?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Abscisic acid (ABA) signaling regulates plant growth and development and participates in response to abiotic stressors. However, details about the PYL-PP2C-SnRK2 gene family, which is the core component of ABA signaling in Camellia sinensis, are unknown. In this work, we identified 14 pyrabactin resistance-likes (PYLs), 84 type 2C protein phosphatase (PP2Cs), and 8 SNF1-related protein kinase 2s (SnRK2s) from C. sinensis. The transcriptomic analysis indicated that PYL-PP2C-SnRK2s were associated with changes of leaf color and the response of C. sinensis to drought and salt stressors. Changes of the expression of Snrk2s were not significant in the process of leaf color change or drought and salt stress response, suggesting that PYLs and PP2Cs may not interact with SnRK2s in C. sinensis during these processes. Finally, Gene Regulatory Network (GRN) construction and interaction networks analysis demonstrated that PYLs and PP2Cs were associated with multiple metabolic pathways during the changes of leaf color.
1. Introduction
Abscisic acid (ABA) is a phytohormone that plays an important role in plant growth and development including seed germination and fruit ripening [Citation1–Citation5]. In addition, ABA signaling pathways improve plant tolerance to multiple abiotic stressors, such as drought, salinity, and cold [Citation6].
Although numerous factors associated with ABA responses had been identified prior to 2009 [Citation7], the model of ABA signaling pathway is limited. Multiple components of the ABA signaling pathway, such as the ABA receptors and binding proteins, were randomly located in different cellular locations [Citation8]. Since 2009, the discovery of PYL-PP2C-SnRK2s as a key component to ABA responses has allowed for a more in depth understanding of ABA signaling. In the PYL-PP2C-SnRK2 gene family, type 2C protein phosphatase (PP2C) and SNF1-related protein kinase 2 (SnRK2) are downstream components of a type of soluble ABA receptor, PYR (pyrabactin resistance)_/PYL (PYR1-like)/RCARs (regulatory components of ABA receptor) [Citation9–Citation12]. ABA signaling as a double negative regulatory model consists of four stages, including PYR/PYL/RCARs, PP2Cs, SnRK2s, and their downstream targets [Citation11,Citation13]. In the absence of ABA, Group A PP2Cs inhibits the activity of SnRKs by dephosphorylation [Citation7,Citation14,Citation15]. ABA signaling is activated by three stages in the presence of ABA. First, the change of structure of PYR/PYL/RCARs is caused by the interaction between ABA, and receptors and then PYR/PYL/RCARs bind PP2Cs to repress phosphatase activities of PP2Cs [Citation10]. Second, kinase activity of SnRK2s released from the PP2C-SnRK2 complex is restored by self-phosphorylation. In the last stage, SnRK2s activated downstream transcription factors, which regulated ABA response genes [Citation11,Citation16,Citation17].
Additionally, the members of the PYL-PP2C-SnRK2 gene family have been characterized and they are involved in growth, development, and a variety of responses to abiotic stressors. Pyrabactin resistance1 (PYR1), an ABA receptor, was first discovered in Arabidopsis [Citation10]. Subsequently, multiple PYLs were identified in Arabidopsis, such as RCAR1 [Citation9]. All PYLs belong to the START/Betvl protein family, which contains a START domain [Citation6]. Overexpression of the PYL genes GhPYL10, GhPYL12, and GhPYL26 from Gossypium spp. in Arabidopsis enhanced the plant tolerance to drought stress [Citation18]. Conversely, the quadruple mutant of pyr1, pyl1, pyl2, and pyl4 in Arabidopsis was insensitive to ABA [Citation10]. In rice, OsPYL2, OsPYL10, and OsPYL11, positive regulators of ABA, play an important role in seed germination, early seedling development, drought tolerance, and cold tolerance [Citation19]. As a core component of ABA signaling, PP2C proteins contain a conserved catalytic domain on the C-terminus. In Arabidopsis, a total of nine Group A PP2Cs were obtained. Of these PP2Cs, AtABll and AtABl2 can regulate the development and stomatal movement of Arabidopsis in the late stage of germination [Citation9]. TaPP2C genes are related to developmental processes and stress responses in Triticum aestivum [Citation20]. Significant ABA hypersensitivity was observed after loss-of-function mutations of Group A PP2Cs, such as ABI1, ABI2, HAB1, HAB2, AHG1, and PP2CA [Citation7,Citation21–Citation25], indicating that they play a negative regulatory role in ABA signaling. As a unique gene family in plants, the SnRK2 family consists of 10 members in Arabidopsis, including SnRK2.1-SnRK2.10. In these SnRKs, AtSnRK2.2, AtSnRK2.3, AtSnRK2.6, AtSnRK2.7, and AtSnRK2.8 can be induced by ABA [Citation26,Citation27]. Overexpression of OsSAPK6 in rice showed high sensitivity to ABA [Citation28].
Camellia sinensis (tea), a highly nutritious woody plant, is widely distributed, especially in subtropical to tropical regions [Citation29]. Although, the function of PYL-PP2C-SnRK2s in plant development and stress responses is well known, the role of PYL-PP2C-SnRK2s in C. sinensis has not yet been studied. In this study, we aimed to identify the PYL-PP2C-SnRK2s family from the whole genome of C.sinensis and investigate the functions of the family in abiotic stress and growth and development. This is the first functional study of the PYL-PP2C-SnRK2s family in C. sinensis and their phylogenetic relationships, chromosome distribution, protein motifs, gene structure, and expression patterns under drought and salt stress as well as in leaves of different colors were investigated. Further, the gene regulatory network (GRN) between PYL-PP2C-SnRK2s (regulators) and their co-expression genes (targets) was constructed. This systematic study will enable us to better understand the role of the core components of ABA signaling in C. sinensis and provide a solid foundation for improving the yield and quality of C. sinensis.
2. Materials and methods
2.1. Identification of PYL-PP2C-SnRK2 genes in C. sinensis
Protein sequences of C. sinensis var. sinensis were downloaded from the previously published genome database (http://tpia.teaplant.org/) [Citation30]. The Arabidopsis Information Resource (TAIR) (https://www.arabidopsis.org/download/index.jsp/) and Rice Genome Annotation Project (RGAP) (http://rice.plantbiology.msu.edu/) were used to obtain PYL, PP2C, and SnRK2 protein sequences of Arabidopsis and rice, respectively [Citation31]. All PYL-PP2C-SnRK2s from Arabidopsis and rice were used as queries to identify tea PYL, PP2C, and SnRK2 genes from the C. sinensis database by employing HMMER software and BLAST. The conserved domains of candidate PYL-PP2C-SnRK2s were verified with the online programs NCBI’s conserved domain database (CCD) and protein families database (PFAM) [Citation32,Citation33]. All PYL-PP2C-SnRK2s identified in C. sinensis are shown in Table S1. The abbreviation of the species name Camellia sinensis (Cs) is the beginning of each gene name and the most prominent Arabidopsis gene from this subfamily was defined as the followed name. Genes not found in Arabidopsis are named after the rice gene. The PYL-PP2C-SnRK2 proteins from C. sinensis, Arabidopsis, and rice were selected to construct a Maximum Likelihood tree using MEGA 7.0 software (bootstrap values for 1,000) [Citation34,Citation35].
2.2. Conserved motifs and gene structure analysis of C. sinensis PYL-PP2C-SnRK2s
Motifs of C. sinensis PYL-PP2C-SnRK2s were identified by using the online program MEME with the maximum number of motifs set to 10 as the parameter. All motifs identified were annotated using InterProScan [Citation36,Citation37]. The gene structure of PYL-PP2C-SnRK2s was analyzed by using the Gene Structure Display Server (GSDS) [Citation38]. The composite picture of gene structure and the phylogenetic tree was generated using Tbtools software [Citation39].
2.3. Chromosomal distribution and gene duplication
The R package Rideogram and Tbtools were used to show the distribution of all C. sinensis PYL-PP2C-SnRK2s on scaffolds.
2.4. Transcriptomic analysis and gene expression patterns
All transcriptomic data were obtained from the publicly available NCBI-SRA database (Drought and salt stressors: ERP012919; different leaf colors: SRP055910). The raw data in SRA format downloaded from the NCBI-SRA database were converted to FASTQ format by using fastq-dump (https://trace.ncbi.nlm.nih.gov/Traces/sra/sra.cgi?view=toolkit_doc&f=fastq-dump). The reference genome was obtained from the previously published genome database CSS(cv.Shuchazao). Subsequently, low quality sequences were removed to generate clean reads by FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/). Three programs were used to analyze clean reads and to obtain the expression levels of all genes based on Fragments Per Kilobase Million (FPKM), HISAT 0.1.5, StringTie (v2.0.4), and Ballgown (a R package) [Citation40–Citation43]. Differentially expressed genes (Log2 fold change >1 or Log2 fold change <−1; P-value < 0.05) in drought and salt stress conditions and in leaves of different colors were identified by performing edgeR (http://www.bioconductor.org/packages/release/bioc/html/edgeR.html) and Ballgown, respectively.
2.5. KEGG pathway enrichment analysis
To further understand the biological functions of genes, a Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis was performed. The significantly enriched pathway analysis uses KEGG as a unit to apply hypergeometric tests to find pathways that are significantly enriched in differentially expressed genes compared to the entire genomic background. This was calculated as follows [Citation44]:
where N is the total number of genes; n is the number of differentially expressed genes in N; m is the number of genes annotated as a particular pathway; M is the number of differentially expressed genes annotated as a particular pathway.
2.6. Gene regulatory network construction KEGG enrichment analysis
An unsupervised GRN was constructed by using the R package GENIE3 with the random forest machine learning algorithm. In this GRN, a total of 229,504 edges were generated and the top 20% of edges (rank by edge core) were defined as high confidence edges [Citation45].
3. Results
3.1. Identification and characteristics of PYL-PP2C-SnRK2s in C. sinensis
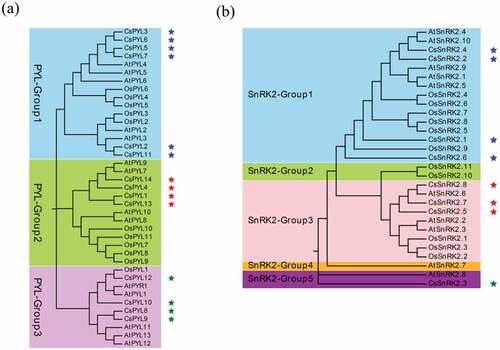
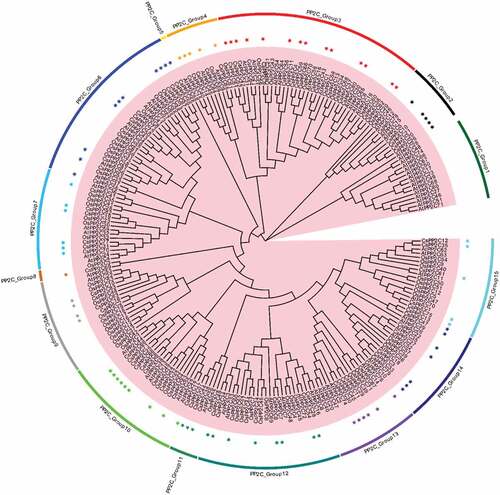
In total, we identified 8 SnRK2s, 84 PP2Cs, and 14 PYLs genes in C. sinensis by performing multiple bioinformatics software (Described in materials and methods) (Table S1). Maximum Likelihood phylogenetic trees reconstructed with the complete PYL-PP2C-SnRK2 protein sequences from C. sinensis, Arabidopsis, and rice showed 3 (PYL_Group1-3), 15 (PP2C_Group1-15), and 5 (SnRK2_Group1-5) subgroups of the PYL, PP2C, and SnRK2 families, respectively ( and ). Multiple orthologous PYL-PP2C-SnRK2s between C. sinensis and Arabidopsis were identified, suggesting that relationships with PYL-PP2C-SnRK2s between C. sinensis and Arabidopsis were closer than that between C. sinensis and rice. These results suggested that the gene phylogeny was in accordance with the species phylogeny. There was no significant difference in the number of PYL-PP2C-SnRK2s in C. sinensis (PYL: 14, PP2C: 84, SnRK2: 8), Arabidopsis (PYL: 14, PP2C: 74, SnRK2: 10), and rice (PYL: 11, PP2C: 72, SnRK2: 11).
3.2. Conserved motifs and gene structure analysis of C. sinensis PYL-PP2C-SnRK2s
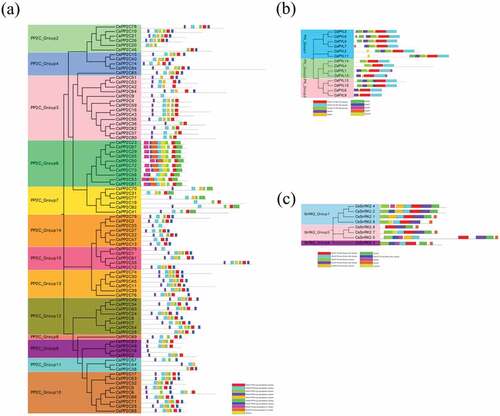
According to the phylogenetic relationships, the conserved motifs of C. sinensis PYL-PP2C-SnRK2 proteins were analyzed using MEME and InterPro databases and ten motifs in each gene family were acquired (). For the C. sinensis PP2C family, the PPM-type phosphatase domain and protein phosphatase 2C families were annotated by protein sequences of CsPP2Cs in motifs 1–7 and 8–9, respectively. Motifs 1–4 were included in all of the subgroups and PP2C_Group 6 has motifs 9 and 10, whereas motif 6 was only present in PP2C_Groups 6 and 7 (). For the C. sinensis PYL family, the protein sequences of motifs 1–3 involved the START-like domain. Motifs 1 and 2 were present in all of the identified CsPYLs. Almost all of the CsPYLs contained motif 3, except for CsPYLs 13, 8, and 9 ()). In the C. sinensis SnRK2 family, motifs 1–5 and 7 were annotated as a protein kinase (-like) domain. All the CsSnRK2s contained motif 1; motifs 2 and 3 not present in CsSnRK2.8, suggesting that all identified CsSnRK2s contained protein kinase domains. In addition, the gene structure of C. sinensis PYLPP2C-SnRK2s was analyzed; exon-intron organizations in the same subgroups were similar (Figure S1). These results indicate that typical family features exist in all PYL-PP2C-SnRK2s obtained from C. sinensis.
3.3. Chromosomal distribution, gene duplication, and syntenic analysis of C. sinensis PP2C-PYL-SnRK2s
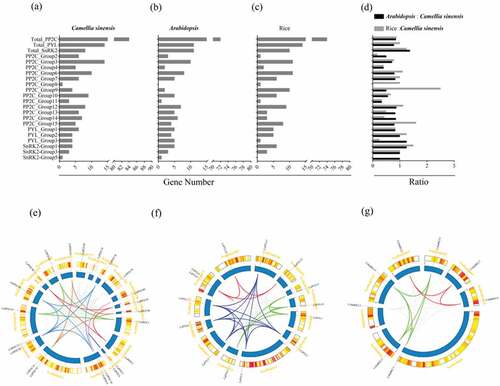
Scaffolds mapped with PP2C-PYL-SnRK2s were shown in the absence of C. sinensis chromosome data. There were a total of 95 scaffolds related to all PP2Cs (80), PYLs (13), and SnRK2s (8). Six shared scaffolds were mapped with PP2C and PYL (Figure S2, Table S2). PP2C-PYL-SnRK2s were unevenly distributed on all scaffolds. Despite the phylogenetic distance of rice, C. sinensis, and Arabidopsis, the number of PP2C-PYL-SnRK2s was similar (–). In addition, the number of most orthologs between C. sinensis and Arabidopsis or rice was similar or even the same in some subgroups, such as PP2C_Group 7 and 8, as well as SnRK2-Group 3 and 5. These results suggest that the number of PP2C-PYL-SnRK2s is conserved during evolution. Duplication events were analyzed with Tbtools software to detect syntenic blocks. Syntenic relationships of scaffolds mapped with all PYLs and SnRK2s and scaffolds containing at least 20 PP2C genes were analyzed (–). We found that genes from the same phylogenetic subgroup were located on multiple scaffolds and most gene replication events occurred on the same scaffold.
Figure 4. Number and location of PP2C-PYL-SnRK2s. The number of PP2C-PYL-SnRK2s identified per subgroup in (a) C. sinensis, (b) Arabidopsis, and (c) rice. (d) The ratio of the total number of PP2C-PYL-SnRK2 genes in all subgroups is shown for Arabidopsis compared to C. sinensis (black) and rice compared to C. sinensis (gray). (e, f, g) All PP2C-PYL-SnRK2 genes are mapped to the C. sinensis scaffolds in a circular diagram using Tbtools. Scaffolds mapped with all PYLs and SnRK2s and scaffolds containing at least 20 PP2C genes are shown. Links represent different genes in the same subgroups
3.4. Expression analysis of PYL-PP2C-SnRK2 genes in response to salt and drought stress
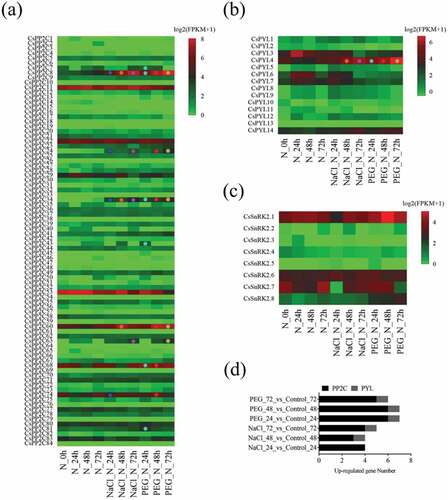
To explore the role of PYL-PP2C-SnRK2 genes in C. sinensis in response to abiotic stress, transcriptomes of leaves treated with 200 mM NaCl and 25% polyethylene glycol (PEG) for different periods of time (24 h, 48 h, and 72h) were downloaded from the publicly available NCBI-SRA database (ERP012919) (, Table S3). Under the NaCl and PEG treatments for 24 h, 0/14 and 1/14 PYLs, 4/84 and 6/84 PP2Cs, and 0/8 and 0/8 SnRK2s showed significant upregulation, respectively (Log2 (fold change) >1; P-value < 0.05) (). After 48 h, 1/14 and 1/14 PYLs, 3/84 and 6/84 PP2Cs, and 0/8 and 0/8 SnRK2s showed significant upregulation, respectively. After 72 h, 1/14 and 1/14 PYLs, 4/84 and 5/84 PP2Cs, and 0/8 and 0/8 SnRK2s showed significant upregulation, respectively. Of note, CsPP2C8 and CsPP2C24 showed up-regulated expression levels at all treatment stages and CsPP2C 24, 60, 68, 74 showed high expression levels at multiple stages (). In addition, only one PYL gene CsPYL4 was induced by salt and drought stress (). These results suggest that PYL-PP2C-SnRK2 genes were involved in the response of C. sinensis to salt and drought stress; overall, six PP2C genes (CsPP2C8, 24, 24, 60, 68, and 74) and one PYL gene (CsPYL4) plays an important role in this process.
Figure 5. Expression profiles of PP2Cs (a), PYLs (b), and SnRK2s (c) in response to salt and drought treatments in C. sinensis. Log2 (FPKM+1) was used to create the heat map. Green indicates low expression, whereas red indicates a high level of expression. Circles of different colors represent differential gene expression during different treatments compared with the controls. (d) The histogram shows the distribution of differential expression of PP2Cs, PYLs, and SnRK2s in different comparisons
3.5. Expression analyses of PYL-PP2C-SnRK2 genes during leaf development and color change
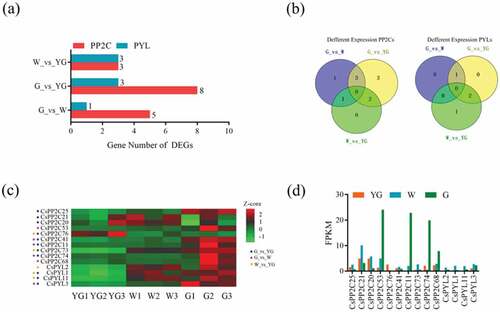
According to previous report, a total of three stages were defined in the development of new C. sinensis shoots, including the yellow-green (YG) stage, albescent (W) stage, and re-greening (G) stage [Citation46]. The changes of leaf color involve a variety of metabolic pathways related to plant growth and development, which was affected by ABA signaling [Citation1–Citation5]. Therefore, PYL-PP2C-SnRK2 genes may be involved in the changes of color in C. sinensis leaves. In order to verify this hypothesis, the transcriptomes of leaves at these three stages were obtained from the publicly available NCBI-SRA database (SRP055910) and expression levels of all mRNAs based on FPKM values was calculated (Figure S3, Table S4). The expression level of all PYL-PP2C-SnRK2s from mRNAs was generated (Table S5). In total, 1,437 (up: 839; down: 598) DEGs in G vs W, 3,457 (up: 1,790; down: 1,667) DEGs in G vs YG, and 1,409 (up: 710; down: 699) in W vs YG were generated (Figure S4, Table S6). There were three PP2Cs and three PYLs in G vs YG, eight PP2Cs and three PYLs in G vs YG, and three PP2Cs, and one PYLs in G vs W generated from all DEGs (). All differentially expressed PP2Cs of W vs YG existed in the other two comparisons () and CsPP2C 11 (CSS038291), 41 (CSS030837), 73 (CSS047922), 74 (CSS049314), 76 (CSS029712) and CsPYL1 (CSS036090), 11 (CSS044035), 3 (CSS050443) were differentially expressed in two comparison groups (). Compared with stage YG, 9/14 and 7/14 genes were upregulated in the W and G stages (). These results suggest that PP2Cs and PYLs are involved leaf color changes. In addition, changes in the expression levels of Snrk2s were not significant, indicating that PP2Cs and PYLs may affect the changes of leaf color via a mechanism not involving SnRK2s.
Figure 6. Expression analyses of PYL-PP2C-SnRK2 genes during leaf development and color change. (a) The histogram shows the number of differential expressed PYL-PP2C-SnRK2 genes in three comparisons, W_vs_YG, G_vs_YG, and G_vs_W. (b) The Venn diagrams show the distribution of differential expression of PP2Cs and PYLs in three comparisons. (c) The expression profile of the differential expression of PP2Cs and PYLs is shown with a heatmap based on the Z-core value. (d) The histogram shows the expression level of all of the differentially expressed PP2Cs and PYLs.
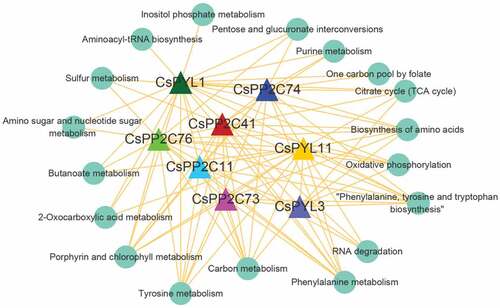
3.6. Regulatory modules of PYL-PP2C-SnRK2 genes related to changes of C. sinensis leaf color
To further explore the regulatory patterns of gene expression during changes of leaf color, a GRN was used to create a directed network of PYL-PP2C-SnRK2 genes and their target genes. Since CsPP2C 11 (CSS038291), 41 (CSS030837), 73 (CSS047922), 74 (CSS049314), 76 (CSS029712) and CsPYL1 (CSS036090), 11 (CSS044035), 3 (CSS050443) were differentially expressed in two comparison groups, they were used to construct the GRN as regulators and 28,682 co-expression mRNAs as target genes. An independent GRN was generated, which included 229,504 edges and the top 20% edges (rank by edge core) (Table S7). All target genes of each regulator were subjected to a KEGG pathway enrichment analysis. Pathways involving at least ten target genes were used to construct an interaction network with regulators (). In this network, four pathways, ‘Carbon metabolism,’ ‘Phenylalanine metabolism,’ ‘Phenylalanine, tyrosine, and tryptophan biosynthesis,’ and ‘Biosynthesis of amino acids’ were affected by all regulators. ‘Tyrosine metabolism’ and ‘Porphyrin and chlorophyll metabolism’ were involved in almost all regulators, except CsPYL3. There were at least four regulators associated with the pathways ‘RNA degradation,’ ‘Citrate cycle (TCA cycle),’ and ‘Oxidative phosphorylation.’ In addition, the pathways ‘Aminoacyl-tRNA biosynthesis’ and ‘Inositol phosphate metabolism’ were only regulated by CsPYL1. These results indicated that PYL-PP2C-SnRK2s may affect the change of C. sinensis leaf color by regulating multiple metabolic pathways.
4. Discussion
The ABA signaling pathway allows a plant to respond to stress conditions, such as drought, salt, and cold stressors [Citation47–Citation49]. The characteristic of the PYL-PP2C-SnRK2 gene family, which is a core regulatory network of the ABA pathway in C. sinensis is unknown. Here, we identified eight SnRK2s, 84 PP2Cs, and 14 PYLs from the C. sinensis genome, which were assigned to four, 13, and three conserved subfamilies, respectively, based on a phylogenetic analysis ( and ). The evolutionary relationships of the PYL-PP2C-SnRK2 gene family between C. sinensis and Arabidopsis are closer than the relationships between C. sinensis and rice, which is consistent with the relatedness of these species. The similarity in the number of PYL-PP2C-SnRK2 genes in C. sinensis, Arabidopsis, and rice indicates conservation. The conserved motif analysis showed that the START-like domain, PPM-type phosphatase domain, and protein kinase domain were contained in all C. sinensis PYLs, PP2Cs, and SnRK2s, respectively. These characteristics identified in C. sinensis are in accordance with other plant species, such as apple and Arabidopsis [Citation48,Citation50]. All PP2C-PYL-SnRK2 genes were unevenly distributed on the scaffolds (Figure S2, TableS2) and replication events of most genes occurred on the same scaffold ().
The ABA signaling pathway is related to drought and salt stress response in plants [Citation47,Citation48]. The overexpression of OsPYL3 and OsPYL9 genes can increase rice tolerance to drought [Citation51], and NtPYLs have been shown to have important functions in the drought tolerance of Nicotiana tabacum [Citation52]. High-throughput next generation sequencing showed that PYL, PP2C, and SRK2E respond to drought stress in Gossypium spp [Citation53]. In banana, the PYL-PP2C-SnRK2 gene family regulated its tolerance to abiotic stress, such as salt and drought stress conditions [Citation54]. In this work, transcriptome data from C. sinensis treated with PEG and NaCl were obtained from the NCBI-SRA database and analyzed. For NaCl treatment, there were two PYLs and 11 PP2Cs that showed significantly upregulation at different stages. A total of three PYLs and 17 PP2Cs showed up-regulated expression in C. sinensis treated PEG. These results suggested that CsPYLs and CsPP2Cs may be associated with the tolerance of C. sinensis to drought and salt stress. In addition, the expression levels of all CsSnRK2s were not significant under NaCl and PEG treatments, indicating that CsSnRK2s may not be affected by drought and salt stress treatments.
Although, the function of the ABA signaling pathway on plant development is well known, no information is available regarding the effect of ABA on leaf color in C. sinensis. Thus, we analyzed the transcriptomes from different colored leaves of C. sinensis. In different colored leaves, the expression levels of multiple PYL-PP2C-SnRKs were significantly different. For example, CsPP2C 11 (CSS038291), 41 (CSS030837), 73 (CSS047922), 74 (CSS049314), 76 (CSS029712) and CsPYL1 (CSS036090), 11 (CSS044035), 3 (CSS050443) showed significantly different expression levels, which indicated that CsPP2Cs and CsPYLs may be involved in the regulation of leaf color changes. In addition, there was no significant change in the expression level of the CsSnRK2s in drought, or salt stress treatments, or among different leaf colors, suggesting that compared with CsSnRK2s, CsPP2Cs and CsPYLs play a more important role in the response of C. sinensis to drought and salt stress as well as leaf color.
The relationship between PYL-PP2C-SnRK2s and their targets was investigated using GRN, which is a directed network of regulators and their target genes. In this work, the regulators CsPP2C 11 (CSS038291), 41 (CSS030837), 73 (CSS047922), 74 (CSS049314), 76 (CSS029712) and CsPYL1 (CSS036090), 11 (CSS044035), 3 (CSS050443) were selected to construct a GRN, and 28,682 co-expression genes were selected as target genes. A KEGG enrichment analysis of target genes showed that CsPP2Cs or CsPYLs are involved in multiple metabolic pathways, such as ‘Phenylalanine metabolism’, ‘Phenylalanine, tyrosine, and tryptophan biosynthesis’, ‘Biosynthesis of amino acids’, and ‘Carbon metabolism’. In plants, phenylalanine metabolism is even more diverse and prevalent pathway. In fact, about 25% of fixed carbon from photosynthesis was used to generate phenylalanine [Citation55] and phenylalanine-derived compounds, which accounts for approximately 40% of plant organic matter [Citation56]. In addition, phenylalanine also affects plant characteristics in many aspects, such as growth and development (lignin) [Citation57], defense (salicylic acid [Citation58], tannins and flavonoids [Citation59]), reproduction (phenylpropanoids and benzenoids) [Citation60] and other metabolic pathways also play an important role in the growth and development of plants, indicating that CsPP2Cs and CsPYLs are essential for plant survival. SnRK2s are important components of the ABA signaling pathway in response to osmotic and salt stress in Arabidopsis [Citation61]. Interestingly, the expression levels of all SnRK2s did not show significant changes among different leaf colors or under salt and drought treatments in C. sinensis. The functional characteristics of SnRKs in C. sinensis need to be further clarified in future studies. This study improves the understanding of PYL-PP2C-SnRK2-mediated ABA signaling in the regulation of leaves change, response to drought and salt stressorse in C. sinensis. The identification of some candidate genes provides a molecular basis for improving C. sinensis quality and yield.
5. Conclusion
In summary, we identified 8 SnRK2s, 84 PP2Cs, and 14 PYLs genes from the whole genome of C. sinensis. The phylogeny of these genes is in accordance with the species phylogeny. Conserved motifs analysis indicates that all identified SnRK2s, PP2Cs, and PYLs contain protein kinase domain, PPM-type phosphatase domain and START-like domain, respectively. The transcriptomic analysis indicates that The PYL-PP2C-SnRK2s are related to C. sinensis responses to drought and salt stressors and participate in the regulation mechanism of leaf color change by multiple metabolic pathways. GRN construction and interaction networks analysis demonstrated that PYLs and PP2Cs were associated with multiple metabolic pathways during the changes of leaf color.
Highlights
The PYL-PP2C-SnRK2 family genes in Camellia sinensis were identified.
The ABA signaling is related to the response of C. sinensis to abiotic stress.
The PYL-PP2C-SnRK2s play an important role in the change of C.sinensis leaf colors.
Author Contributions
P. X., Y. W. and G. H. designed the experiments; P. X., X. Z. and H. S. performed the experiments and analyzed the data; X. Z. and X. L. contributed the methodology; P. X., and X. Z. wrote the original draft and Y. W. and G. H. revised and edited the paper.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
The supplemental data for this article can be accessed here.
Additional information
Funding
References
- Feng CZ, Chen Y, Wang C, et al. Arabidopsis RAV1 transcription factor, phosphorylated by SnRK2 kinases, regulates the expression of ABI3, ABI4, and ABI5 during seed germination and early seedling development. Plant J. 2014;80(4):654–668.
- Tijero V, Teribia N, Munoz P, et al. Implication of abscisic acid on ripening and quality in sweet cherries: differential effects during pre- and post-harvest. Front Plant Sci. 2016;7:602.
- Li D, Mou W, Luo Z, et al. Developmental and stress regulation on expression of a novel miRNA, Fan-miR73, and its target ABI5 in strawberry. Sci Rep. 2016;6:28385.
- Mou W, Li D, Bu J, et al. Comprehensive analysis of ABA effects on ethylene biosynthesis and signaling during tomato fruit ripening. PloS One. 2016;11(4):e0154072.
- Jia H, Jiu S, Zhang C, et al. Abscisic acid and sucrose regulate tomato and strawberry fruit ripening through the abscisic acid-stress-ripening transcription factor. Plant Biotechnol J. 2016;14(10):2045–2065.
- Cutler SR, Rodriguez PL, Finkelstein RR, et al. Abscisic acid: emergence of a core signaling network. Annu Rev Plant Biol. 2010;61:651–679.
- Hirayama T, Shinozaki K. Perception and transduction of abscisic acid signals: keys to the function of the versatile plant hormone ABA. Trends Plant Sci. 2007;12(8):343–351.
- McCourt P, Creelman R. The ABA receptors – we report you decide. Curr Opin Plant Biol. 2008;11(5):474–478.
- Ma Y, Szostkiewicz I, Korte A, et al. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science. 2009;324(5930):1064–1068.
- Park SY, Fung P, Nishimura N, et al. Abscisic acid inhibits type 2C protein phosphatases via the PYR/PYL family of START proteins. Science. 2009;324(5930):1068–1071.
- Umezawa T, Sugiyama N, Mizoguchi M, et al. Type 2C protein phosphatases directly regulate abscisic acid-activated protein kinases in Arabidopsis. Proc Natl Acad Sci U S A. 2009;106(41):17588–17593.
- Vlad F, Rubio S, Rodrigues A, et al. Protein phosphatases 2C regulate the activation of the Snf1-related kinase OST1 by abscisic acid in Arabidopsis. Plant Cell. 2009;21(10):3170–3184.
- Fujii H, Zhu JK. Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc Natl Acad Sci U S A. 2009;106(20):8380–8385.
- Hubbard KE, Nishimura N, Hitomi K, et al. Early abscisic acid signal transduction mechanisms: newly discovered components and newly emerging questions. Genes Dev. 2010;24(16):1695–1708.
- Yan A, Chen Z. The pivotal role of abscisic acid signaling during transition from seed maturation to germination. Plant Cell Rep. 2017;36(5):689–703.
- Peterson FC, Burgie ES, Park SY, et al. Structural basis for selective activation of ABA receptors. Nat Struct Mol Biol. 2010;17(9):1109–1113.
- Yang W, Zhang W, Wang X. Post-translational control of ABA signalling: the roles of protein phosphorylation and ubiquitination. Plant Biotechnol J. 2017;15(1):4–14.
- Chen Y, Feng L, Wei N, et al. Overexpression of cotton PYL genes in Arabidopsis enhances the transgenic plant tolerance to drought stress. Plant Physiol Biochem. 2017;115:229–238.
- Chen Z, Lan K, Yun Z, et al. Endosperm-specific OsPYL8 and OsPYL9 act as positive regulators of the ABA signaling pathway in rice seed germination. Funct Plant Biol. 2017;44(6).
- Yu X, Han J, Wang E, et al. Genome-wide identification and homoeologous expression analysis of PP2C genes in wheat (Triticum aestivum L.). Front Genet. 2019;10:561.
- Allen GJ, Kuchitsu K, Chu SP, et al. Arabidopsis abi1-1 and abi2-1 phosphatase mutations reduce abscisic acid-induced cytoplasmic calcium rises in guard cells. Plant Cell. 1999;11(9):1785–1798.
- Saez A, Apostolova N, Gonzalez-Guzman M, et al. Gain-of-function and loss-of-function phenotypes of the protein phosphatase 2C HAB1 reveal its role as a negative regulator of abscisic acid signalling. Plant J. 2004;37(3):354–369.
- Kuhn JM, Boisson-Dernier A, Dizon MB, et al. The protein phosphatase AtPP2CA negatively regulates abscisic acid signal transduction in arabidopsis, and effects of abh1 on AtPP2CA mRNA. Plant Physiol. 2006;140(1):127–139.
- Sun HL, Wang XJ, Ding WH, et al. Identification of an important site for function of the type 2C protein phosphatase ABI2 in abscisic acid signalling in Arabidopsis. J Exp Bot. 2011;62(15):5713–5725.
- Rubio S, Rodrigues A, Saez A, et al. Triple loss of function of protein phosphatases type 2C leads to partial constitutive response to endogenous abscisic acid. Plant Physiol. 2009;150(3):1345–1355.
- Hrabak EM, Chan CW, Gribskov M, et al. The Arabidopsis CDPK-SnRK superfamily of protein kinases. Plant Physiol. 2003;132(2):666–680.
- Boudsocq M, Barbier-Brygoo H, Lauriere C. Identification of nine sucrose nonfermenting 1-related protein kinases 2 activated by hyperosmotic and saline stresses in Arabidopsis thaliana. J Biol Chem. 2004;279(40):41758–41766.
- Chang Y, Nguyen BH, Xie Y, et al. Co-overexpression of the constitutively active form of OsbZIP46 and ABA-activated protein kinase SAPK6 improves drought and temperature stress resistance in rice. Front Plant Sci. 2017;8:1102.
- Wang XC, Zhao QY, Ma CL, et al. Global transcriptome profiles of Camellia sinensis during cold acclimation. BMC Genomics. 2013;14:415.
- Xia EH, Li FD, Tong W, et al. Tea plant information archive: a comprehensive genomics and bioinformatics platform for tea plant. Plant Biotechnol J. 2019;17(10):1938–1953.
- Kawahara Y, de la Bastide M, Hamilton JP, et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice (NY). 2013;6(1):4.
- Marchler-Bauer A, Derbyshire MK, Gonzales NR, et al. CDD: NCBI’s conserved domain database. Nucleic Acids Res. 2015;43(Database issue):D222–6.
- Finn RD, Coggill P, Eberhardt RY, et al. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 2016;44(D1):D279–85.
- Hall BG. Building phylogenetic trees from molecular data with MEGA. Mol Biol Evol. 2013;30(5):1229–1235.
- Pattengale ND, Alipour M, Bininda-Emonds OR, et al. How many bootstrap replicates are necessary? J Comput Biol. 2010;17(3):337–354.
- Brown P, Baxter L, Hickman R, et al. MEME-LaB: motif analysis in clusters. Bioinformatics. 2013;29(13):1696–1697.
- Mulder N, Apweiler R. InterPro and InterProScan: tools for protein sequence classification and comparison. Methods Mol Biol. 2007;396:59–70.
- Hu B, Jin J, Guo AY, et al. GSDS 2.0: an upgraded gene feature visualization server. Bioinformatics. 2015;31(8):1296–1297.
- Chen C, Xia R, Chen H, et al. TBtools, a Toolkit for Biologists integrating various HTS-data handling tools with a user-friendly interface. bioRxiv. 2018;289660.
- Pertea M, Kim D, Pertea GM, et al. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650–1667.
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357–360.
- Pertea M, Pertea GM, Antonescu CM, et al. StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol. 2015;33(3):290–295.
- Frazee AC, Pertea G, Jaffe AE, et al. Ballgown bridges the gap between transcriptome assembly and expression analysis. Nat Biotechnol. 2015;33(3):243–246.
- Walley JW, Sartor RC, Shen Z, et al. Integration of omic networks in a developmental atlas of maize. Science. 2016;353(6301):814–818.
- Robinson JT, Thorvaldsdottir H, Winckler W, et al. Integrative genomics viewer. Nat Biotechnol. 2011;29(1):24–26.
- Li CF, Xu YX, Ma JQ, et al. Biochemical and transcriptomic analyses reveal different metabolite biosynthesis profiles among three color and developmental stages in ‘Anji Baicha’ (Camellia sinensis). BMC Plant Biol. 2016;16(1):195.
- Umezawa T, Nakashima K, Miyakawa T, et al. Molecular basis of the core regulatory network in ABA responses: sensing, signaling and transport. Plant Cell Physiol. 2010;51(11):1821–1839.
- Ben-Ari G. The ABA signal transduction mechanism in commercial crops: learning from Arabidopsis. Plant Cell Rep. 2012;31(8):1357–1369.
- Larkindale J, Vierling E. Core genome responses involved in acclimation to high temperature. Plant Physiol. 2008;146(2):748–761.
- Shao Y, Qin Y, Zou Y, et al. Genome-wide identification and expression profiling of the SnRK2 gene family in Malus prunifolia. Gene. 2014;552(1):87–97.
- Tian X, Wang Z, Li X, et al. Characterization and functional analysis of pyrabactin resistance-like abscisic acid receptor family in rice. Rice (N Y). 2015;8(1):28.
- Bai G, Xie H, Yao H, et al. Genome-wide identification and characterization of ABA receptor PYL/RCAR gene family reveals evolution and roles in drought stress in Nicotiana tabacum. BMC Genomics. 2019;20(1).
- Hasan MM, Ma F, Islam F, et al. Comparative transcriptomic analysis of biological process and key pathway in three cotton (Gossypium spp.) species under drought stress. Int J Mol Sci. 2019;20(9):2076.
- Hu W, Yan Y, Shi H, et al. The core regulatory network of the abscisic acid pathway in banana: genome-wide identification and expression analyses during development, ripening, and abiotic stress. BMC Plant Biol. 2017;17(1):145.
- None. Shikimic acid; metabolism and metabolites. Haslam E, editor. Chichester, UK: Wiley; 1993. p. 387. 26(6) 855-0.
- Razal RA, Ellis S, Singh S, et al. Nitrogen recycling in phenylpropanoid metabolism. Phytochemistry. 1996;41(1):31–35.
- Bonawitz ND, Chapple C. The genetics of lignin biosynthesis: connecting genotype to phenotype. Annu Rev Genet. 2010;44:337–363.
- Vlot AC, Dempsey DA, Klessig DF. Salicylic Acid, a multifaceted hormone to combat disease. Annu Rev Phytopathol. 2009;47:177–206.
- Vogt T. Phenylpropanoid biosynthesis: invited review. Mol Plant. 2009;ssp106.
- Dudareva N, Klempien A, Muhlemann JK, et al. Biosynthesis, function and metabolic engineering of plant volatile organic compounds. New Phytol. 2013;198(1):16–32.
- Kawa D, Meyer AJ, Dekker HL, et al. SnRK2 protein kinases and mRNA decapping machinery control root development and response to salt. Plant Physiol. 2019;pp.00818.2019.