ABSTRACT
Lysin motif (LysM) is a highly conserved carbohydrate binding module that is widely present in proteins from both prokaryotes and eukaryotes. LysM domains from many LysM-containing proteins can be taken out of their natural context and retain their ability to bind peptidoglycan. Therefore, LysM has enormous potential for applications in both industry and medicine. This potential has stimulated an intensive search for LysM modules with different evolutionary origins. The p60 protein (Lm-p60) is an NlpC/P60-containing peptidoglycan hydrolase secreted by Listeria monocytogenes. The N-terminus of Lm-p60 contains 2 LysM modules separated by an SH3 module. Our recent study of Lm-p60 demonstrates that the N-terminal half of Lm-p60, comprised of 2 LysM and 1 SH3 module, is able to recognize and bind peptidoglycan. The LysM domain of Lm-p60 contains only 2 LysM modules, which is the minimum number of LysM modules in most NlpC/P60-containing proteins, but it shows strong affinity for peptidoglycan. Moreover, these 2 LysM modules have only 38.64% similarity to each other. These data allowed us to conclude that the 2 LysM modules from Lm-p60 have different evolutionary origins, suggesting that they are suitable candidate peptidoglycan-binding modules for protein engineering in order to create a protein with a high binding affinity to peptidoglycan.
What is Listeria monocytogenes p60 protein?
Listeria monocytogenes p60 protein (Lm-p60) is a major extracellular protein present in all L. monocytogenes isolates. It was first identified by Kuhn and Goebel (1989).Citation1 Lm-p60 was named because of its apparent molecular weight after SDS-PAGE electrophoresis. Previous immunoblot analysis of culture supernatants of 6 Listeria species with polyclonal antibodies against Lm-p60 indicated the presence of the Lm-p60-related proteins in all of the tested culture supernatants, demonstrating that Lm-p60 is an important extracellular protein for all 6 Listeria species.Citation2 In early studies of L. monocytogenes virulence factors, Lm-p60 was suggested to be involved in the invasion of L. monocytogenes and defined as an invasion-associated protein (Iap). Therefore, the Lm-p60-encoding gene was designated as iap.Citation1 Several years later, Lm-p60 was shown to be an essential housekeeping protein for all Listeria species and shown to possess murein hydrolase activity required for cell division.Citation3 The ability of Lm-p60 to hydrolyze the bacterial cell wall has been verified by many studies.Citation4-6
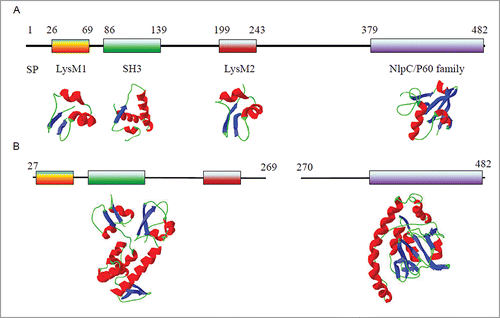
BLAST analysis of the Lm-p60 sequence has shown that Lm-p60 is highly similar to LytF, which is found in Bacillus subtilis and has been identified as a γ-D-glutamate-meso-diaminopimelatemuropeptidase.Citation7 Therefore, Lm-p60 is predicted to hydrolyze the amide bond (or isopeptide bond) between the γ-carboxylic group of the D-glutamine moiety and the ϵ-amino group of the meso-2,6-diaminopimelic acid moiety of the stem peptide in L. monocytogenes murein (peptidoglycan).Citation4 The peptidoglycan cleavage site of Lm-p60 is different from that of lysozyme (). A conserved domain search indicated that Lm-p60 contains 2 LysM modules, one SH3 module and one NlpC/P60 family module. The domain organization of Lm-p60 is shown in . Two LysM modules with an intervening SH3 module are located in the N-terminal half of Lm-p60, while the NlpC/P60 family domain is located in the C-terminus of Lm-p60. The NlpC/P60 domain is found in a very large family of cell wall endopeptidases that widely exist in viruses, bacteria, archaea and eukaryotes.Citation8,9 The biochemical function of NlpC/P60-containing proteins is highly conserved, but the physiological roles of these proteins are diverse. Almost all NlpC/P60-containing enzymes that have been characterized are γ-D-glutamate-meso-diaminopimelate endopeptidases.Citation10,11 Between the LysM2 module and the NlpC/P60 family module lies a low complexity amino acid sequence; the low complexity suggests that this sequence is a linker and that Lm-p60 could be split into 2 structural domains in this linker region. Our experimental evidence verified that splitting the 2 structural domains in the peptide between amino acid residues 269 and 270 allowed the domains to function independently.Citation12,13 The N-terminal half of Lm-p60 () functions as the substrate-binding domain and can specifically recognize and bind the bacterial cell wall. The C-terminal half, which contains only the NlpC/P60 family domain, functions as the catalytic domain. When the cell wall hydrolytic activity of this individually expressed domain was detected using a renaturing SDS-PAGE procedure, the C-terminal domain displayed weak lytic activity.
Figure 1. Structure of L. monocytogenes murein, showing the predicted cleavage sites for Lm-p60 and the cleavage sites for lysozyme.
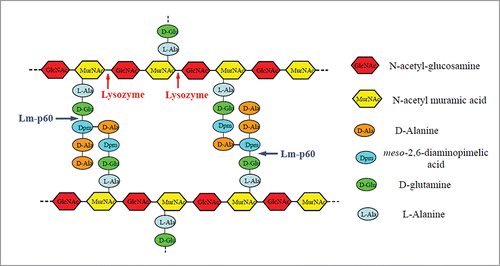
Figure 2. The domain organization of Lm-p60 and the predicted structures of Lm-p60 domains. (A) Schematic representation of the domain organization of Lm-p60 and the predicted structures of the corresponding domains. SP, signal peptide. (B) Two truncated terminal domains characterized in the literature.Citation13
The potential applications of LysM in industry and medicine
The Lysin motif (LysM) is a widespread protein module. It was first identified in the bacterial lysin of the Bacillus phage.Citation14 Currently, LysM is found in many proteins that can bind peptidoglycan, chitin or lipooligosaccharides. A search of the recently updated Pfam database showed that the LysM-containing protein family (PF01476) contains 11733 protein sequences from 1760 species of viral, eukaryotic and prokaryotic origin (http://pfam.xfam.org/family/PF01476#tabview=tab0). The sequence length of a single LysM typically varies from 44 to 65 amino acid residues. Multiple copies (up to 12) of LysM are frequently found in one protein. Multiple LysMs within 1 protein are often separated by intervening sequences and are mainly present in one terminus of the protein. LysM has a highly conserved βααβ structure in which 2 β strands form an anti-parallel β sheet and then 2 α helices are folded onto the same side of the β sheet ().
LysM is a highly conserved carbohydrate binding module that specifically binds N-acetylglucosamine. Carbohydrate recognition is very important for many essential biological processes, including bacterial cell wall degradation,Citation9,15 bacterial and viral infection response,Citation16 pathogen defense,Citation17,18 immune response modulationCitation19,20 and signaling.Citation21-24 Therefore, LysM domains can be found almost in all living organisms (except archaea). Since the identification of LysM, the functions and structures of LysM domains originating from different LysM-containing proteins have been intensively studied. Recently, many studies of LysM domains have focused on their possible applications in medicine and industry, which include cell immobilization, bacterial cell surface protein display, microbe detection, fusion to proteins for protein purification, mediation of symbiosis and novel vaccine development. For example, LysM domains have been used to display various heterologous proteins on the bacterial cell surfaceCitation25,26 and to develop pneumococcal and influenza vaccines.Citation27,28
Characterization of LysMs from Lm-p60
Lm-p60 is a modular protein of the NlpC/P60 endopeptidase family containing the catalytic domain of NlpC/P60. NlpC/P60-containing proteins are often found with peptidoglycan-binding domains. SH3 and LysM modules are often found in NlpC/P60 endopeptidases. Most NlpC/P60-containing proteins in the Pfam database have more than 2 LysM modules.Citation24,29,30 For instance, CwlS (cell wall-lytic enzyme associated with cell separation), an NlpC/P60 endopeptidase from Bacillus subtilis, possesses 4 LysM modules,Citation30 and Enterococcus faecalis AtlA, a peptidoglycan hydrolase involved in cell division, contains 6 LysM modules. Furthermore, the 6 LysM modules found in E. faecalis AtlA display 65-100% similarity to each other.Citation24 These studies of the LysM domain have shown that a single LysM module is able to bind short oligosaccharide ligands and that multiple LysM modules act additively to increase the binding affinity of the LysM domain to long glycan chains. Lm-p60 has only 2 LysM modules, which is the minimum number of LysM modules for most NlpC/P60-containing proteins. However, a study of the LysM domains of CwlS demonstrated that 3 LysM modules are necessary for proper binding to long peptidoglycan fragments.Citation30 This suggests that the LysMs from Lm-p60 may have a higher affinity for peptidoglycan than LysMs from other LysM-containing proteins. Furthermore, the 2 LysM modules in Lm-p60 display only 38.64% similarity to each other, and our unpublished data suggest that these 2 LysMs contribute to the binding affinity of Lm-p60 and peptidoglycan differently. All of these data suggest that the evolutionary origins of the 2 LysMs from Lm-p60 differ. Studying LysM modules that may have different evolutionary origins is important for engineering LysM domains with high binding affinity to peptidoglycan. It has been directly shown that the LysMs from Lm-p60 are a good resource for the engineering of proteins with high binding affinity to peptidoglycan.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Funding
This work was funded in part by the National Science Foundation of China (31170073), the Jiangsu Key Laboratory of Zoonosis, the Priority Academic Program Development of Jiangsu Higher Education Institutions (PAPD), and the Natural Science Foundation of Jiangsu Province (Grant No.BK20150445).
References
- Kuhn M, Goebel W. Identification of an extracellular protein of Listeria monocytogenes possibly involved in intracellular uptake by mammalian cells. Infect Immun 1989; 57:55-61; PMID:2491841
- Bubert A, Kuhn M, Goebel W, Kohler S. Structural and functional properties of the p60 proteins from different Listeria species. J Bacteriol 1992; 174:8166-71; PMID:1459966
- Wuenscher MD, Kohler S, Bubert A, Gerike U, Goebel W. The iap gene of Listeria monocytogenes is essential for cell viability, and its gene product, p60, has bacteriolytic activity. J Bacteriol 1993; 175:3491-501; PMID:8099071
- Lenz LL, Mohammadi S, Geissler A, Portnoy DA. SecA2-dependent secretion of autolytic enzymes promotes Listeria monocytogenes pathogenesis. Proc Natl Acad Sci U S A 2003; 100:12432-7; PMID:14527997; http://dx.doi.org/10.1073/pnas.2133653100
- Machata S, Hain T, Rohde M, Chakraborty T. Simultaneous deficiency of both MurA and p60 proteins generates a rough phenotype in Listeria monocytogenes. J Bacteriol 2005; 187:8385-94; PMID:16321943; http://dx.doi.org/10.1128/JB.187.24.8385-8394.2005
- Pilgrim S, Kolb-Maurer A, Gentschev I, Goebel W, Kuhn M. Deletion of the gene encoding p60 in Listeria monocytogenes leads to abnormal cell division and loss of actin-based motility. Infect Immun 2003; 71:3473-84; PMID:12761132; http://dx.doi.org/10.1128/IAI.71.6.3473-3484.2003
- Margot P, Pagni M, Karamata D. Bacillus subtilis 168 gene lytF encodes a γ-D-glutamate-meso-diaminopimelate muropeptidase expressed by the alternative vegetative sigma factor, σD. Microbiol 1999; 145:57-65; PMID:10206711; http://dx.doi.org/10.1099/13500872-145-1-57
- Anantharaman V, Aravind L. Evolutionary history, structural features and biochemical diversity of the NlpC/P60 superfamily of enzymes. Genome Biol 2003; 4:R11; PMID:12620121; http://dx.doi.org/10.1186/gb-2003-4-2-r11
- Bateman A, Rawlings ND. The CHAP domain: a large family of amidases including GSP amidase and peptidoglycan hydrolases. Trends Biochem Sci 2003; 28:234-7; PMID:12765834; http://dx.doi.org/10.1016/S0968-0004(03)00061-6
- Xu Q, Abdubek P, Astakhova T, Axelrod HL, Bakolitsa C, Cai X, Carlton D, Chen C, Chiu HJ, Chiu M, et al. Structure of the γ-D-glutamyl-L-diamino acid endopeptidase YkfC from Bacillus cereus in complex with L-Ala-γ-D-Glu: insights into substrate recognition by NlpC/P60 cysteine peptidases. Acta Crystallogr Sect F Struct Biol Cryst Commun 2010; 66:1354-64; PMID:20944232; http://dx.doi.org/10.1107/S1744309110021214
- Xu Q, Chiu HJ, Farr CL, Jaroszewski L, Knuth MW, Miller MD, Lesley SA, Godzik A, Elsliger MA, Deacon AM, et al. Structures of a bifunctional cell wall hydrolase CwlT containing a novel bacterial lysozyme and an NlpC/P60 DL-endopeptidase. J Mol Biol 2014; 426:169-84; PMID:24051416; http://dx.doi.org/10.1016/j.jmb.2013.09.011
- Guo ML, Gu H, Xu Q, Zuo JR. Study on the domain function of Listeria monocytogenes p60 protein. N Biotechnol 2014; 31:S197-S8.
- Yu M, Zuo J, Gu H, Guo M, Yin Y. Domain function dissection and catalytic properties of Listeria monocytogenes p60 protein with bacteriolytic activity. Appl Microbiol Biotechnol 2015; 99:10527-37; PMID:26363556; http://dx.doi.org/10.1007/s00253-015-6967-5
- Garvey KJ, Saedi MS, Ito J. Nucleotide sequence of Bacillus phage Ø29 genes 14 and 15: homology of gene 15 with other phage lysozymes. Nucleic Acids Res 1986; 14:10001-8; PMID:3027653; http://dx.doi.org/10.1093/nar/14.24.10001
- Birkeland NK. Cloning, molecular characterization, and expression of the genes encoding the lytic functions of lactococcal bacteriophage φLCE: A dual lysis system of modular design. Can J Microbiol 1994; 40:658-65; PMID:7922887; http://dx.doi.org/10.1139/m94-104
- Balzarini J. Large-molecular-weight carbohydrate-binding agents as HIV entry inhibitors targeting glycoprotein gp120. Curr Opin HIV AIDS 2006; 1:355-60; PMID:19372833; http://dx.doi.org/10.1097/01.COH.000-0239846.36076.2c
- Miya A, Albert P, Shinya T, Desaki Y, Ichimura K, Shirasu K, Narusaka Y, Kawakami N, Kaku H, Shibuya N. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci U S A 2007; 104:19613-8; PMID:18042724; http://dx.doi.org/10.1073/pnas.0705147104
- Bolton MD, Van Esse HP, Vossen JH, De Jonge R, Stergiopoulos I, Stulemeijer IJ, van den Berg GC, Borrás-Hidalgo O, Dekker HL, de Koster CG, et al. The novel Cladosporium fulvum lysin motif effector Ecp6 is a virulence factor with orthologues in other fungal species. Mol Microbiol 2008; 69:119-36; PMID:18452583; http://dx.doi.org/10.1111/j.1365-2958.2008.06270.x
- de Jonge R, van Esse HP, Kombrink A, Shinya T, Desaki Y, Bours R, van der Krol S, Shibuya N, Joosten MH, Thomma BP. Conserved fungal LysM effector Ecp6 prevents chitin-triggered immunity in plants. Science 2010; 329:953-5; PMID:20724636; http://dx.doi.org/10.1126/science.1190859
- Rabinovich GA, Toscano MA. Turning'sweet'on immunity: galectin-glycan interactions in immune tolerance and inflammation. Nat Rev Immunol 2009; 9:338-52; PMID:19365409; http://dx.doi.org/10.1038/nri2536
- Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. LysM domain receptor kinases regulating rhizobial Nod factor-induced infection. Science 2003; 302:630-3; PMID:12947035; http://dx.doi.org/10.1126/science.1090074
- Nakahara S, Raz A. On the role of galectins in signal transduction. Methods Enzymol 2006; 417:273-89; PMID:17132511; http://dx.doi.org/10.1016/S0076-6879(06)17019-6
- Radutoiu S, Madsen LH, Madsen EB, Felle HH, Umehara Y, Grønlund M, Sato S, Nakamura Y, Tabata S, Sandal N, et al. Plant recognition of symbiotic bacteria requires two LysM receptor-like kinases. Nature 2003; 425:585-92; PMID:14534578; http://dx.doi.org/10.1038/nature02039
- Mesnage S, Dellarole M, Baxter NJ, Rouget JB, Dimitrov JD, Wang N, Fujimoto Y, Hounslow AM, Lacroix-Desmazes S, Fukase K, et al. Molecular basis for bacterial peptidoglycan recognition by LysM domains. Nat Commun 2014; 5:4269; PMID:24978025; http://dx.doi.org/10.1038/ncomms5269
- Hu S, Kong J, Kong W, Guo T, Ji M. Characterization of a novel LysM domain from Lactobacillus fermentum bacteriophage endolysin and its use as an anchor to display heterologous proteins on the surfaces of lactic acid bacteria. Appl Environ Microbiol 2010; 76:2410-8; PMID:20173067; http://dx.doi.org/10.1128/AEM.01752-09
- Zadravec P, Štrukelj B, Berlec A. Heterologous surface display on lactic acid bacteria: non-GMO alternative? Bioengineered 2015; 6:179-83; PMID:25880164; http://dx.doi.org/10.1080/21655979.2015.1040956
- Audouy SA, van Selm S, van Roosmalen ML, Post E, Kanninga R, Neef J, Estevão S, Nieuwenhuis EE, Adrian PV, Leenhouts K, et al. Development of lactococcal GEM-based pneumococcal vaccines. Vaccine 2007; 25:2497-506; PMID:17081660; http://dx.doi.org/10.1016/j.vaccine.2006.09.026
- Saluja V, Amorij JP, van Roosmalen ML, Leenhouts K, Huckriede A, Hinrichs WL, Frijlink HW. Intranasal delivery of influenza subunit vaccine formulated with GEM particles as an adjuvant. AAPS J 2010; 12:109-16; PMID:20058113; http://dx.doi.org/10.1208/s12248-009-9168-2
- Steen A, Buist G, Horsburgh GJ, Venema G, Kuipers OP, Foster SJ, Kok J. AcmA of Lactococcus lactis is an N-acetylglucosaminidase with an optimal number of LysM domains for proper functioning. FEBS J 2005; 272:2854-68; PMID:15943817; http://dx.doi.org/10.1111/j.1742-4658.2005.04706.x
- Wong JEMM, Alsarraf HMAB, Kaspersen JD, Pedersen JS, Stougaard J, Thirup S, Blaise M. Cooperative binding of LysM domains determines the carbohydrate affinity of a bacterial endopeptidase protein. FEBS J 2014; 281:1196-208; PMID:24355088; http://dx.doi.org/10.1111/febs.12698
