Abstract
A functional metagenomics based approach exploiting the microbiota of suppressive soils from an organic field site has succeeded in the identification of a clone with the ability to inhibit the growth of Bacillus subtilis DSM10. Sequencing of the fosmid identified a putative β-lactamase-like gene abgT. Transposon mutagenesis of the abgT gene resulted in a loss in ability to inhibit the growth of B. subtilis DSM10. Further analysis of the deduced amino acid sequence of AbgT revealed moderate homology to esterases, suggesting that the protein may possess hydrolytic activity. Weak lipolytic activity was detected; however the clone did not appear to produce any β-lactamase activity. Phylogenetic analysis revealed the protein is a member of the family VIII group of lipase/esterases and clusters with a number of proteins of metagenomic origin. The abgT gene was sub-cloned into a protein expression vector and when introduced into the abgT transposon mutant clones restored the ability of the clones to inhibit the growth of B. subtilis DSM10, clearly indicating that the abgT gene is involved in the antibacterial activity. While the precise role of this protein has yet to fully elucidated, it may be involved in the generation of free fatty acid with antibacterial properties. Thus functional metagenomic approaches continue to provide a significant resource for the discovery of novel functional proteins and it is clear that hydrolytic enzymes, such as AbgT, may be a potential source for the development of future antimicrobial therapies.
Introduction
In recent years, metagenomic approaches have increasingly been employed to access the uncultured and uncultivable majority of microorganisms present in the environment, which is estimated to be more than 99% of the existing prokaryotes in soil systems.Citation1 Metagenomic analyses, such as large-scale shotgun sequencing, have to date been applied to various environments and have led to the discovery of novel species, and even the elucidation of novel pathways and community-specific metabolism.Citation2-7 Apart from sequencing-based approaches, metagenomic libraries can also be screened for the presence and expression of specific functions and activities, also known as gain-of-function, and a range of novel biocatalysts, many with potential industrial applications, as well as small molecules with novel bioactivities, have already been discovered using such functional metagenomics based techniques.Citation8-16 With its extremely high, but still largely underexplored and unexploited, microbial diversity and abundance, soil is one of the richest potential sources for novel biocatalysts and novel natural products.Citation17-20 In recent years, functional metagenomic analyses has led to the discovery of a wide range of enzymes and bioactive metabolites, e.g. antimicrobial agents, from the soil microbiota.Citation21-23 Soils that suppress plant disease, or so called suppressive soils, are known to exert their antagonistic function through their soil microbiota;Citation24 particularly through the presence of phytopathogenic antagonistic (antibiotic) activity.Citation25 Indeed it has recently been reported that upon attack by fungal pathogens plants exploit the microbial consortia from soil to protect themselves against infection.Citation26 Thus suppressive soils which show a natural suppression of growth of certain plant pathogens are likely to be a valuable source for the discovery of novel bioactive compounds.
Metagenomic approaches have also been employed to assess the bacterial natural product biosynthetic diversity of different soil microbial communities.Citation27 From this work it appears that prokaryotic natural product biosynthesis biodiversity is potentially much larger than we have previously thought from culture dependent based approaches. Indeed a wide variety of natural products including indigo, indirubin,Citation28,29 turbomycin A, turbomycin B,Citation30 violacein,Citation31 palmitoylputrescine,Citation32 norcardamine, terragines A-E33 and tetarimycin A34 among others, have in the past been isolated from environmentally derived metagenomic libraries. In addition with the advent of improved heterologous host expression systems in Streptomyces spp., in different Proteobacteria such as Agrobacterium tumefaciens, Escherichia coli, Caulobacter vibrioides, Pseudomonas putida, Burkholderia graminis and Ralstonia metallidurans and in Saccharomyces cerevisiae,Citation18,35-37 an increasing number of bioactive small molecules with potential biopharmaceutical applications are being discovered on an on-going basis. Examples include the pentacyclic polyketide erdacin,Citation35 the fluostatinsCitation36 the antitumor substance BE-54017,Citation14 the indolotryptoline antiproliferative agent (borregomycin A) and the dihydroxyindolocarbazole anticancer/antibiotics (borregomycins B-D)Citation38 together with the antibiotics fasamycins A and B.Citation39
Thus given the potential of suppressive soils containing microbiota with antimicrobial and anti-phytopathogenic activities, we employed a functional metagenomics-based approach in an effort to identify novel antibiotic activities from the microbiome of soils from an organic field site. This approach resulted in the cloning of a β-lactamase-like gene, abgT, with an ability to inhibit the growth of B. subtilis DSM10.
Results
Screening of the soil library
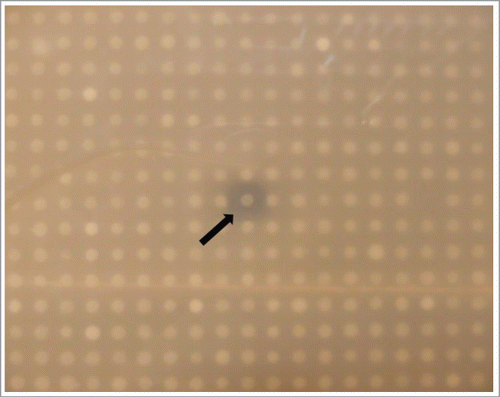
Approximately 14000 clones were screened for antimicrobial activity against Pseudomonas aeruginosa PA01 and Bacillus subtilis DSM10, and a single clone, TO-T-020-P12, was found to clearly inhibit the growth of B. subtilis DSM10 (). This clone was subjected to further analysis and was found to consistently produce a zone of inhibition against B. subtilis DSM10 in overlay assays. The clone did not, however, appear to have the ability to inhibit P. aeruginosa PA01. Furthermore the clone TO-T-020-P12 did not inhibit growth of B. subtilis DSM10 in the absence of arabinose, which was responsible for inducing fosmid replication to multiple copy numbers. The TO-T-020-P12 fosmid was isolated and a fresh stock of E. coli EPI300™ T1 cells was re-transformed with the fosmid and activity was confirmed. E. coli EPI300™ T1 clones containing pCC1FOS with the fosmid control DNA supplied with the library production kit were used as a negative control and consistently failed to produce any activity against either B. subtilis DSM10 or P. aeruginosa PA01.
Sequencing and annotation of fosmid
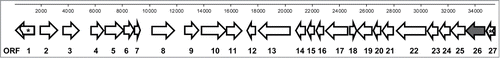
The TO-T-020-P12 fosmid was sequenced by Roche 454 pyrosequencing, and assembly of the resulting sequences produced a contig of 42137 bp, with a G-C content of 60.5%. Further examination of the sequence for vector contamination yielded an insert size of 35277 bp (GenBank accession no. JX846920). The insert sequence was analyzed for putative ORFs using both the FGENESB and the MetaGene programmes. While each program predicted a slightly different number of ORFs, in general there was substantial agreement between the main ORFs present (Table S1). Each ORF was putatively annotated using blastP searches for homologous proteins and the predicted annotations were further refined by manual examination with the final annotation selected based on evidence that the proteins appeared to be a better fit and more closely-related. A total of 27 ORFs were putatively annotated (, ). The genes appeared to encode for a range of proteins with potentially diverse functions including proteins involved in hydrolysis, oxidation/reduction, biosynthesis, membrane transport, transcriptional regulation and a number of proteins of as yet unknown function. The putative annotations for each ORF listed in reflect the top matches after blastP searches. The vast majority of the species associated with these proteins appear to be environmental isolates, particularly from soil and aquatic habitats, and include members of the following bacterial phyla; Proteobacteria, Firmicutes, Acidobacteria, Bacteroidetes and Verrucomicrobia. A number of the putative ORFs were found to be associated with the bacterium Ellin514, with 4 adjacent putative proteins (Genes 9–12) appearing to share homology with proteins from this bacterium. This strain, also known as Pedosphaera parvula Ellin514, is an obligate aerobic bacterium, which was isolated from pasture soil in Victoria, Australia, and belongs to the Verrucomicrobia phylum whose members are common in terrestrial environments.Citation40 Attempts were made to further determine the phylogenetic origin of the metagenomic DNA using MEGAN, however the results did not predict any taxonomic origin below the Proteobacteria phylum level, and with only 32% of the ORFs assigned to this phylum it is difficult to postulate more precisely the origin of the metagenomic DNA in clone TO-T-020-P12.
Table 1. Putative annotation of the open reading frames from fosmid TO-T 020 P12
Figure 2. Schematic representation of putative open reading frames. Putative open reading frames were identified in the 35277 bp DNA insert of fosmid clone TO-T-020-P12. The arrows represent the position and direction of transcription of each open reading. The numbers correspond to the ORFs listed in . The abgT gene (ORF 26 -) is highlighted in a darker color . Arrows containing * represent partial open reading frames.
Transposon mutagenesis and identification of insertion site
Two mutants, TO-T-020- P12 E2 and TO-T-020-P12 F2, were obtained following screening of transposon mutants which had lost the ability to inhibit growth of B. subtilis DSM10. The transposon insertion sites were determined by bidirectional sequencing and both mutants were found to have insertions at the 3′ end of ORF 26 approximately 55 bp from the stop codon. Putative annotation of the fosmid revealed that ORF 26 showed similarity to a β-lactamase protein (). This ORF was subsequently designated as gene abgT and the results indicated that transcription of this gene had been disrupted and therefore that it was potentially playing a role in the observed antibacterial activity.
Analysis of the abgT gene
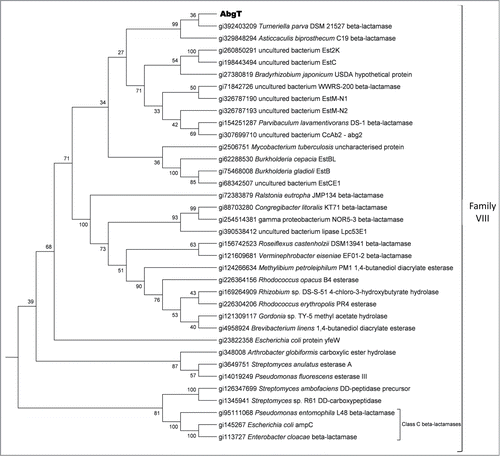
The abgT gene which is 1335 bp yields a predicted protein of 444 amino acids, with a molecular mass of 47.2 kDa. The protein has a theoretical pI of 9.22 and an instability index of 30.65, suggesting that it is a stable protein. The ORF is located toward the 3′ end of the insert sequence and is adjacent to a partial ORF with similarity to a pyruvate/2-oxoglutarate dehydrogenase (). The gene appears to have a GTG start codon with an almost perfect Shine-Delgarno consensus sequence located 6 bp upstream. Analysis of the promoter region suggests that this gene may be under the control of its own promoter, with putative -10 (tggcatact) and -35 (gtgtcg) regions beginning 226 bp upstream from the proposed start codon. Further analysis of the protein revealed a potential signal peptide of 25 amino acids (VTAANSLSRILLALALGVTPHVAQA), suggesting that the protein may be secreted. The predicted amino acid sequence of the AbgT protein contains a β-lactamase conserved domain (pfam00144) and has highest identity, of up to 55%, to several proteins classified as putative β-lactamases, with highest identity to a β-lactamase from Turneriella parva DSM 21527. Alignments of the AbgT protein sequence with other β-lactamase like proteins, Class C β-lactamases and representative of the different lipase family groups was performed and subsequent phylogenetic analyses revealed that AbgT is a member of the family VIII group of lipases/esterases ().Citation41,42 This group contains the Class C β-lactamases and a number of β-lactamase like proteins, though the exact function of many of these proteins has yet to be biochemically determined.
Lipase and β-lactamase activities
To determine if the protein possessed any potential lipase activity the original clone TO-T- 020-P12 and Tn5 mutant TO-T-020-P12 E2 were incubated on plates containing 1% tributyrin. Clone TO-T-020-P12 produced lipolytic activity (), though the observed activity was quite weak compared to a metagenomic lipase clone which had previously been isolated in our laboratory and determined to have high levels of activity.Citation13 Transcriptional disruption of the abgT gene resulted in a loss of lipolytic activity as the mutant strain TO-T-020-P12 E2 failed to produce any zone of clearing on tributyrin, even after prolonged incubation. As the protein displayed similarity to β-lactamase type proteins the chromogenic nitrocefin assay was used to determine if there was any significant β-lactamase activity present. Both the original clone TO-T-020-P12 and the mutants TO-T-020-P12 E2 and TO-T-020-P12 F2 were tested for activity, using ampicillin resistant cells as a positive control. Cells containing ampicillin resistance genes produced a red color when tested with the nitrocefin as expected. However, the metagenomic clones failed to produce a similar reaction under the conditions tested and therefore it was concluded that AbgT does not possess true β-lactamase activity (Figure S1). The overlay assays with B. subtilis DSM10 were also performed in conjugation with the nitrocefin assay to confirm that antibacterial activity was observed as expected under the growth conditions used.
Figure 4. Lipolytic and antibacterial activity of the abgT gene. (A) Clone TO-T-020-P12 produced weak lipolytic activity on agar plates containing 1% tributyrin. When the abgT gene was disrupted by Tn5 insertion in TO-T 020 P12 E2 lipolytic activity was lost. Metagenomic lipase Lpc53E1 was previously characterized in our laboratory and was used for comparison of lipolytic activity. (B) Random mutagenesis of fosmid TO-T 020 P12 produced 2 mutants unable to inhibit the growth of B. subtilis DSM10. Both mutants regained the ability to inhibit growth after complementation of the disrupted abgT gene. (1) TO-T- 020-P12 (2) TO-T-020-P12 E2 (3) TO-T-020-P12 F2 (4) TO-T-020-P12 with pBadMyc-HisA-AB-RH1 (5) TO-T-020-P12 E2 with pBadMyc-HisA-AB-RH1 (6) TO-T-020-P12 F2 with pBadMyc-HisA-AB-RH1.
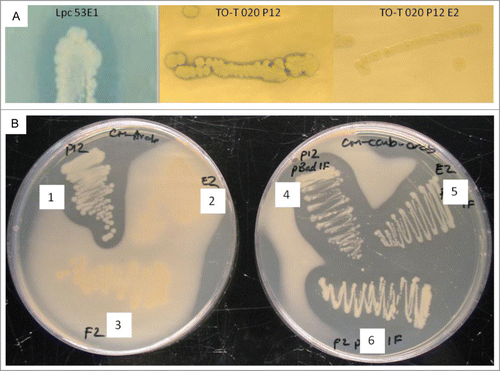
Restoration of antibacterial activity in the abgT disrupted mutants
To confirm that the β-lactamase-like gene was indeed involved in the observed antibacterial activity the abgT gene was cloned into the expression vector pBadMyc-HisA to create pBadMyc-HisA-AB-RH1. The original clone TO-T 020 P12 and both mutants TO-T-020-P12 E2 and TO-T 020 P12 F2 were transformed with the recombinant vector and the clones were subsequently overlaid with B. subtilis DSM10 (). The clone TO-T-020-P12 showed clear ability to inhibit the growth of B. subtilis DSM10, but as the results indicate when the abgT gene was disrupted in the TO-T-020-P12 E2 and TO-T-020-P12 F2 mutants both were unable to inhibit the growth of B. subtilis DSM10 strain (; 2 and 3). However, complementation of the TO-T-020-P12 E2 and TO-T-020-P12 F2 mutants with the vector borne abgT restored the ability of each mutant to inhibit growth (; and ). As a control, each strain was also transformed with the pBadMyc-HisA vector lacking an insert and the presence of the vector did not affect the inability of the mutants to inhibit B. subtilis DSM10 growth. Thus the abgT gene product is clearly responsible for the observed anti- B. subtilis DSM10 activity. To confirm that a protein of the correct size was being produced from the expression vector the His-tagged AbgT protein was purified from the total soluble protein fraction of cell lysate derived from arabinose induced E. coli EPI300™ T1 cells containing pBadMyc-HisA-AB-RH1. Staining of the SDS-PAGE gel revealed a protein with a mass just below 50kDa, which correlates well with the predicted in silico molecular mass of AbgT ().
Figure 5. Purification of His6-AbgT protein. His6- AbgT was purified from the total soluble protein fraction of cell lysate of E coli EPI300TM T1 transformed with the recombinant vector pBadMyc-HisA-AB-RH1. The expressed antibacterial protein was purified from the supernatant by Ni-NTA spin column and resolved by SDS-PAGE on a 10% gel. (A) Anti-bacillus activity of the purified Abg protein from cell lysate (1; purified fraction of Ni-NTA column, 2; protein fraction from cell lysate, 3; supernatant fraction, 4; wash from Ni-NTA elution, 5; control with cell free lysate from EP1300 cells). (B) Lane 1 stained with Invision His-tag In-gel stain showing the mass of the purified protein at approximately 50 kDa. (C) Lane 1 showing the purified protein stained with Coomassie brilliant blue.
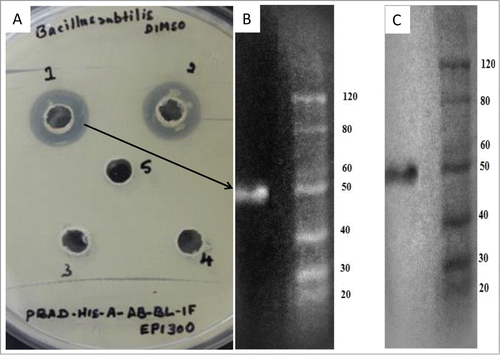
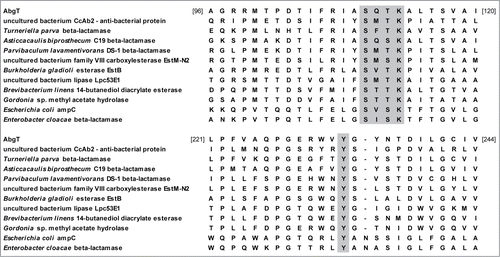
Figure 6. Partial alignment of AbgT with related members of the family VIII group of lipases. The alignment includes; closely related putative β-lactamase proteins from Turneriella parva, Asticcacaulis biprosthecum and Parvibaculum lavamentivorans; antibacterial protein from CcAb2; esterase and hydrolase enzymes from uncultured bacteria (EstM-N2 and Lpc53E1), Burkholderia gladioli, Brevibacteroum linens and Gordonia sp; class C β-lactamases from Escherichia coli and Enterobacter cloacae. Conserved sites S-X-X-K and Y are shaded. The numbers represent amino acids of the AbgT protein.
Discussion
We report here on the cloning of the abgT gene, isolated following the functional screening of a soil metagenomic library; the product of which clearly inhibits the growth of B. subtilis DSM10. The AbgT protein while not possessing any β-lactamase activity itself was found to possess a high level of identity with several putative β-lactamases, with highest identity to a β-lactamase from Turneriella parva DSM 21527. Strains of T. parva have previously been isolated from contaminated culture medium, tap water and from the uterus of a sow.Citation43 AbgT also displayed similarity to a number of other β-lactamase proteins, including β-lactamases from Asticcacaulis biprosthecum C19, Verrucomicrobiae bacterium DG1235 and Muricauda ruestringensis DSM 13258.
Functional screening of fosmid libraries associated with the marine sponge Cymbastela concertrica has identified a novel antibacterial protein, Abg 2, which appears to also have similar characteristics to the AbgT protein.Citation44 Both showed antibacterial activity against Bacillus strains, contain β-lactamase conserved domains and show similarity to the same proteins in the SwissProt database (). Despite these similarities there was only moderate pairwise homology (33%) between these 2 proteins at the amino acid level. In addition the predicted AbgT protein sequence displayed a weak similarity to some lipase/esterase enzymes including a lipase/esterase from Hydrogenophaga sp. PBC and a putative esterase from Methylomicrobium alcaliphilum. Furthermore analysis of the predicted AbgT protein sequence using the SwissProt database suggested a protein that potentially possessed hydrolytic activity (). The protein showed modest homology to an esterase from Burkholderia gladioli and to the YfeW protein from E. coli and Salmonella species and appears to be similar to type C β-lactamases, which share sequence characteristics with members of the group VIII family of lipases. Recent work in our laboratory has identified a halo-tolerant lipase from the metagenome of a marine sponge which also belongs to this group.Citation13 Members of this group are known to possess a conserved S-X-X-K amino acid motif in their active site, with a conserved Y amino acid downstream, and analysis of the AbgT protein sequence revealed that it too contains such conserved regions () and shows similarity to EstB from Burkholderia gladioli, an esterase which also contains a β-lactamase fold. No significant β-lactamase activity was detected for this protein and yet catalytic activity was shown to be located within the conserved β-lactamase S-X-X-K motif.Citation45,46 We therefore suggest that the AbgT protein may produce hydrolytic activity mediated around the S-X-X-K catalytic site. Furthermore, a protein secreted by the accessory glands of the female sand-fly Phlebotomus papatasi has been shown to have both lipase-like activity and antibacterial activity against both Gram-positive and Gram-negative bacteria.Citation47
Table 2. Protein similarity searches for AbgT amino acid sequence
While the exact mechanism by which AbgT produces the observed effects remains to be elucidated it is tempting to speculate that hydrolytic activity may lead to the production of free fatty acids with antibacterial properties. Free fatty acids can be released from lipids by the hydrolytic action of enzymes and have been shown to possess a diverse range of biological activities, including antibacterial properties.Citation48 Indeed, free fatty acids are thought to play an important role in the innate immune system, defending against potential pathogens particularly in the skin and mucous membranes.Citation49 The exact mechanism of the antimicrobial action of free fatty acids remains unclear; however, the main target appears to be the cell membrane. There has been a suggestion that free fatty acids may disrupt the electron transport chain or interfere with oxidative phosphorylation, or may lead to increased fluidity of the membrane which can ultimately lead to instability and cell lysis.Citation48 Moreover in a recent study it has been shown that specific fatty acids can inhibit the growth of Staphylococcus aureus by disrupting the cytoplasmic membrane which allows metabolites and low molecular weight proteins to leak from the cell.Citation50 Thus the AbgT protein may act by releasing fatty acids that lead to disruption of the cell membrane in B. subtilis DSM10, resulting in cell death. Another alternative is that the AbgT protein may function in a similar manner to a putative lipase which the Brady group have recently isolated following the functional screening of a Ralstonia metallidurans hosted soil metagenomic library.Citation37 They propose that the anti B. subtilis activity displayed by this putative lipase may result from its ability to cleave ester bonds present in the bacterial cell wall.
While the generation of antibacterial free fatty acids is a plausible explanation for the inhibition of the growth of B. subtilis DSM10 it should also be noted that a recent study discovered 2 novel esterases from a soil metagenome that were capable of reactivating the antibiotic activity of chloramphenicol from its acetylated derivatives formed by the action of chloramphenicol acetyl tranferases.Citation51 As chloramphenicol was present in the media and the B. subtilis DSM10 strain was sensitive to this antibiotic, the AbgT protein may produce a similar effect. However there was no sequence similarity between AbgT and the 2 esterases identified in the above study and in addition, these esterases were members of the group IV family of lipases rather than the VIII family.
In summary, the data suggests that the AbgT protein is involved in growth inhibition of B. subtilis DSM10 and it seems likely that the effects may be mediated by hydrolytic activity. These results suggest not only that functional metagenomic based approaches can provide a significant resource for the discovery of novel functional proteins but that hydrolytic enzymes involved in the release of free fatty acids may be a target for the development of new biomedical therapies.
Materials and Methods
Sampling
Suppressive soil samples were collected from an organic field trial site at the Teagasc Oak Park research facility (Co. Carlow, Ireland; http://www.agresearch.teagasc.ie/oakpark/) that had been in yearly crop rotation for 7 y and had only been fertilized with organic farmyard manure and compost. Samples were taken from a triticale field shortly after harvest. Bulk samples were taken from the surface to a depth of approx. Ten cm using sterile, DNA-free tools (treated with 5% sodium hypochlorite for 30 min prior to washing and autoclaving) and stored in sterile, DNA-free plastic containers. Samples were transported to the laboratory on ice, aseptically fractionated and stored at 4°C until further processing.
Extraction and purification of High Molecular Weight DNA
Metagenomic DNA was extracted from the soil as previously describedCitation52 with the following modifications. Soil was sieved through one stainless steel mesh (mesh size 4 mm). Fifteen grams of freshly sieved soil was mixed with 20 ml preheated (70°C) lysis buffer in 50 ml tubes and incubated at 70°C for 2 hours with thorough mixing by hand every 30 min. After cooling for 30 min at room temperature, the tubes were centrifuged at 4000 × g for 20 min at 4°C and the supernatant was transferred to a fresh tube. The soil pellet was re-suspended in fresh preheated lysis buffer (20 ml) and the extraction procedure was repeated. Supernatants from both extractions were pooled and DNA was precipitated with 0.7 vol isopropanol at −20°C overnight. Precipitated DNA was pelleted by centrifugation at 5000 × g for 60 min at 4°C and the pellet was washed once with 10 ml of 70% ethanol and air-dried. One ml of TE buffer (10 mM Tris-HCl, 1 mM EDTA, pH8) was added to the pellet and the high-molecular weight (HMW) DNA was dissolved slowly by incubation at 4°C for several hours with gently regular agitation by hand. The crude HMW-DNA was purified and size-separated by pulsed-field gel electrophoresis (PFGE; at 6 V / cm, 1–25 switch time, 120° angle, 17 hrs, 14°C) in 0.5x TBE (45 mM Tris-HCl, 45 mM boric acid, 1 mM EDTA, pH 8.3) on a 1% PFGE grade agarose gel (Bio-Rad Laboratories). For size comparison, each gel contained a PFGE DNA Molecular marker (MidRange II PFG Marker, New England Biolabs). The marker lane and approx. 0.5 cm of the sample DNA lane were cut off and stained for 30 min in 1x SYBR Safe dye (Invitrogen) for visualization. Two gel-slices (approx. One cm) containing unstained DNA of about 30–50 kb and 50–70 kb were cut out and the DNA was electro-eluted out of the gel and concentrated as described by BradyCitation52 with the following modifications: Electro-elution was performed for 3 h with replacement of running buffer after 90 min; DNA was concentrated to a final volume of approx. 100 μl with VivaSpin 6 (MWCO 50000) concentrators (Sartorius). Washed and concentrated DNA was dislodged from the membranes by placing the filter upside-down in a clean 50 ml tube and centrifuging for 5 min at 5000 × g. Size and purity was checked by PFGE with the conditions stated above. DNA concentration was determined using the NanoDrop ND-1000 spectrophotometer.
Construction of metagenomic fosmid library
HMW DNA was end-repaired and metagenomic libraries were constructed using the CopyControl™ Fosmid Library Production Kit pCC1FOS (Epicentre Biotechnologies), according to the manufacturer's instructions. Titers were calculated and approximately 4000–5000 E. coli EPI300™ T1 fosmid clones were plated onto 20 × 20 cm Luria-Bertani (LB) agar supplemented with 12.5 μg/ml chloramphenicol. The clones were picked with the Genetix Qpix2 XT robotic system (Molecular Devices) into 384-well plates containing LB media with the following supplements: 6.3 g/l K2HPO4, 1.8g/l KH2PO4, 0.5 g/l sodium citrate dihydrate, 0.09 g/l MgSO4.7H2O, 6% glycerol. After incubation at 37°C for 18 - 20 hrs, the libraries were stored at -80°C. Twelve clones from the HMW DNA library were randomly picked for determination of average insert size. Isolated fosmids were digested with NotI for 3–4 hr and analyzed by Pulsed Field Gel Electrophoresis with the following conditions: 1% agarose in 0.5% TBE, 6 V/cm, 1–25 switch time, 120° angle, 11.5 hrs, 14°C. Induction of the fosmid to multiple copies was performed using 10% arabinose (filter sterilized). Fosmid DNA was extracted using the GeneJET™ Plasmid Miniprep kit (Fermentas) and DNA was eluted in 30 μl of water. To ensure efficient library production the fosmid control DNA supplied with the kit was also subjected to the same reaction conditions as the isolated HMW DNA. E. coli EPI300™ T1 clones containing pCC1FOS with cloned fosmid control DNA were subsequently used as a control when screening the soil metagenomic libraries for antimicrobial activities.
Screening of fosmid libraries for antimicrobial activities
Fosmid clones were replicated into fresh 384-well plates containing LB medium, supplemented with 12.5 μg/ml chloramphenicol, and incubated at 37°C overnight. The clones were arrayed on 20 × 20 cm agar plates containing LB medium, 12.5 μg/ml chloramphenicol and 0.01% arabinose using the QPix2 XT robotic system. Plates were incubated at 37°C overnight, followed by further incubation at 25°C for 3–5 d after which the clones were exposed to UV light for 1 min and then overlaid with soft (0.5%) agar containing the following test strains: Pseudomonas aeruginosa PA01 and Bacillus subtilis DSM10. P. aeruginosa PA01 and B. subtilis DSM10 were grown to OD600 of approx. One.0 and 1.5 respectively and diluted 1:50 for B. subtilis DSM10 and 1:100 for P. aeruginosa PA01 in soft LB agar before being carefully poured over the metagenomic clones. The plates were incubated overnight and examined for zones of growth inhibition.
Sequencing and annotation of fosmid
Fosmids were sequenced by Roche 454 pyrosequencing. Sequencing and assembly were carried out by the University of Liverpool, Center for Genomic Research. Contigs from the assembly were analyzed for putative open reading frames (ORFs) using the FGENESB-Bacterial Operon and Gene Prediction Program (www.softberry.com) and the MetaGene program.Citation53 The Basic Local Alignment Search Tool (BLAST) at NCBI (http://www.ncbi.nlm.nih.gov/BLAST) was used to search DNA and protein databases for homologous DNA and protein sequences.Citation54 Potential ORFs were analyzed using blastP searches against the non-redundant protein sequences database and the SwissProt database. Conserved functional or structural protein domains were identified using the Conserved Domain Database at the NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml).Citation55 Sequences upstream of predicted start codons were examined for the presence of bacterial promoters using the BPROM-Prediction of Bacterial Promoters Program (www.softberry.com). The theoretical parameters of putative proteins were calculated using the ExPASy ProtParam tool.Citation56 Protein sequences were also examined for signal peptides using the SignalP 4.0 server.Citation57 Multiple sequence alignments and phylogenetic analyses were conducted using MEGA version 4.1 software.Citation58 Evolutionary trees were constructed using the Neighbor-Joining method with a bootstrap test of 1000 replicates. Attempts were made to predict the potential phylogenetic origin of the fosmid DNA using MEGAN.Citation59
Transposon mutagenesis of fosmids
Fosmids were subjected to mutagenesis using the EZ-Tn5TM<TET-1> Insertion kit (Epicentre Biotechnologies), according to the manufacturer's instructions. The mutant mini-library was plated on LB medium containing 12.5 μg/ml chloramphenicol and 10 μg/ml tetracycline. The clones were handpicked into liquid media in 96-well plates, incubated overnight and subsequently replicated onto solid LB medium containing 12.5 μg/ml chloramphenicol and 0.01% arabinose to induce the fosmids to multiple copies. The plates were then incubated overnight at 37°C, followed by 2 further overnight incubations at 25°C and overlaid with the B. subtilis DSM10 as described above. Mutants that appeared to have lost the ability to inhibit growth were selected for further analysis. The insertion site of the EZ-Tn5 transposon was mapped using bidirectional sequencing with primers TET-1 FP-1 (5′ GGG TGC GCA TGA TCC TCT AGA GT 3′) and TET-1 RP-1 (5′ TAA ATT GCA CTG AAA TCT AGA AAT A 3′).
Detection of lipolytic and β-lactamase activities
Lipolytic activity was detected by plating the clones on LB medium containing 1% tributyrin (Sigma-Aldrich). The plates were incubated at 37°C overnight, followed by further incubation at 25°C for up to 7 d during which the plates were examined regularly for zones of clearing. The chromogenic β-lactamase substrate nitrocefin (Calbiochem) was used to detect β-lactamase activity. Briefly, a 1 mM working solution of nitrocefin was prepared according to the manufacturer's instructions. One drop of the nitrocefin solution was placed directly on the surface of a colony and the development of a red color was used as an indication of a positive result. In addition the colonies were also tested for activity by emulsifying the colony into one drop of the nitrocefin solution on the surface of a glass slide.
Sub-cloning of the abgT gene and complementation of the transposon mutant
The abgT gene was amplified from the fosmid TO-T 020 P12 using the primers AB-BL F Pag (5′ TAT TAC TCA TGA CCG CTG CGA ACT CAC TG 3′) and AB-BL R IF Hind (5′ ATC ATT AAG CTT TCT TCC GTG GCG GAC 3′). Restriction sites were mis-primed (underlined above) into the 5′ end of the primers to allow insertion of the gene into the expression vector pBadMyc-HisA (Invitrogen). The gene was inserted under the control of the arabinose inducible PBAD promoter and the stop codon of the open reading frame was removed to allow a C-terminal fusion with the poly-histidine region of the vector. PCR was performed using Pfu DNA polymerase and the PCR product (∼1.3 kb) was purified from a 1% agarose gel and subsequently digested using the restriction enzymes PagI and HindIII at 37°C for 90 min. The pBadMyc-HisA vector was digested using NcoI and HindIII using similar conditions. The digested products were purified and ligated overnight at 16°C with T4 DNA ligase (Fermentas). E. coli EPI300™ T1 cells were transformed with the ligation mixture and plated on LB medium containing 100 μg/ml carbenicillin. The presence of the insert was confirmed by colony PCR and restriction digestion. Transposon mutant clones were subsequently transformed with the purified recombinant vector.
Purification of His-tagged AbgT protein
E. coli EPI300™ T1 cells containing the recombinant pBadMyc-HisA vector were grown in LB medium containing 100 μg/ml carbenicillin until mid-log phase and then induced with 0.01% arabinose for 4 h. The cells were harvested by centrifugation, re-suspended in lysis buffer (50 mM of NaH2PO4, 300 mM of NaCl, 10 mM imidazole, pH 8.0) and incubated on ice for 20 minutes. Cells were then disrupted by sonication in ice with 3 5 second pulses at high intensity and the supernatant was collected after centrifugation. The His-tagged protein was purified by Ni-NTA spin column with varying concentration of imidazole (20- 250 mM) used in the elution buffer to maximise the amount of purified product in a single elute. The purified protein was dialyzed and lyophilized before being resolved by SDS–PAGE.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
1018493_Supplemental_Material.docx
Download MS Word (59.5 KB)Funding
This work was supported in part by grants awarded by the Irish Department of Agriculture, Fisheries and Food (FIRM/RSF/CoFoRD; FIRM 08/RDC/629); the European Commission (FP7-PEOPLE-2013-ITN, 607786) and the Marine Institute (Beaufort award).
Supplemental Material
Supplemental data for this article can be accessed on the publisher's website.
References
- Amann RI, Ludwig W, Schleifer KH. Phylogenetic identification and in situ detection of individual microbial cells without cultivation. Microbiol Rev 1995; 59: 143-69; PMID:7535888.
- Beja O, Koonin EV, Aravind L, Taylor LT, Seitz H, Stein JL, Bensen DC, Feldman RA, Swanson RV, DeLong EF. Comparative genomic analysis of archaeal genotypic variants in a single population and in two different oceanic provinces. Appl Environ Microbiol 2002; 68: 335-45; PMID:11772643; http://dx.doi.org/10.1128/AEM.68.1.335-345.2002
- Hallam SJ, Putnam N, Preston CM, Detter JC, Rokhsar D, Richardson PM, DeLong EF. Reverse methanogenesis: testing the hypothesis with environmental genomics. Science 2004; 305: 1457-62; PMID:15353801; http://dx.doi.org/10.1126/science.1100025
- Rondon MR, August PR, Bettermann AD, Brady SF, Grossman TH, Liles MR, Loiacono KA, Lynch BA MacNeil IA, Minor C, et al. Cloning the soil metagenome: a strategy for accessing the genetic and functional diversity of uncultured microorganisms. Appl Environ Microbiol 2000; 66: 2541-7; PMID:10831436; http://dx.doi.org/10.1128/AEM.66.6.2541-2547.2000
- Treusch AH, Kletzin A, Raddatz G, Ochsenreiter T, Quaiser A, Meurer G, Schuster SC, Schleper C. Characterization of large-insert DNA libraries from soil for environmental genomic studies of archaea. Environ Microbiol 2004; 6: 970-80; PMID:15305922; http://dx.doi.org/10.1111/j.1462-2920.2004.00663.x
- Tyson GW, Chapman J, Hugenholtz P, Allen EE, Ram RJ, Richardson PM, Solovyev VV, Rubin EM, Rokhsar DS, Banfield JF. Community structure and metabolism through reconstruction of microbial genomes from the environment. Nature 2004; 428: 37-43; PMID:14961025; http://dx.doi.org/10.1038/nature02340
- Venter JC, Remington K, Heidelberg JF, Halpern AL, Rusch D, Eisen JA, Wu D, Paulsen I, Nelson KE, Nelson W, et al. Environmental genome shotgun sequencing of the sargasso sea. Science 2004; 304: 66-74; PMID:15001713; http://dx.doi.org/10.1126/science.1093857
- Cowan D, Meyer Q, Stafford W, Muyanga S, Cameron R, Wittwer P. Metagenomic gene discovery: past, present and future. Trends Biotechnol 2005; 23: 321-9; PMID:15922085; http://dx.doi.org/10.1016/j.tibtech.2005.04.001
- Kennedy J, Marchesi JR, Dobson AD. Metagenomic approaches to exploit the biotechnological potential of the microbial consortia of marine sponges. Appl Microbiol Biotechnol 2007; 75: 11-20; PMID:17318533; http://dx.doi.org/10.1007/s00253-007-0875-2
- Kim YJ, Choi GS, Kim SB, Yoon GS, Kim YS, Ryu YW. Screening and characterization of a novel esterase from a metagenomic library. Protein Expr Purif 2006; 45: 315-23; PMID:16061395; http://dx.doi.org/10.1016/j.pep.2005.06.008
- Lorenz P, Eck J. Metagenomics and industrial applications. Nat Rev Microbiol 2005; 3: 510-6; PMID:15931168; http://dx.doi.org/10.1038/nrmicro1161
- Lorenz P, Liebeton K, Niehaus F, Eck J. Screening for novel enzymes for biocatalytic processes: accessing the metagenome as a resource of novel functional sequence space. Curr Opin Biotechnol 2002; 13: 572-7; PMID:12482516; http://dx.doi.org/10.1016/S0958-1669(02)00345-2
- Selvin J, Kennedy J, Lejon DP, Kiran S, Dobson AD. Isolation identification and biochemical characterization of a novel halo-tolerant lipase from the metagenome of the marine sponge haliclona simulans. Microb Cell Fact 2012; 11: 72; PMID:22657530; http://dx.doi.org/10.1186/1475-2859-11-72
- Chang FY, Brady SF. Cloning and characterization of an environmental DNA-derived gene cluster that encodes the biosynthesis of the antitumor substance BE-54017. J Am Chem Soc 2011; 133: 9996-9; PMID:21542592; http://dx.doi.org/10.1021/ja2022653
- Iqbal HA, Feng Z, Brady SF. Biocatalysts and their small molecule products from metagenomic studies. Curr Opin Chem Biol 2012; 16: 109-16; PMID:22455793; http://dx.doi.org/10.1016/j.cbpa.2012.02.015
- Felczykowska A, Dydecka A, Bohdanowicz M, Gasior T, Sobon M, Kobas J, Bloch S, Nejman-Falenczyk B, Wegrzyn G. The use of fosmid metagenomic libraries in preliminary screening for various biological activities. Micro Cell Factories 2014; 13:105; PMID:25048369; http://dx.doi.org/10.1186/s12934-014-0105-4
- Daniel R. The soil metagenome - a rich resource for the discovery of novel natural products. Curr Opin Biotechnol 2004; 15: 199-204; PMID:15193327; http://dx.doi.org/10.1016/j.copbio.2004.04.005
- Craig JW, Chang FY, Kim JH, Obiajulu SC, Brady SF. Expanding small-molecule functional metagenomics through parallel screening of broad-host-range cosmid environmental DNA libraries in diverse proteobacteria. Appl Environ Microbiol 2010; 76: 1633-41; PMID:20081001; http://dx.doi.org/10.1128/AEM.02169-09
- Daniel R. The metagenomics of soil. Nat Rev Microbiol 2005; 3: 470-8; PMID:15931165; http://dx.doi.org/10.1038/nrmicro1160
- Handelsman J, Rondon MR, Brady SF, Clardy J, Goodman RM. Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem Biol 1998; 5: R245-249; PMID:9818143; http://dx.doi.org/10.1016/S1074-5521(98)90108-9
- Chung EJ, Lim HK, Kim JC, Choi GJ, Park EJ, Lee MH, Chung YR, Lee SW. Forest soil metagenome gene cluster involved in antifungal activity expression in escherichia coli. Appl Environ Microbiol 2008; 74: 723-30; PMID:18065615; http://dx.doi.org/10.1128/AEM.01911-07
- Kim SJ, Lee CM, Han BR, Kim MY, Yeo YS, Yoon SH, Koo BS, Jun HK. Characterization of a gene encoding cellulase from uncultured soil bacteria. FEMS Microbiol Lett 2008; 282: 44-51; PMID:18355282; http://dx.doi.org/10.1111/j.1574-6968.2008.01097.x
- van Elsas JD, Speksnijder AJ, van Overbeek LS. A procedure for the metagenomics exploration of disease-suppressive soils. J Microbiol Methods 2008; 75: 515-22; PMID:18778739; http://dx.doi.org/10.1016/j.mimet.2008.08.004
- Steinberg C, Edel-Hermann V, Alabouvette C, Lemanceau P. Soil suppressiveness to plant disease. In: van Elsas JD, Jansson JK, Trevors JT, editors. Modern Soil Microbiology. Boca Raton: Taylor & Francis; 2006; 455-78.
- van Elsas JD, Costa R, Jansson J, Sjoling S, Bailey M, Nalin R, Vogel TM, van Overbeek L. The metagenomics of disease-suppressive soils - experiences from the METACONTROL project. Trends Biotechnol 2008; 26: 591-601; PMID:18774191; http://dx.doi.org/10.1016/j.tibtech.2008.07.004
- Mendes R, Kruijt M, de Bruijn I, Dekkers E, van der Voort M, Schneider JH, Piceno YM, DeSantis TZ, Andersen GL, Bakker PA et al. Deciphering the rhizosphere microbiome for disease-suppressive bacteria. Science 2011; 332: 1097-100; PMID:21551032; http://dx.doi.org/10.1126/science.1203980
- Reddy BV, Kallifidas D, Kim JH, Charlop-Powers Z, Feng Z, Brady SF. Natural product biosynthetic gene diversity in geographically distinct soil microbiomes. Appl Environ Microbiol 2012; 78: 3744-52; PMID:22427492; http://dx.doi.org/10.1128/AEM.00102-12
- Lim HK, Chung EJ, Kim JC, Choi GJ, Jang KS, Chung YR, Cho KY, Lee SW. Characterization of a forest soil metagenome clone that confers indirubin and indigo production on escherichia coli. Appl Environ Microbiol 2005; 71: 7768-77; PMID:16332749; http://dx.doi.org/10.1128/AEM.71.12.7768-7777.2005
- Guan C, Ju J, Borlee BR, Williamson LL, Shen B, Raffa KF, Handelsman J. Signal mimics derived from a metagenomic analysis of the gypsy moth gut microbiota. Appl Environ Microbiol 2007; 73: 3669-76; PMID:17435000; http://dx.doi.org/10.1128/AEM.02617-06
- Gillespie DE, Brady SF, Bettermann AD, Cianciotto NP, Liles MR, Rondon MR, Clardy J, Goodman RM, Handelsman J. Isolation of antibiotics turbomycin A and B from a metagenomic library of soil microbial DNA. Appl Environ Microbiol 2002; 68: 4301-6; PMID:12200279; http://dx.doi.org/10.1128/AEM.68.9.4301-4306.2002
- Brady SF, Chao CJ, Handelsman J, Clardy J. Cloning and heterologous expression of a natural product biosynthetic gene cluster from eDNA. Org Lett 2001; 3: 1981-4; PMID:11418029; http://dx.doi.org/10.1021/ol015949k
- Brady SF, Clardy J. Palmitoylputrescine, an antibiotic isolated from the heterologous expression of DNA extracted from bromeliad tank water. J Nat Prod 2004; 67: 1283-6; PMID:15332842; http://dx.doi.org/10.1021/np0499766
- Wang GY, Graziani E, Waters B, Pan W, Li X, McDermott J, Meurer G, Saxena G, Andersen RJ, Davies J. Novel natural products from soil DNA libraries in a streptomycete host. Org Lett 2000; 2: 2401-4; PMID:10956506; http://dx.doi.org/10.1021/ol005860z
- Kallifidas D, Kang HS, Brady SF. Tetarimycin A, an MRSA-active antibiotic identified through induced expression of environmental DNA gene clusters. J Am Chem Soc 2012; 134: 19552-5; PMID:23157252; http://dx.doi.org/10.1021/ja3093828
- King RW, Bauer JD, Brady SF. An environmental DNA-derived type II polyketide biosynthetic pathway encodes the biosynthesis of the pentacyclic polyketide erdacin. Angew Chem Int Ed Engl 2009; 48: 6257-61; PMID:19621341; http://dx.doi.org/10.1002/anie.200901209
- Feng Z, Kim JH, Brady SF. Fluostatins produced by the heterologous expression of a TAR reassembled environmental DNA derived type II PKS gene cluster. J Am Chem Soc 2010; 132: 11902-3; PMID:20690632; http://dx.doi.org/10.1021/ja104550p
- Iqbal HA, Craig JW, Brady SF. Antibacterial enzymes from the functional screening of metagenomic libraries hosted in ralstonia metallidurans. FEMS Microbiol Letts 2014; 354: 19-26; PMID:24661178; http://dx.doi.org/10.1111/1574-6968.12431
- Chang FY, Brady SF. Discovery of indolotryptoline antiproliferative agents by homology-guided metagenomic screening. Proc Natl Acad Sci USA 2013; 110: 2473-83; PMID:23302687.
- Feng Z, Chakraborty D, Dewell SB, Reddy BV, Brady SF. Environmental DNA encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. Am Chem Soc 2012; 134: 2981-7; PMID:22224500; http://dx.doi.org/10.1021/ja207662w
- Kant R, van Passel MW, Sangwan P, Palva A, Lucas S, Copeland A, Lapidus A, Glavina del Rio T, Dalin E, Tice H, et al. Genome sequence of "pedosphaera parvula" Ellin514, an aerobic verrucomicrobial isolate from pasture soil. J Bacteriol 2011; 193: 2900-1; PMID:21460084; http://dx.doi.org/10.1128/JB.00299-11
- Arpigny JL, Jaeger KE. Bacterial lipolytic enzymes: classification and properties. Biochem J 1999; 343: 177-83; PMID:10493927; http://dx.doi.org/10.1042/0264-6021:3430177
- Yu EY, Kwon MA, Lee M, Oh JY, Choi JE, Lee JY, Song BK, Hahm DH, Song JK. Isolation and characterization of cold-active family VIII esterases from an arctic soil metagenome. Appl Microbiol Biotechnol 2011; 90: 573-81; PMID:21318360; http://dx.doi.org/10.1007/s00253-011-3132-7
- Levett PN, Morey RE, Galloway R, Steigerwalt AG, Ellis WA. Reclassification of leptospira parva hovind-hougen et al. 1982 as turneriella parva gen. nov., comb. nov. Int J Syst Evol Microbiol 2005; 55: 1497-9; PMID:16014471; http://dx.doi.org/10.1099/ijs.0.63088-0
- Yung PY, Burke C, Lewis M, Kjelleberg S, Thomas T. Novel antibacterial proteins from the microbial communities associated with the sponge cymbastela concentrica and the green alga ulva australis. Appl Environ Microbiol 2011; 77: 1512-5; PMID:21183639; http://dx.doi.org/10.1128/AEM.02038-10
- Petersen EI, Valinger G, Solkner B, Stubenrauch G, Schwab H. A novel esterase from burkholderia gladioli which shows high deacetylation activity on cephalosporins is related to β-lactamases and DD-peptidases. J Biotechnol 2001; 89: 11-25; PMID:11472796; http://dx.doi.org/10.1016/S0168-1656(01)00284-X
- Wagner UG, Petersen EI, Schwab H, Kratky C. EstB from burkholderia gladioli: a novel esterase with a β-lactamase fold reveals steric factors to discriminate between esterolytic and β-lactam cleaving activity. Protein Sci 2002; 11: 467-78; PMID:11847270; http://dx.doi.org/10.1110/ps.33002
- Belardinelli M, Fausto AM, Guerra L, Buonocore F, Bongiorno G, Maroli M, Mazzini M. Lipase and antibacterial activities of a recombinant protein from the accessory glands of female phlebotomus papatasi (diptera: psychodidae). Ann Trop Med Parasitol 2005; 99: 673-82; PMID:16212801; http://dx.doi.org/10.1179/136485905X51472
- Desbois AP, Smith VJ. Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl Microbiol Biotechnol 2010; 85: 1629-42; PMID:19956944; http://dx.doi.org/10.1007/s00253-009-2355-3
- Drake DR, Brogden KA, Dawson DV, Wertz PW. Thematic review series: skin lipids. antimicrobial lipids at the skin surface. J Lipid Res 2008; 49: 4-11; PMID:17906220; http://dx.doi.org/10.1194/jlr.R700016-JLR200
- Parsons JB, Yao J, Frank MW, Jackson P, Rock CO. Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from staphylococcus aureus. J Bacteriol 2012; 194: 5294-304; PMID:22843840; http://dx.doi.org/10.1128/JB.00743-12
- Tao W, Lee MH, Yoon MY, Kim JC, Malhotra S, Wu J, Hwang EC, Lee SW. Characterization of two metagenome-derived esterases that reactivate chloramphenicol by counteracting chloramphenicol acetyltransferase. J Microbiol Biotechnol 2011; 21: 1203-10; PMID:22210605; http://dx.doi.org/10.4014/jmb.1107.07034
- Brady SF. Construction of soil environmental DNA cosmid libraries and screening for clones that produce biologically active small molecules. Nat Protoc 2007; 2: 1297-305; PMID:17546026; http://dx.doi.org/10.1038/nprot.2007.195
- Noguchi H, Park J, Takagi T. MetaGene: prokaryotic gene finding from environmental genome shotgun sequences. Nucleic Acids Res 2006; 34: 5623-30; PMID:17028096; http://dx.doi.org/10.1093/nar/gkl723
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol 1990; 215 403-10; PMID:2231712; http://dx.doi.org/10.1016/S0022-2836(05)80360-2
- Marchler-Bauer A, Lu S, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, et al. CDD: a conserved domain database for the functional annotation of proteins. Nucleic Acids Res 2011; 39: D225-9; PMID:21109532; http://dx.doi.org/10.1093/nar/gkq1189
- Gasteiger E, Hoogland C, Gattiker A, Duvaud S, Wilkins MR, Appel RD, Bairoch A. Protein identification and analysis tools on the ExPasy server. In: Walker JM, editor. The Proteomics Protocols Handbook. New York: Taylor & Francis; 2005; 571-607.
- Petersen TN, Brunak S, von Heijne G, Nielsen H. SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat Methods 2011; 8: 785-6; PMID:21959131; http://dx.doi.org/10.1038/nmeth.1701
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol Biol Evol 2007; 24: 1596-9; PMID:17488738; http://dx.doi.org/10.1093/molbev/msm092
- Huson DH, Auch AF, Qi J, Schuster SC. MEGAN analysis of metagenomic data. Genome Res 2007; 17: 377-86; PMID:17255551; http://dx.doi.org/10.1101/gr.5969107