ABSTRACT
Tonsillitis is the inflammation of the tonsils due to infection, many patients ultimately have to undergo tonsillectomy. In order to improve the accuracy of diagnosis and even create a new treatment for tonsillitis, we constructed a prokaryotic expression single-chain antibody fragment library against Streptococcus pneumoniae with immunoglobulin heavy chain variable region (VH), κ light chain (Vκ), and λ light chain (Vλ) genes by using human tonsil tissue. Plasmid DNA sequencing showed that single-chain antibodies were complete and constructed correctly. The binding activity of recombinant clones was detected by enzyme-linked immunosorbent assay (ELISA), results showed that the binding activity and specificity of anti-S. pneumoniae single-chain fragment variable (scfv) is proved to be successful. The single-chain antibody may be an attractive strategy for tonsillitis etiologic diagnosis and therapy.
Chronic tonsillitis is one of the most common diseases evolved from long-term bacterial or virus infection, many patients may ultimately undergo tonsillectomy [Citation1]. Not only have is there huge damage in local, but it can also induce other organs diseases like acute nephritis, rheumatic arthritis, or cardiac diseases. Currently, the pathogenesis of chronic tonsillitis is not completely clear. It is generally believed that its occurrence and development are associated with recurrent acute tonsillitis, bacteria in the tonsillar reproduction and autoimmune reaction [Citation2].
In recent decades, chronic tonsillitis research just identifies the proceed with identifying the bacterial category, but the reproduction of bacterial in tonsils cannot completely reflect the relationship with infection. By examining the bacterial specific antibody in human tonsillar lymphoid cells, the infectious bacteria can be reflected [Citation3–Citation5].
The progress in antibody engineering has been directed toward the expression of antibody fragments in bacterial and phage display systems, leading to increasing of various applications in biology, clinical diagnosis, and therapy. Phage-displayed antibody library has been widely used to derive high-affinity target-specific antibodies. Single-chain fragment variable (scfv) antibody is a fragment of full antibody in which heavy chain (VH) and light chain variable region (VL) are linked to each other by a peptide linker and can be displayed on the surface of bacteriophages enabling identification of antibodies with unique specificity in phage display technology.
The development of antibody-research process has three steps: polyclonal antibody, monoclonal antibody, and genetic engineering antibody [Citation6]. Genetic engineering antibodies not only possess high specificity but also have wide clinical applications. The single-chain antibody is a kind of genetic engineering antibody and has many advantages like lower molecular weight, stronger tissue penetration, and faster body clearance [Citation7,Citation8]. It has the potential value for clinical diagnosis and treatment [Citation9–Citation13].
Until now, the primary treatment strategy of chronic tonsillitis is tonsillectomy, but postoperative pain, risk of bleeding, and risk of anesthesia are the major problems in clinical care. This study aims to construct a prokaryotic expression single-chain antibody fragment library against Streptococcus pneumoniae with immunoglobulin genes, which variable regions are VH, κ light chain (Vκ), and λ light chain (Vλ) and extract from antibody-producing cells in human tonsil tissue. Then identified the binding activity and specificity of the antibody. We hope to fill in the gaps in the research on the related single-chain antibody of chronic tonsillitis and provide a new idea for the diagnosis and treatment of chronic tonsillitis.
Materials and methods
Specimens and strains
Source of specimen
The patient has more than three episodes of inflammation of the tonsils within 1 year, without the history of rheumatoid arthritis, myocarditis, nephritis, upper respiratory tract infection, and long-term low fever. The research project has passed the ethical review, and the patient has signed the informed consent form.
Strain sources
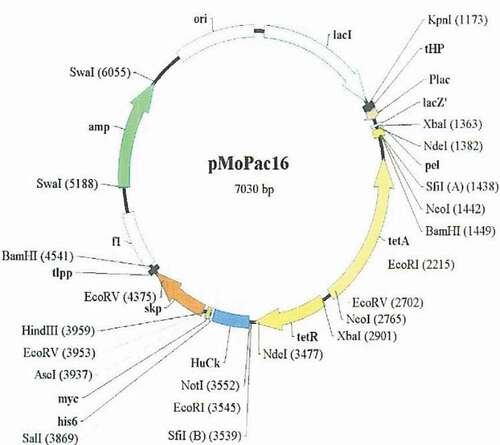
Staphylococcus aureus (ATCC 25923), Streptococcus A (ATCC 32209), B Streptococcus (ATCC 32204), and S. pneumoniae (ATCC 31108) were purchased from the Department of Microbiology, Sun Yat-sen University. Plasmid source: expression vector pMo Pac16 is from Dr. Andrew Hayburst, a cell molecular biology research institute, the University of Texas at Austin. Source of Escherichia coli: E. coli Rosetta (DE3) is from the antibody group of the R & D Department of Shenzhen Lvshiyuan Biotechnology Co., Ltd.
Lymphocyte isolation
The tonsillar tissue was obtained during operation and was placed in the precold RPMI 1640 complete medium. Then, the sample was ground to allow lymphocytes to pass through a 35–40 µm mesh into serum-free RPMI 1640 medium. Lymphocytes were isolated by lymphocyte separation medium.
Preparation of inactivated intact bacterial antigen
S. aureus, alpha Streptococcus, beta Streptococcus, and S. pneumoniae were added to sterilized lysogeny broth (LB) medium supplemented with 10% fetal bovine serum (FBS), respectively. After incubating at 37°C for 18–24 h with shaking, the bacterial cell density was measured. After centrifugation, the pellet was washed with PBS and fixed in 0.5% glutaraldehyde.
Detection of specific IgG in serum by indirect ELISA
Each well of 96-well microtiter plates was coated with 50 µL of 5 × 108/mL inactivate the bacteria (S. aureus, alpha Streptococcus, beta Streptococcus, and S. pneumoniae) in 0.1 M bicarbonate buffer pH 9.6 (0.06 M NaHCO3, 0.04 M Na2CO3) for 24 h at 37°C. The plates were washed with PBS which containing 0.05% Tween 20 to remove unbound antigen. Then incubated with 220 µL block solution (1 g/L bovine serum albumin in PBS) per well for overnight at 4°C and washed with PBST. A total of 50 µL of 1 mg/mL rabbit IgG was added to each well of ELISA plates coated by the Staphylococcus total antigen, respectively for overnight at 4°C then washed with PBST.
A total of 50 µL of the patient’s sera fold diluted (1:10,000, 1:20,000, 1:40,000, 1:60,000, 1:80,000, 1:1,20,000, and 1:1,40,000) in PBS containing 5% skimmed milk powder was added to each well in duplicate, respectively, following incubation for 1 h at 37°C, the plates were washed with PBST. A total of 50 µL 1:1000 diluted the biotin rat antihuman IgG (γ) antibody in antibody diluent (10 g/L bovine serum albumin) was added to each well, incubated for 1 h at 37°C, washed with PBS. A total of 50 µL 1:4000 diluted the streptavidin-horse radish peroxidase was added to each well and incubated for 1 h at 37°C and washed with PBS. A totao of 100 µL tetramethylbenzidine was added to each well and incubated for 30 min in the dark at room temperature. The resultant color reaction was stopped by addition of 50 µL 1 M HCl per well. The absorbance values at 405 nm (A405) were determined using an ELISA plate reader.
Labeling of the total bacterial antigen of Streptococcus pneumoniae with biotin
The inactivated S. pneumoniae, which was fixed in 0.5% glutaraldehyde, then washed and suspended with PBS, adjust the concentration to 25 × 10 × 106/mL. Preparation of 10 mM NHS-LC-Biotin solution: weigh 0.454 mg NHS-dPEG4-biotin (molecular weight is 454.54), dissolved in 100 uL dimethyl sulfoxide; 100 uL 10 mM NHS-LC-biotin solution was added to 1 mL of S. pneumoniae suspension, and incubated for 30 min at room temperature, then overnight at 4°C. After washing S. pneumoniae by centrifugation, the pellet was suspended with PBS.
Immunomagnetic beads for the selection of tonsillar lymphocytes with the specific binding of Streptococcus pneumoniae antigen
The human lymphocytes were washed with PBS by centrifugation at 3000 × g for 5 min. A total of 100 µL of the S. pneumoniae suspension labeled with biotin was added to each tube of lymphocytes and incubated for 30 min at 4°C. After washing with PBS by centrifugation at 3000 × g for 5 min, cells pellet was resuspended with 400 uL PBS and added 100 uL immunomagnetic beads, incubated for 10 min at 4°C, placed the tube on the magnetic separator to concentrate the magnetic beads for 5 min, removed the supernatant. Added 1 mL PBS to resuspended magnetic beads. Then centrifuged at 3000 × g for 5 min, and the lymphocytes pellet were used for extracting RNA.
Extraction of total RNA
Total RNA was isolated from lymphocyte by TRIZOL reagent, according to the manufacturer’s instructions (Invitrogen). RNA pellets were washed by reprecipitation in ethanol and quantitated by absorbance at 260 nm. Full-length cDNAs were generated from 10 μg of total RNA, using SuperScript II Reverse Transcriptase kit (Invitrogen™, Cat.#18064022) according to the manufacturer’s instructions
Amplification of VH, Vλ, Vκ, and scfv gene
Referring to Pansri [Citation14], the primer was designed by the software Prmier Premier 5 and modified as shown in the attachment, synthesized by Sigma Aldrich Company. The genes for variable regions of the VH, Vκ, and Vλ were amplified using each of the primers described in – separately. PCR amplification reactions were carried out using the Taq polymerase (NEB, USA). After checking the amplification, all heavy or light chain PCR products were detected by electrophoresis on a 1% agarose gel and purified using QIAQuick gel extraction kit (QIAGEN). The gene splicing by overlap extension-PCR method was used to recombine VH linker, Vκ linker, and Vλ linker. VH linker, Vκ linker, and Vλ linker chains were connected using Hsback and Hsfor as primers to overlap and splice into a complete scfv gene. The scfv gene and plasmid pMo Pac16 () with the same restriction endonuclease digestion, digestion system for 50 uL system, according to the operating instructions of Bio Labs company. The scfv gene and plasmid pMo Pac16 after the enzyme digestion were linked by the T4 DNA ligase in the following reaction system. The reaction conditions were 16°C for overnight.
Table 1. scfv sequence alignment on ClustaIW.
Table 2. The results of scfv sequence analysis on IGBLAST.
The recombinant plasmid was extracted by E.Z.N.A.® Plasmid Mini Kit I (Omega, D642). The extracted recombinant plasmid identified by restriction endonuclease enzyme SfiI digestion. A total of 10 recombinant plasmids identified by restriction enzyme digestion, sent to Guangzhou branch of biotech Co., Ltd., sequencing, CmSuite8 software, VBASE2 database for DNA sequence and amino acid analysis, and sequence comparison through ClustaIW and IGBLAST. Then, induced expression of anti-S. pneumoniae scfv was conducted.
Radioactivity autoradiography and the DNA glass milk recovery method was used to identify and recover DNA.
Detection of scfv binding activity of soluble anti-S. pneumoniae antibody by ELISA
The ELISA method is the same as mentioned above. ELISA plate was coated with S. pneumoniae whole bacteria antigen. The soluble scfv supernatant of 67 McAbs and culture supernatant of one empty plasmid (negative control) was added to each plate, then 37°C incubated for 1 h. After washing with PBST and adding I antibody and II antibodies, the reaction was terminated after the colorable reaction. The steps are the same as before. The absorbance (OD) value of each hole was measured by 450 nm. The negative control and blank control were set up. The detection results of the P/N ratio were larger than 2.1 is positive. Then, detected the specificity of positive soluble S. pneumoniae scfv by ELISA method.
Results
Detection of serum specific IgG by indirect ELISA
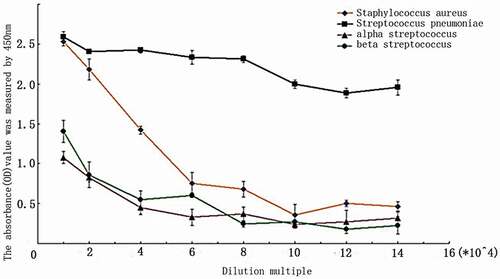
The patient’s serum was diluted by 1:10,000, 1:20,000, 1:40,000, 1:60,000, 1:80,000, 1:1,00,000, 1:1,20,000, and 1:1,40,000 and detected by indirect ELISA. The results showed that the serum specific IgG antibody was mainly against S. pneumoniae ().
VH, Vλ, andVκ gene amplification
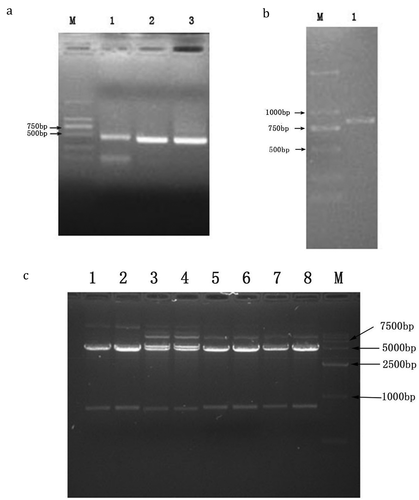
Extraction of total RNA from human tonsil lymphocytes specific binding to S. pneumoniae after the separation of immunomagnetic beads and reverse transcriptional synthesis of the first chain of cDNA; cDNA employed as a template to amplify the immunoglobulin rearranged variable genes by using the upstream and downstream primers described in –. The analysis result of electrophoresis shows that DNA fragments were visible clearly at about 400 bp (), which was consistent with the expected length.
Figure 3. (a) Amplification of VH linker, Vλ linker, and Vκ linker gene by PCR, M: DNA marker DL2000. (1) PCR product of VH linker. (2) PCR product of Vκ linker. (3) PCR product of Vλ linker. (b) Purified PCR products of scfv fragment. M, DNA marker DL2000 1: purified PCR products of scfv. (c) The identification of recombinant clones by SfiI. M: DNA marker DL15000. Channel 1–8, the identification of pMo Pac16-scFv by Sfi.
scfv gene amplification
Using Hsback and Hsfor as primers, VH linker and Vλ linker, VH linker and Vκ linker were connected by the connection peptide linker. The overlap extension (SOE) was carried out by PCR, and the amplified product was purified by 1% agarose gel electrophoresis and then analyzed by electrophoresis. A clear bright band appears at around 800 bp (), and the scfv gene is consistent with the expected length.
Identification of recombinant plasmids by enzyme digestion
The transformed bacterial cells were seeded on the Ampicillin LB plate until appropriate and homogeneous colonies were found. Recombinant plasmid pMo Pac16-scfv was extracted from E. coli, and the plasmid was analyzed by 1% agarose gel electrophoresis after Sfi I enzyme digestion. Each lane appeared clear and bright bands in the corresponding position about 800 bp and 5000 bp (). The results showed that the scfv gene had been successfully cloned to plasmid pMo Pac16. The band at 800 bp was expected the length of the theoretical scfv. Third and fourth lane about the location of the 5000 bp visible 2 clear and bright bands, the lower band is considered to be pMo Pac16-scfv after being cut by Sfi I enzyme, the upper band is an incomplete pMo Pac16-scFv after Sfi I enzyme digestion, which was in accordance with the theory. In the result, the impurity band which more than 7500 bp at first to eighth lanes is considered part of supercoiled plasmid or open-loop plasmid. Because in the electrophoresis of the super helix plasmid, the swimming speed is not exactly proportional to the size of the electrophoresis, and the open loop plasmids cause the electrophoretic swimming speed to be slow due to the partial dissolving of the chain.
DNA sequencing and diversity analysis of scfv gene
10 correct recombinant plasmids were selected and sent to Sangon Biotech Co., Ltd for sequencing. Analysis of DNA sequence and amino acid sequence through CmSuite8 software and VBASE2 database, and sequence alignment through ClustaIW and IGBLAST. The results of DNA sequencing showed that 10 single-chain antibodies were complete and constructed correctly. Figures S1 and S2 show the results of one of the scfv genes (sequence number 2) DNA, amino acid sequence analysis and IGBLAST analysis. The total length of the scfv gene was 732 bp and encoded 244 amino acids. The VH gene is 357 bp length and encodes 119 amino acids, the VL gene is 330 bp length and encodes 110 amino acids, the linker gene is 45 bp, and the encoding 15 peptide sequence (Gly4 Ser) 3, coincide with expectations. The results of the VBASE2 database analysis showed that both VH and VL contained four framework regions and three complementary determinant areas (complementarity determining region). IGBLAST results showed that the VH gene was highly homologous to IGHV3-30-3*01 (91.2%), the VL gene is completely homologous with IGLV6-57*02 (100%). The results of ClustaIW showed that the other sequences were obviously different except scfv sequence 6 and sequence 9, as shown in . The V gene of each sequence was highly homologous to the human IgG V gene, all other scfv sequences have no homology light chain and heavy chain, see . The results showed that the built scfv library was rich in diversity.
Detection of binding activity of soluble anti-S. pneumoniae scfv by ELISA
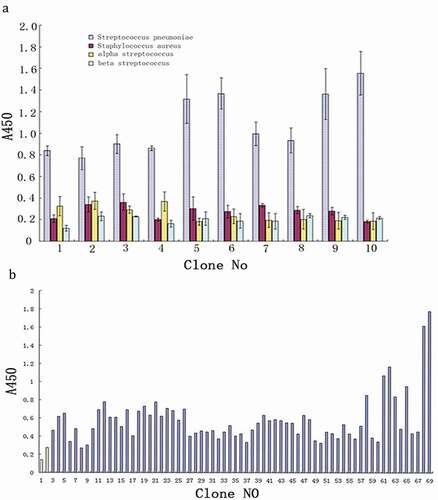
A total of 67 clones induced expression of soluble scfv of anti-S. pneumoniae and the empty plasmid transformants as negative control were selected randomly. Whole bacteria of S. pneumoniae was used as the envelope antigen, and the anti-S. pneumoniae scfv was detected in the ELISA method, and the blank control was set up. The detection results of the P/N ratio was more than or equal to 2.1 were considered as positive. As shown in , except for samples 6, 8, 9, 17, 27, 32, 37, 49, 50, 53, 55, 55, and 55(total of 13, accounting for 19.4%), the rest were positive, indicating that scfv had good binding activity.
Detection of the specificity of soluble anti-S. pneumoniae scfv by ELISA
Ten monoclonal strains with the highest activity were selected to induce the expression of positive soluble S. pneumoniae scfv. S. aureus, alpha Streptococcus, beta Streptococcus, and S. pneumoniae were used as inclusion antigens, respectively. The binding activity of positive S. pneumoniae scfv to four bacteria was detected by ELISA and established the negative control (empty plasmid transformant) and blank control. As shown in , the results showed that the positive scfv of S. pneumoniae was significantly higher than that of S. aureus, alpha Streptococcus, beta Streptococcus (OD value were mote than two times), with good specificity.
Discussion
In this study, immunomagnetic beads sorting method was used to extract antibody-producing cells from human tonsils, which was specifically combined with S. pneumoniae, and extracted RNA from it. By reverse transcription-polymerase chain reaction (RT-PCR), VH, Vλ, and Vκ genes were successfully amplified. The anti-S. pneumoniae scfv gene was constructed by SOE then was connected with pMo Pac16 and converted to Rosetta (DE3), finally successful constructed the human tonsillar antibody producing cell anti-S. pneumoniae single-chain antibody prokaryotic expression library, sequence alignment analysis by ClustaIW and IGBLAST showed that the single-chain antibody library have abundant diversity, which laid the foundation for screening high-affinity human tonsil antibody-producing cells against S. pneumoniae single-chain antibody. The induced expression of S. pneumoniae single-chain antibody by ELISA analysis showed that it had high binding activity (54 of the 67 clones were positive, accounting for 80.6%) and specificity (combined with the whole cell antigen of S. pneumoniae was significantly higher than that of S. aureus, alpha Streptococcus, beta Streptococcus OD value was greater than others in two times).
Chronic tonsillitis is a common clinical disease, but little progress was made in the pathogenesis of chronic tonsillitis, and the treatment way mainly relies on surgery. This study simulates tonsil local lymphocytes produce antibodies process in tonsil disease immune response after the invasion of pathogenic bacteria, successfully constructed human single-chain antibody library related to chronic tonsillitis. Compared with the previous bacterial culture and identification of chronic tonsillitis, our study of the local tonsil cells antibody can specifically reflect the invasion of chronic tonsillitis pathogenic bacteria. Furthermore, single-chain antibody as a diagnostic antibody for detection of pathogens is more rapid than traditional methods of pathogen culture and identification. In addition, single-chain antibodies as diagnostic antibodies are more specific and sensitive than conventional monoclonal antibodies [Citation15]. Compared with the traditional antibacteria, antiviral treatment, single-chain antibody treatment is comparatively better due to its directed therapy, targeted, high specificity, nonspecific binding, very few side effect, simple preparation process, low cost, and its high potential clinical application. Even so, it is undeniable that the scfv also has its limitations, such as low expression, low affinity [Citation16], and poor stability.
In this study, we constructed a scfv antibody specific to the tonsil antibody by high-throughput scfv library direct screening technique. Compared with traditional technology like phage display and ribosome display technology, this method which we used for screening were similar to single B cells monoclonal antibody technology, directly screening from human single B cells level, the advantage is can make antibodies affinity higher and specific. Removing a lot of unrelated antibody gene can greatly reduce the storage capacity of the antibody library and get high-affinity scfv. Not only reduce the workload and construction cost but also make the screened antibody can better reflect the real status in the patient’s natural infection condition. B cells monoclonal antibody technology specifically select B cells mainly use the random isolation method (micromanipulation, laser capture microdissection, or fluorescence-activated cell sorting) or the antigen-selective isolation method (microengraving/cell-based micro-array chip systems) [Citation17]. Compared with the single B cells monoclonal antibody technology, the operation procedure is simpler, faster, and cheap. In addition, this experiment screened the whole cell as the antigen, which could make the antigen maintain its complete and natural conformation and could simulate the natural invasion progress of S. pneumoniae in the tonsil. Subsequently, we extracted the total RNA of tonsil lymphocytes screened by immunomagnetic bead sorting technique, based on the genetic theory of antibody diversity [Citation18–Citation20], a specific primer was designed, and the coding sequence of the restriction fragment and restriction site was designed in the primer, RT-PCR technology was used to amplify VH, Vλ, and Vκ genes. Then the overlap extension splicing method was used, and the 15-peptide sequence (Gly4 Ser) 3 (Linker) which was designed by Huston [Citation9] was used to connect VH and Vλ, VH and V κ, then the scfv was constructed. The renaturation peptide (Gly4Ser) three refolding process appeared high efficiency, it maybe because its construction is long and soft. When refolding, it can effectively reduce the steric hindrance between VH and VL and Vκ, which is more conducive to the correct folding of scfv domains. We chose E. coli as an expression system of single-chain antibody. Because it is simple and rapid in cultivation and expression process and convenient for subsequent research or clinical application. The soluble expression was chosen to make the active single-chain antibody can be directly obtained and convenient for the subsequent detection of immune activity. We induced the expression of pMo Pac16-scFv/Rosetta (DE3) to obtain soluble anti-S. pneumoniae single-chain antibody. The binding activity of recombinant clones was detected by ELISA, and 10 positive scfv with the highest binding activity were selected for specific detection by ELISA, the results showed that the single-chain antibody had good binding activity and specificity. At this point, the antipneumococcal antibody library which has high affinity and specificity was successfully constructed. Our study would contribute for chronic tonsillitis etiological research, in the meantime, it is a first step for created a new way to using the human antibody to diagnose and treat chronic tonsillitis and maybe can change the current situation that surgery was almost deemed as the only way for chronic tonsillitis treatment.
In addition, this research still has great development space, such as by immunofluorescence, immunoblotting, and immune blocking test was carried out for further identification of single-chain antibody can increase the biological activity; ELISA-coated antigen types, further detection of specific scfv antibody against S. pneumoniae. The screened antigens can be changed from S. pneumoniae whole body antigen to capsular polysaccharide or other pathogenic antigens, so that single-chain antibody can be more specific and reduce antigen cross reaction; reduction of antigenic cross reaction; it can expand the study of pathogenic bacteria and prepare specific antibodies against Streptococcus hemolytic Streptococcus, and also can expand the study of pathogenic bacteria and prepare the specific antibodies against beta-hemolytic Streptococcus, which is significant for the treatment of heart and kidney diseases caused by the immune and inflammatory reaction induced by the bacteria.
Conclusions
This study constructed a prokaryotic expression human single-chain antibody fragments library against S. pneumoniae, the diversity of the library is rich. Detection results show the binding activity and specificity of anti-S. pneumoniae scfv acquired by the inducible expression is fairly good. The human single-chain antibody maybe can create a new strategy for tonsillitis etiologic diagnosis and therapy.
Supplemental Material
Download MS Word (81.1 KB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
supplemental data for this article can be accessed here.
References
- Windfuhr JP, Toepfner N, Steffen G, Waldfahrer FBerner R. 2016. Clinical practice guideline: tonsillitis ii. surgical management. European Archives Of Oto-rhino-laryngology 273 (4):989–1009.
- Windfuhr JP, Toepfner N, Steffen G, et al. Clinical practice guideline: tonsillitis I. Diagnostics and nonsurgical management[J]. Eur Arch Otorhinolaryngol. 2016;273(4):973–987.
- Khadilkar MN, Ankle NR. Anaerobic bacteriological microbiota in surface and core of tonsils in chronic tonsillitis[J]. J Clin Diagn Res. 2016;10(11):MC1–MC3.
- Dias EP, Rocha ML, Carvalho MO, et al. Detection of Epstein-Barr virus in recurrent tonsillitis[J]. Braz J Otorhinolaryngol. 2009;75(1):30–34.
- Proenca-Modena JL, Buzatto GP. Respiratory viruses are continuously detected in children with chronic tonsillitis throughout the year. Int J Pediatr Otorhinolaryngol. 2014;78(10):1655–1661.
- Kohler G. Milstein C.Continuous cultures of fused cells secreting antibody of predefined specificity[J]. Nature. 1975;256(5517):495–497.
- Elvin JG, Couston RG. van der Walle CF. Therapeutic antibodies: market considerations, disease targets and bioprocessing[J]. Int J Pharm. 2013;440(1):83–98.
- Yokota T, Milenic DE, Whitlow M, et al. Rapid tumor penetration of a single-chain Fv and comparison with other immunoglobulin forms[J]. Cancer Res. 1992;52(12):3402–3408.
- Qamsari ES, Sharifzadeh Z, Bagheri S. Isolation and characterization of anti c-met single chain fragment variable (scFv) antibodies [J]. J Immunotoxicol. 2017;14(1):23–30.
- Chowdhury S, Viner JL, Beers R, et al. Isolation of a high-affinity stable single-chain Fv specific for mesothelin from DNA-immunized mice by phage display and construction of a recombinant immunotoxin with antitumor activity [J]. Proc Natl Acad Sci U S A. 1998;95(2):669–674.
- Saeed M, van Brakel M, Zalba S. Targeting melanoma with immunoliposomes coupled to anti-MAGE A1 TCR-like single-chain antibody [J]. Int J Nanomedicine. 2016;11:955–975.
- Chari RVJ. Targeted delivery of chemotherapeutics: tumor-activatedprodrug therapy[J]. Adv Drug Deliv Rev. 1998;31(1–2):89–104.
- Gattenl¨Ohner S, J¨Orissen H, Huhn M, et al. A human recombinant autoantibody-based immunotoxin specific for the fetal acetylcholine receptor inhibits rhabdomyosarcoma growth in vitro and in a murine transplantation model[J]. J Biomed Biotechnol. 2010;2010:11, Article ID 187621
- Pansri P, Jaruseranee N, Rangnoi K, et al. A compact phage display human scFv library for selection of antibodies to a wide variety of antigens[J]. BMC Biotechnol. 2009;9:6.
- Sapats S, Gould G, Trinidad L, et al. An ELISA for detection of infectious bursal disease virus and differentiation of very virulent strains based on single chain recombinant chicken antibodies[J]. Avian Pathol. 2005;34:449–455.
- Paul S, Nishiyama Y, Planque S, et al. Theory of proteolytic antibody occurrence[J]. Immunol Lett. 2006;103(1):8–16.
- Chi X. Single B cell monoclonal antibody technologies and applications[J]. Chin J Biotechnol. 2012;28(6):651–660.
- Thorn SN. Large rate Auelerations in antibodies catalysis by Strategic use of haptenic charge[J]. Nature. 1995;373:228.
- Market E, Papavasiliou FN. V(D)J recombination and the evolution of the adaptive immune system[J]. PLoS Biol. 2003;1(1):E16.
- Diaz M, Casali P. Somatic immunoglobulin hypermutation[J]. Curr Opin Immunol. 2002;14(2):235–240.