficacy against H. armigera in Bt cotton at large scale in Pakistan. GM Crops Food. 2020;12(1):1–17. doi:https://doi.org/10.1080/21645698.2020.1799644.
- Ahmad S, Cheema HMN, Khan AA, Khan RSA, Ahmad JN. Resistance status of helicoverpa armigera against Bt cotton in Pakistan. Transgenic Res. 2019;28(2):199–212. doi:https://doi.org/10.1007/s11248-019-00114-9.
- DiTommaso A, Averill KM, Hoffmann MP, Fuchsberg JR, Losey JE. Integrating insect, resistance, and floral resource management in weed control decision-making. Weed Sci. 2016;64(4):743–56. doi:https://doi.org/10.1614/WS-D-16-00052.1.
- Mushtaq R, Shahzad K, Mansoor S, Shah ZH, Alsamadany H, Mujtaba T, Al-Zahrani Y, Alzahrani HA, Ahmed Z, Bashir A. Exploration of cotton leaf curl virus resistance genes and their screening in gossypium arboreum by targeting resistance gene analogues. AoB Plants. 2018;10(6):ply067. doi:https://doi.org/10.1093/aobpla/ply067.