?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Diabetes mellitus is a group of metabolic disorder of glucose metabolism. The proper management of blood glucose level is an indicator in the treatment of this complex pathology. The aim of the present study was to evaluate the inhibitory potential of Vicia faba crude seed extracts on the activities of α-amylase. Phytochemical screening, FTIR and HPLC analysis confirmed the presence of a phenolic compound with percentage yield of 9.87% in the acetone extract. Acetone extract of seed had highest inhibitory potential against porcine α-amylase (IC50 value of 2.94 mg/mL). Kinetic analysis revealed that the acetone and methanol extract displayed mixed mode of inhibition toward α-amylase. In-silico analysis was agreement with in-vitro studies in which phenolic compounds (catechin, epicatechin, gallic acid, and proanthocyanidin) showed more negative free energy against standard drug (acarbose) and bound with catalytic residues of α-amylase. These results might be due to the synergistic action of constituents present in seed extract or acting separately.
Introduction
India is rich in valuable natural resources of fruits, vegetables, cereals, herbs and medicinal plants many of which remain unexploited and are limited for regional or rural use only. Vicia faba plant is one such example, and it belongs to pulses category. Faba beans as annual legume and its botanical name is Vicia faba L.Citation1,2 It is commonally known as as Bakala in Hindi language or local language.Citation3 Faba beans are nutritionally rich and staple pulses of less privileged society of India. The frequency of diabetes cases increasing globally and 300 million people may effect by 2025 worldwide.Citation4 Diabetes is a result of deficiency either in endogenous insulin production by the pancreatic β cells (otherwise known as type-1 diabetes) or hindered insulin secretion and or action (type-2 diabetes). Type-1 diabetes is an autoimmune disease due to T-cell mediated destruction of the pancreatic β cells. In type-2 diabetes, there is a gradual development of insulin resistance and β cell dysfunction, associated with obesity and an imbalance lifestyle.Citation5 Pancreatic α-amylase (E.C. 3.2.1.1) is an enzyme in the digestive system and catalyzes the hydrolysis of starch to a mixture of smaller oligosaccharides consisting of maltose, maltotriose and a number of α (l-6) and α (1–4) oligoglucans. The activity of HPA (human pancreatic α-amylase) in the small intestine is related to increase in postprandial glucose levels, and the control of which could be a major strategy in treatment of type-2 diabetes.Citation6 Inhibitors of pancreatic α-amylase delay carbohydrate digestion causing a reduction in the rate of glucose absorption and decrease the postprandial serum glucose levels.Citation7 Some synthetic drugs are available in the market such as acarbose and miglitol which inhibit α-glucosidase and α-amylase. They are not site-specific, and produce more side effects. The major side effects of these inhibitors are gastrointestinal problems viz. bloating, abdominal problem, diarrhea and flatulence.Citation8 Previous studies suggest the anti-diabetic and free radical scavenging property of faba beans points out hypoglycemic and antioxidant effect.Citation9 The presence of vicine or divicine in faba beans seeds,Citation10 may be one cause of antidiabetic activity. However, the antidiabetic potency of Vicia faba seed and their mechanism of inhibition have not yet been extensively studied till now. Therefore, the objective of above study is to evaluate the antidiabetic potential of Vicia faba crude seed extract and its further characterization.
Experimental methods
Plant material
Vicia faba seeds were collected from local farmers of Darbhanga district Bihar, India.
Chemicals and reagents
Porcine Alpha-amylase was purchased from Sigma-Aldrich Co., St Louis, USA, while starch soluble (extra pure) were obtained from Himedia laboratories. Other chemicals and reagents were of analytical grade and water used were distilled.
Preparation of plant extract
Vicia faba seeds were washed with water to remove all contaminants and again washed with 70% ethanol. Vicia faba seed was dried in an oven at 50°C for 24 h and ground to fine powder. Thirty g of powdered sample was dissolved in 100 ml of solvent and kept for 48 h under shaking at 130 rpm and temperature 40°C. Solvents used were methanol, ethanol, acetone, hexane, chloroform and water. After the extract preparation samples were transferred into rotatory vacuum evaporator and kept at 50°C till all the solvent was evaporated. Dried extract was kept in a refrigerator at 4°C for their future use in phytochemicals analysis. Dried extracts were weighed,and stock solution was made in 10% dimethylsulphoxide.
Phytochemical screening
Phytochemical compositions of the seeds extract were determined using the methods described by Trease and Evans, 2002Citation11 and Sofowora, 1993.Citation12 Filtrates were treated with Mayer's reagent (Potassium Mercuric Iodide). Formation of a yellow colored precipitate indicates the presence of alkaloids. Extracts were diluted with distilled water up to 10 ml and mixed properly. Formation of foam indicate the presence of saponins. 0.25 g of extract was shaken with 2 ml of water. If foam produced persists for 10 min it indicated the presence of saponins. Extracts were treated with 3–4 drops of ferric chloride solution. Formation of bluish black color indicated the presence of phenols or tannins. 1% gelatin solution containing sodium chloride was added to the extract. Formation of white precipitate showed the presence of tannins. Extracts were treated with few drops of sodium hydroxide solution. Formation of intense yellow color, which later became colorless on addition of dilute acid, indicated the presence of flavonoids. 5 ml (1 mg/ml) of the fraction with few drops of chloroform was added and then 3 ml of concentrated H2SO4 was added in the mixture. Change of reddish brown color revealed the presence of terpenoids. The extracts were treated with few drops of conc. Nitric acid. Formation of yellow color marked the presence of proteins. The test solution was mixed with few drops of Benedict's reagent (alkaline solution containing cupric citrate complex) and boiled in a water bath. The formation of reddish brown precipitate to showed a positive result for the presence of carbohydrate.
α -amylase inhibitory assay
The enzyme inhibition studies were performed using a modified procedure of McCue and Shetty, 2004.Citation13 The set of tubes containing 500 μl of extract (1.25–10 mg/ml) was kept along with 500 μl of 0.02 M sodium phosphate buffer (pH 6.9) containing α-amylase solution (0.5 mg/ml) in water bath for 10 min at 25°C . 500 μl of 1% starch solution made in 0.02 M sodium phosphate buffer of pH 6.9 was added at regular interval and further incubated at 25°C for 10 min. The reaction was stopped by adding 500 μl of dinitrosalicylic acid (DNS) reagent. The tubes were kept in boiling water bath for 5 min and cooled to room temperature. 5 ml distilled water was added and the absorbance was measured at 540 nm using a spectrophotometer. Control was made using the same protocol by only replacing the extract with distilled water. The α-amylase inhibitory activity was evaluated. Concentrations of extracts resulting in 50% inhibition of enzyme activity (IC50) were determined. The IC50 value was defined as the concentration of the extract, containing the α-amylase inhibitor that inhibited 50% of the PPA activity and calculated by given formula below:
Mode of α-amylase inhibition
The inhibitory activity of seed extract on α-amylase by the seed extract was performed according to the modified method described by Ali et al. 2006.Citation14 500 μL of the extract (5 mg/mL) was added with 500 μL of an α-amylase solution and preincubated for 10 min at 25°C in one set of tubes. α-amylase was preincubated with 500 μl of phosphate buffer (pH 6.9) in another set of tubes. 500 μL of the soluble starch solution at increasing concentrations (0.30–5.0 mg/mL) was added to both sets of reaction mixtures to start the reaction. The resulting mixture was again incubated for 10 min at 25°C in boiling water bath for 5 min after the addition of 1000 μL of DNS to terminate the reaction. The amount of reducing sugars was determined by spectrophotometrically at 540 nm using a standard maltose curve and converted to reaction rates. A double reciprocal plot (1/(V) versus 1/( S) where is V is reaction speed and (S) is substrate concentration was plotted. The mode of inhibition of the crude seed extract on α-amylase activity was determined by analysis of the double reciprocal (Lineweaver-Burk) plot using Michaelis-Menten kinetics.Citation15
Fractionation of acetone seed extract
The crude extract of Vicia faba seed was purified successively, with a silica gel (280-300 mesh) column. Fractions were collected by gradient elution with chloroform-methanol (50:1, 25:1,10:1, 4:1, 2:1). The fraction which had efficient α- amylase inhibitory activity was subjected to FTIR analysis and HPLC analysis. Phenol concentrations in every fraction were estimated by Lowry's method.Citation16 Total carbohydrate content in the fractions was determined by the phenol -sulfuric acid method.Citation17
HPLC analysis condition
The Waters HPLC apparatus with photodiode array detector and the C18 column was used for method development. Millipore filtration assembly was used for mobile phase filtration. The sample was filtered by syringe filter (0.45 µm). A mobile phase consisting of a mixture of acetonitrile (A), H2O (B), (0.1% acetic acid) in a ratio of (60:40 v/v) was filtered (0.22µm) and, degassed by ultrasonicator. Flow rate was maintained at 0.5 ml/min. Isocratic elution was used to study different fractions.
Fourier transform infrared spectroscopy (FTIR)
Alpha ATR-FTIR, Bruker, USA was used for the FTIR analysis. Samples were scanned in the wave number range of 400–4000 cm−1. FTIR study revealed that the chemical bonds absorbed radiation in the middle infrared (MIR) region between 400 and 4000 cm−1, and each functional group of a molecule had specific absorption frequencies in the IR spectrum. Fourier transform infrared spectra (FTIR) can generally use to determine the structure of biological composition and structure of molecular functional group. It can be determined by analyzing the position, width, and intensity of acquired spectra in a complex biological system based on individual algorithms.
Statistical data analysis
All experiments were performed in 3 different sets with each set in triplicates. The data are expressed as mean ± SD. Values of p ≤ 0.05 which are considered as significant.
In-silico analysis
Preparation of protein
The three-dimensional (3D) structure of (PDB ID: 1PIF) pig α-amylase was retrieved from RCSB protein data bank. Energy minimization was done by using Chimera 1.10.2.
Selection of ligands
Based on literatureCitation18-20 the active constituents of Vicia faba seed extract i.e., tannins (Gallic acid, Proanthocyanidins, Catechin, Epicatechin) were selected. Acarbose was selected as a standard drug as reference molecule.
Ligand preparation
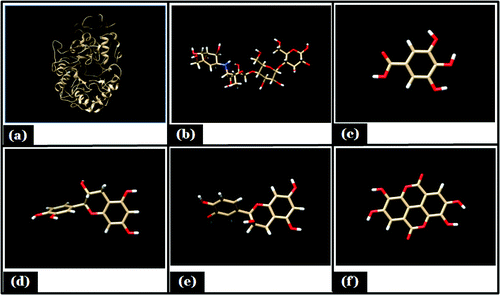
Three dimensional structures of different ligands were retrieved from PubChem Compound data base (http://pubchem.ncbi.nlm.nih.gov). The ID of different ligands were as follows: Gallic acid [CID370], Acarbose [CID10256], Catechein [CID73160], Epicatechein [CID3084390], Proanthocyanidin [CID 1080651]. Their energy forms were minimized, by pyRX virtual screening tool Citation21 and were converted to PDB format by the Open Babel 2.3.1.Citation22 Minimized energy structure of receptor and ligands are shown as ().
Docking of ligand-receptor procedure
Receptors (α amylase (1pif.pdb)) were used for the AudoDock Tools 1.5.6 for docking. Docking was performed by using Autodock 4.2.0 in the interface of MGLTool 1.5.4.Citation23,24 Receptor preparation includes all the necessary steps before docking such as (i) removal of solvent molecules and ions and extraction of cocrystallized ligand (ii) addition of polar hydrogens (iii) assignment of AD4 atom type and finally (iv) assignment of Gasteiger charges. AutoGrid was used to generate grid maps. The grid maps represent the ligand in real docking target site were calculated with gid box dimension of 60 × 60 × 60 Å and spacing of 0.375 Å by taking the center of the ligand as the center of the grid. Docking of the ligand was performed with default parameters except keeping 30 runs. Further interaction analysis was done using the autodock tools and visualized by the Chimera1.10.2.Citation25 To study the intermolecular interactions between the targets and the compounds LigPlot12 were used to plot their interactions from 3D to 2D. AutoDock is based on semiempirical free energy force field to confirms or evaluates actual conformations during docking simulations. The optimized conformation represents possible binding modes of the ligand within the site: ΔG=ΔGvdw+ΔGhbond+ΔGelec+ΔGtor+ΔGdesolv. The first 3 terms indicate van der Waals force, hydrogen bonding, and electrostatics interaction respectively. The term ΔGtor is for rotation and translation energy and ΔGdesolv is for desolvation energy upon binding and the hydrophobic effect The binding energy (kcal/mol), that indicates how strongly a ligand binds to the receptor, its calculation was based on the scoring function used in the AutoDock tools.
Results and discussion
Dry weight analysis
The sequential extraction was performed and phenolic component was found to be highest with percentage yield of 9.87% in acetone extract, followed by methanol 8.30%, and water (7.36%).
Phytochemical screening
The chemoprofiling of seed extract in different solvents (hexane, chloroform, ethanol, water, acetone, methanol, and ethyl acetate) indicated the presence of flavonoids, tannins, reducing sugar, protein, and alkaloids in all extracts (). Saponins were detected only in the water extract while tannin was absent in hexane extract.
Table 1. Chemoprofiling of Vicia faba seed extracts in different solvents.
Inhibitory potency of Vicia faba seed extracts against α-amylase activity
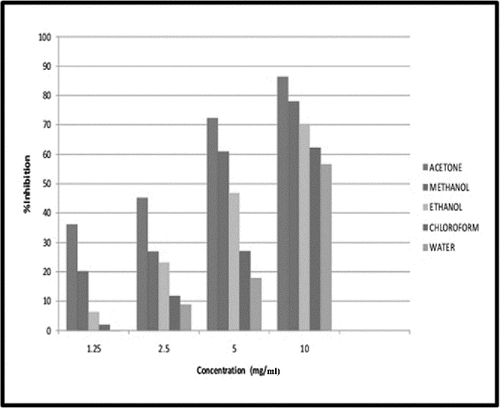
showed the α-amylase inhibition potential of the Vicia faba seed extracts (10mg/ml) was determined Maximum inhibition percentage was observed in acetone extract (86.3%) followed by methanol (77.9%), ethanol (70.36%), chloroform (62.3%), and water (56.6%) extract. At lower concentration of seed extract (1.25 mg/ml), least inhibition percentage was found to have 0.012 % (water extract) and 1.97% (chloroform). The inhibition of α-amylase by all the extracts at lower concentrations (0.73–1.25 mg/ml) showed no significant difference from one another but at higher concentrations (3–5 mg/ml), the inhibitory potential of acetone and methanol extract was significantly unique (P < 0.05) in comparison to other extracts studied. Extrapolation of α-amylase effectiveness from the dose-response curve showed that acetone and methanol extract contained the most efficient α-amylase inhibitor with an IC50 value of 2.94 mg/ml (). This could be one of the several strategies adopted in diabetes mellitus treatment. The inhibition of digesting enzymes α-amylase may lead to lower down postprandial glucose level.Citation26 This is an effort to search for alternative drugs from medicinal plants with efficient potential and lesser side effects than synthetic drugs.Citation27-29
Table 2. IC50 values for α-amylase inhibitory potential of Vicia faba seed extracts.
Mode of enzyme inhibition and their kinetic analysis
showed that the enzyme follows Michelis- Mentens kinetics. To find out the type of inhibition Lineweaver-Burk plot was made for α -amylase activity was estimated using starch as substrate and mixed type of the enzyme inhibition was observed (). It was evident from that Km and Vmax value of acetone, and methanol extract were found to be 0.56mM, 0.14mM/min (Km,Vmax) and 0.50mM, 0.16 mM/min (Km,Vmax) respectively ().Whereas Km and Vmax of reference molecule were found to be 0.40 mM 0.20 mM/min respectively. Thus it was evident that apparent Km value increased in the case of acetone extract with decreased Vmax value. Kinetic analysis showed that inhibition is a combination of competitive and non competitive enzyme inhibition. These findings supported the reports indicating that excessive inhibition of pancreatic α- amylase could result in the bacterial fermentation of undigested carbohydrates and therefore mild α-amylase inhibitory activity is desirable.Citation30 Mixed type of inhibition in LB plot confirmed that the active components in the extract might not compete with the substrate for binding to the active site rather the inhibitors may bind to a separate site on the enzyme to delay the conversion of disaccharides to monosaccharides.Citation29,31 This mixed type inhibition kinetics is specific of some medicinal plants, due to the variety of phytochemicals present in the extract and may be acting in different ways.Citation32 The findings made are in confirmatory of other studies on α-amylase and α-glucosidase inhibitors of different medicinal plants. The potential inhibitors from plants mainly belong to flavonoids class which has features of inhibiting α-amylase and α -glucosidase activities.Citation33
Figure 3. Mode of inhibition of α-amylase by acetone and methanol seed extract (Michaelis- Menten plot).
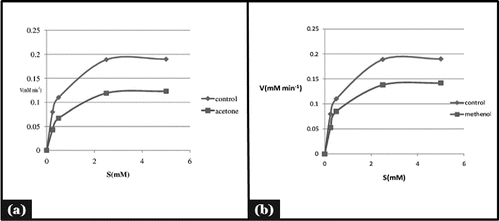
Figure 4. Mode of inhibition of α-amylase by acetone and methanol seed extract (Lineweaver-Burk plot).
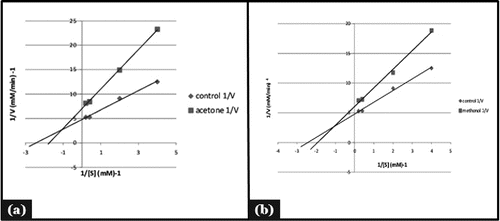
Table 3. Kinetic analysis of Vicia faba seed extract with respect to reference molecule.
FTIR analysis
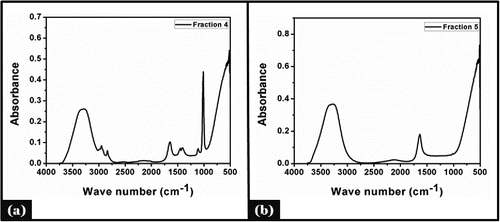
The acetone extract was applied to silica gel column chromatography by gradient elution with chlororform-methanol to get most potent inhibitor fraction. All the fractions were tested for their inhibitory activity (data not shown) and the extract was found to be effective % of inhibition. The fraction was then subjected to HPLC.All the extract fraction were also checked for total phenol and total sugar contents (). FTIR analysis showed a broad band at 3200-3320 cm−1 which belonged to stretching vibration of a phenolic hydroxyl group (-OH). Appearance of band at wave number 2900-3000 cm−1 () indicated presence of vibration stretching of aromatic (C-H) group, also the medium band at 1632-1654 cm−1 representing presence of aromatic ketone group in the all of the fractions () but the sharp band at 1020 cm−1 belonged to ether (C-O) group and the medium band at 1300-1400 cm−1 represents presence of bending vibration of (C-H) group belonging to methyl group (). Clear bands in the 3200-3305 range were observed for the phenolic compounds in all fraction () and characteristics bands of carbonyl group observed at 1100-1118 cm−1. Literature also revealed that the tannic acid contains some aromatic esters which show signal characteristics bands of carbonyl groups: C=O stretching vibration at 1730-1705 cm−1 and C-O at 1100-1300 cm−1.Citation34,35
Table 4. Total sugar.total phenol content of acetonic seed extract fraction.
HPLC analysis
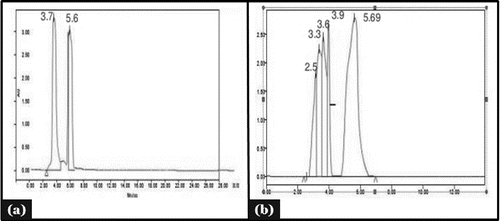
HPLC was done for confirmation of a phenolic compound in acetone fraction (F5). Tannic acid as a standard molecule (Gallotannin) with its retention time (5.69 minutes) and fraction 5 showed similar retention time 5.69 minutes at 274 nm (). HPLC data showed that the phenolic compound was present in the extract.
Molecular docking studies
The molecular docking tool can be exploited to study the interaction between a small molecule and a receptor at the atomic level, which may give the insight to characterize the behavior of small molecules in the binding site of target proteins as well as to elucidate fundamental biochemical processes.Citation36 On the basis of in-vitro studies (phytochemical screening, FTIR characterization,and HPLC) polyphenols was selected as a ligand for molecular docking studies. Three interacting catalytic binding sites of porcine α-amylase (catalytic triads of Asp197, Glu233, Asp300) were identified.Citation37 The docked stable confirmation showed binding energy in the range of −2.29 to −6.56 Kcal/mol (). It was observed that the structure catechin exhibits binding energy of −6.56 Kcal/mol and the acarbose as a standard drug exhibits binding energy of −2.29 Kcal/mol. The binding energies (ΔGBE) of the all best-docked molecule were given in (). The 2D view of protein–ligand interactions was generated by lig plot.Citation38 Lig plot () and autodock tool interaction analysisCitation39 () showed different residues such as Glu233, Arg195, His201, Ile235, ASP197 present on the catalytic site of α-amylase interacted with ligands. A standard drug acarbose exhibit hydrogen binding with amino acid residues Asp197, Asp300, Glu233 which directly participate in the catalytic activity of this enzyme. The docking studies showed that the van der Waals, electrostatic and desolvation energies play an important role in binding. The hydrophobic interactions were formed by Val234, lys200, Val163, leu165, Ser191, Asp197 in acarbose with porcine α amylase molecule. All the selected polyphenolic compound showed more negative binding energy in comparison to a standard drug as acarbose. Above results were in agreement with the literature, it showed that non-covalent interactions occur between polyphenols and enzymes.Citation40 Hydroxyl groups and galloyl groups are present in the molecular structure of polyphenol.Citation41 The phenolic groups are responsible for hydrogen bonding with the polar amino acid of an enzyme. There are many hydrophobic amino acids residues found in enzymes (protein). Galloyl groups in polyphenols are responsible for hydrophobicityCitation42 and therefore, polyphenols can interact with enzymes through hydrophobic association.Citation41 The galloyl moiety may play a crucial role in interaction with mammalian α-amylase its positions in aromatic ring mostly affect the effectiveness.Citation43-45 The combined effects of hydrophobic interaction and hydrogen bond formation between polyphenols and the porcine-α amylase could contribute to control postprandial hyperglycemia in type 2 diabetic patients. The knowledge of interaction between phenolic compounds and digestive enzymes may be an initial step toward the development of drug, nutraceuticals or functional foods.
Figure 7. Lig plot showing hydrogen bonding and hdrophobic interaction in different ligand, (A) Acarbose; (B) Epicatechein; (C) Gallic acid; (D) Proanthocyanidin; (E) Catechin with porcine α- amylase.
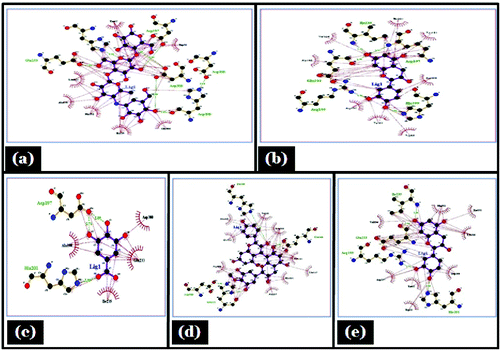
Figure 8. Showing autodock tool analysis of different types of interaction in different ligands. (A) Catechin; (B) Epicatechin; (C) Acarbose; (D) Proanthocyanidin; (E) Gallic acid with porcine α- amylase.
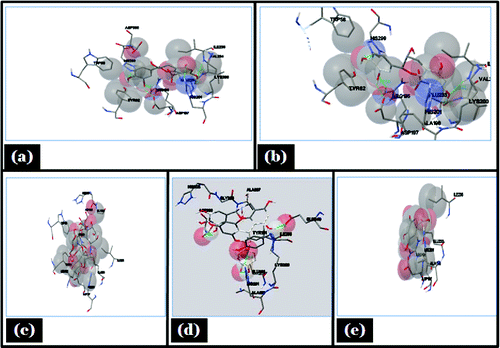
Table 5. Docking analysis of different ligands with porcine α amylase.
Conclusion
Acetone and methanol extract of Vicia faba seed displayed most effective inhibition against the α-amylase enzyme. This enzyme activity showed that seed extract suggested the mixed type of inhibition with α-amylase. However, the further in-silico study confirmed that polyphenolic compound binds with the catalytic residues of α-amylase by hydrogen bonding and hydrophobic interaction. This inhibition of enzyme might be either due to synergistic effect of the phytochemical constituents of polyphenols present in it or acting separately.
Disclosure of potential conflicts of interest
No potential conflicts of interest were disclosed.
Acknowledgement
The authors thank the School of Biochemical Engineering, Varanasi, India for providing infrastructure for research work.
Funding
The author Dhiraj Kumar Choudhary thanks the School of Biochemical Engineering, Government of India, New Delhi, for providing research support grant for the completion of the work.
References
- Singh AK, Bhat BP, Sundaram PK, Gupta AK, Singh D. Planting geometry to optimize growth and productivity faba bean (Vicia faba L.) and soil fertility. J Environ Biol 2013; 34(1):117-22; PMID:24006817
- Hanelt P, Mettin D. Biosystematics of the genus Vicia L. (Leguminosae). Annu Rev Ecol Syst 1989; 20:199-223; https://doi.org/10.1146/annurev.es.20.110189.001215
- Harlan JR, Ethiopia. A centre of diversity. Econ Bot 1969; 23:309-14; https://doi.org/10.1007/BF02860676
- Gupta OP, Phatak S. Pandemic trends in prevalence of diabetes mellitus and associated coronary heart disease in india their causes and prevention. Int J Diabetes Dev Countries 2003; 23:37-50
- Zimmet P, Alberti K, Shaw J. Global and societal implications of the diabetes epidemic. Nature 2001; 414:782-7
- Eichler HG, Korn A, Gasic S, Prison W, Businger J. The effect of new specific α-amylase inhibitor on postprandial glucose and insulin excursions in normal subjects and Type 2 (noninsulin dependent) diabetic patients. Diabetologia 1984; 26(4):278-81
- Tarling CA, Woods K, Zhang R, Brastianos HC, Brayer GD, Andersen RJ, Withers SG. The search for novel human pancreatic α-amylase inhibitors: High throughput screening of terrestrial and marine natural product extracts. Chem Bio Chem 2008; 9:433-8; https://doi.org/10.1002/cbic.200700470
- Cheng AYY, Fantus IG. Oral antihyperglycemic therapy for type 2 diabetes mellitus. Canadian Med Assoc J 2005; 172(2):213-26; https://doi.org/10.1503/cmaj.1031414
- Fatima S, Kapoor, R. In vivo and in vitro glycemic effects of certain legumes. J Food Sci Technol-Mysore 2006; 3:263-6
- Bjerg B, Knudsen J, Olsen O, Poulsen M, Soerensen H. Quantitative analysis and inheritance of vicine and convicine content in seeds of Vicia faba L. Z. Pflanzen- Zücht 1985; 94:135-148
- Trease GE, Evans WC. Pharmacognosy; 15th Edn. Saunders: Elsevier (A Division of Reed Elsevier India Pvt. Limited). 2002:214-393.
- Sofowora A. Medicinal Plants and Traditional Medicine in Africa; 2nd Edn.Spectrum Books Limited: Ibadan, Nigeria, 1993; 1-153 pp
- McCue PP, Shetty K. Inhibitory effects of rosmarinic acid extracts on porcine pancreatic amylase in vitro. Asia Pacific J Clin Nutr 2004; 13(1):101-6; PMID:15003922
- Ali H, Houghton PJ, Soumyanath A. α-Amylase inhibitory activity of some Malaysian plants used to treat diabetes; with particular reference to Phyllanthus amarus. J Ethnopharmacol 2006; 107(3):449-55; PMID:16678367; https://doi.org/10.1016/j.jep.2006.04.004
- Nelson DL, Cox MM. Lehninger principles of biochemistry, Freeman WH, New York, NY, USA, 5th edition. 2008
- Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. Protein measurement with the folin phenol reagent. J Biol Chem 1951; 193:265-75; PMID:14907713
- Saha SK, Brewer CF. Determination of the concentrations of oligosaccharides, complex type carbohydrates and glycoproteins using the phenol sulphuric acid method. Carbohydr Res 1994; 254:157-63; PMID:8180982; https://doi.org/10.1016/0008-6215(94)84249-3
- Tiwari S, Bala M. J Herbs Spices Medicinal Plants 2009; 15(2):203-15; https://doi.org/10.1080/10496470903139496
- Manu S, Sandhya T, Jagdish P, Surender S, Yogendra K M S, Madhu B. Hippophae leaf extract concentration regulates antioxidant and prooxidant effects on DNA. Journal of Dietary Supplements 2010. 7(60); PMID:22435574
- Kawakami K, Aketa S, Nakanami M, Iizuka S, Hirayama M. Major water-soluble polyphenols, proanthocyanidins, in leaves of persimmon (Diospyros kaki) and their α-amylase inhibitory activity. Biosci Biotechnol Biochem 2010; 74:1380-5; https://doi.org/10.1271/bbb.100056
- Dallakyan S, Olson AJ. Small-molecule library screening by docking with PyRx. Methods Mol Biol 2015; 1263:243-50; PMID:25618350; https://doi.org/10.1007/978-1-4939-2269-7_19
- O'Boyle NM, Banck M, James CA, Morley C, Vandermeersch T, Hutchison GR. Open Babel: An open chemical toolbox. J Cheminf 2011; 3:33; https://doi.org/10.1186/1758-2946-3-33
- He, Fahu et al. Solution structure of the cysteine–rich domain in Fn14, a member of the tumor necrosis factor receptor superfamily. Protein Science 2009. 18(3):650–656.
- Morris, GM, Huey, R, Lindstrom, W, Sanner, MF, Belew, RK, Goodsell, DS and Olson, AJ. Autodock4 and AutoDockTools4: automated docking with selective receptor flexiblity. J. Computational Chemistry 2009. 16: 2785-91.
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J Comput Chem 2004; 25(13):1605-12; PMID:15264254; https://doi.org/10.1002/jcc.20084
- Mccue PY, Kwon I, Shetty K. Anti-amylase, anti-glucosidase and anti-angiotensin I-converting enzyme potential of selected foods. J Food Biochem 2005; 29(3):278-94.https://doi.org/10.1111/j.1745-4514.2005.00020.x
- Matsui T, Ogunwande IA, Abesundara KJ, Matsumoto K. Anti-hyperglycemic potential of natural products. Mini-Rev Medicinal Chem 2006; 6(3):349-56; PMID:16515474; https://doi.org/10.2174/138955706776073484
- Matsuda H, Morikawa T, Yoshikawa M. Antidiabetogenic constituents from several natural medicines. Pure Applied Chem 2002; 74(7):1301-8; https://doi.org/10.1351/pac200274071301
- Ogunwande, IA, Matsui T, Fujise T, Matsumoto K. α-Glucosidase inhibitory profile of Nigerian medicinal plants in immobilized assay system. Food Sci Technol Res 2007; 13(2):169-72; https://doi.org/10.3136/fstr.13.169
- Apostolidis E, Kwon YI, Shetty K, Inhibitory potential of herb, fruit, and fungal-enriched cheese against key enzymes linked to type2 diabetes and hypertension. Innovative Food Sci Emerging Technologies 2007; 8(1):46-54; https://doi.org/10.1016/j.ifset.2006.06.001
- Mogale AM, Lebelo LS, Thovhogi N, de Freitas AN, Shai LJ. α-amylase and α-glucosidase inhibitory effects of sclerocarya birrea [(A. Rich.) Hochst.] subspecies caffra (Sond) Kokwaro (Anacardiaceae) stem-bark extracts. African J Biotechnol 2011; 66(10):15033-9
- Mills S, Bone K. Principles and Practice of Phytotherapy: Modern herbal medicine. Edinburgh, Churchill Livingstone 2000; 387 pp
- Kwon YI, Apostolidis E, Shetty, K. Evaluation of pepper (Capsicum annuum) for management of diabetes and hypertension. J Food Biochem 2007; 31(3):370-85; https://doi.org/10.1111/j.1745-4514.2007.00120.x
- Silverstein RM, Bassler GC, Morrill TC. Spectrometric Identification of Organic Compounds; 7th ed., John Wiley & Sons Inc. USA, 1981; 95-8 pp
- Stuart BH. Experimental methods in infrared spectroscopy fundamentals and applications, John Wiley & Sons, Ltd, Chichester, UK 2005:76-77 pp
- Conkey Mc, Sobolev BJ, Edelman VM. The performance of current methods in ligand protein docking. Curr Sci 2002; 83:845-55
- Uitdehaag JCM, Mosi R, Kalk KH, Van der veen BA, Dijkhuizen L, WIthers SG, Dijkstra BW. X-ray structures along the reaction pathway of cyclodextrin glycosyltransferase elucidate catalysis in the α- amylase family. Nat Struct Biol 1999; 6:432-6
- Wallace AC, Laskowski RA, Thornton JM. LIGPLOT. a program to generate schematic diagrams of protein-ligand interactions. Protein Eng 1996; 8:127-34; https://doi.org/10.1093/protein/8.2.127
- Michel F, Sanner P. A Programming Language for Software Integration and Development. J. Mol. Graphics Mod 1999; 17:57-61
- Siebert KJ, Troukhanova NV, Lynn PY. Nature of polyphenols protein interactions. J Agricultural Food Chem 1996; 44:80-5; https://doi.org/10.1021/jf9502459
- He Q, Lv Y, Yao K. Effects of tea polyphenols on the activities of α-amylase,pepsin, trypsin and lipase. Food Chem 2006; 101:1178-82; https://doi.org/10.1016/j.foodchem.2006.03.020
- He Q, Shi B, Ya K. Interactions of gallotannins with proteins, amino acids, phospholipids and sugars. Food Chem 2006; 95:250-4; https://doi.org/10.1016/j.foodchem.2004.11.055
- Koh LW, Wong LL, Loo YY, Kasapis S, Huang D. Evaluation of different teas against starch digestibility by mammalian glycosidases. J Agricultural Food Chem 2010; 58:148-54; PMID:20050703; https://doi.org/10.1021/jf903011g
- Toda M, Kawabata J, Kasai T. Inhibitory effects of ellagiandgallotannins on rat intestinal α-glucosidase complexes. Biosci Biotechnol Biochemistry 2001; 65:542-7; https://doi.org/10.1271/bbb.65.542
- Gao H, Huang YN, Xu PY, Kawabata J. Inhibitory effect on α-glucosidase by the fruits of Terminalia chebula Retz. Food Chemistry 2007; 105:628-34; https://doi.org/10.1016/j.foodchem.2007.04.023