ABSTRACT
The effects of adding biochar rice husk (R), white popinee (WP), bamboo (BB), or coconut (CT) on microbial community in fermentation broths from glucose were investigated. The added biochars acted as biofilm carriers on which Sporolactobacillus spathodeae, Clostridium sensu stricto 11 sp., Clostridium sensu stricto 12 sp., Clostridium sensu stricto 1 sp., and Clostridium sensu stricto 5 sp. were enriched. Fermentation reactions substantially increased the amounts of acid-producers in biofilm. The homoacetogens, Clostridium carboxidivorans and Clostridium drakei, were identified in the biofilm in the first two batches of fermentation with biochars as electron conductors between acid-producers and homoacetogens to assist homoacetogenesis. The heterotrophic bacteria overcompeted the acid-producers in the biofilm in long-term fermentation.
1. Introduction
Acid-producing strains produce volatile fatty acids (VFA), hydrogen (H2) gas and carbon dioxide (CO2) gas from complicated organic molecules via fermentation reactions [Citation1,Citation2]. In fermentation, dark fermentation presents an effective pathway to obtain valuable chemicals from organic waste [Citation3,Citation4]. Enhanced fermentation process is of great practical interests [Citation5,Citation6]. The efficiency of dark fermentation can be enhanced by adding carbon-based materials, including biochar [Citation7,Citation8]. For instance, biochars that are added to fermenting broths can increase the yield of H2 from the organic wastes [Citation9,Citation10].
Lu et al. [Citation11] added nine biochars to a microbial consortium and noted that biochars with excess functional groups and high crystallinity would lead to the preferential production of acetate over butyrate at the expense of the production of H2 and CO2. The authors posited that homoacetogenesis was effective in the fermenting consortium as the biochar acted as an electron conductor for interspecies electron transfer between the acid producers and the homoacetogens. Lu et al. [Citation12] subsequently performed sequential dark fermentation tests on a biochar-amended microbial consortium with repeatedly replenished glucose. The authors noted that the biofilm on the biochars preferentially produced acetate in the second batch fermentation but not in subsequent fermentation batches. Such an occurrence is of great practical interest for the long-term operation of biochar-amended fermentation to produce VFA.
The microbial communities in dark fermenting broths with or without the addition of biochar were monitored. The biased enrichment of the consortium with functional strains by the addition biochar was reported for the first time. This study reported data that providing in-depth understanding to the evolution of fermentation processes from organic substances based on microbial community information.
2. Experimental
2.1. Materials and fermentation test
Four biochars, rice husk (R), white popinee (WP), bamboo (BB), and coconut (CT) were formed by slow pyrolysis at 550°C for 4 h in N2 atmosphere. The fermentative consortium that was collected from anaerobic digestion processes in Luxing Environmental Protection Corp. (Shangdong Province, China) was the original consortium used in this study. This consortium was similar to those used by Lu et al. [Citation12], which was pretreated by boiling at 100°C for one hr for ease of comparison.
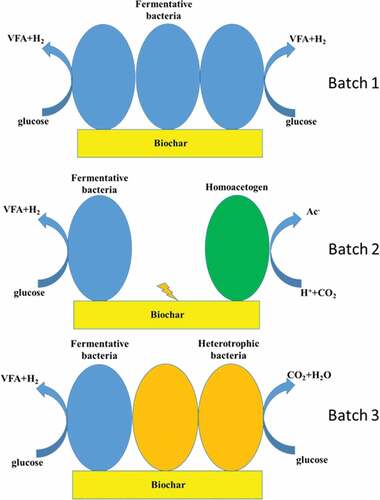
The original consortium was added to 100 mL serum flasks that were fed with 1070 mg/L glucose at pH 7 and 600 mg/L biochar to a working volume of 80 mL. Before each test, the headspace of the serum flask was purged with N2 gas. The fermentation tests were then performed at 36°C with intensive shaking and regular sampling. At the end of fermentation, the biomass to which biochars had been added was collected as sediment after 2 h of free settling. The supernatant was separately collected and the cells in the supernatants were regarded as being in suspended growth. Some of the biomass with the amended biochar samples were ultrasonicated at 50 W for 5 min in an ice bath and the suspensions were collected after another 2 h of settling. The fermentation tests were repeated four times, named as batch 1‒4, respectively, in this study. Batch 1 and batch 2 were used to simulate the early stage of fermentation process and Batch 3 and batch 4 were used to simulate the long-term fermentation process.
2.2. DNA extraction, PCR, high-throughput sequencing
The genomic DNA of granule samples was extracted using a Fast DNA Spin Kit (MP Biomedicals, MA. USA). The bacterial gene sequence was amplified using ABI GeneAmp 9700 (ABI, USA) with Transstart Fastpfu DNA polymerase by primers 338F and 806R. Each sample was extracted and amplified in triplicate to ensure the quality of the obtained data. The amplified products were purified using an AxyPrep DNA extraction kit (Axygen, USA). The amplified genomic samples were used to construct an amplicon library, which was sequenced using Illumina PE250 (Illumina Inc., San Diego, USA). The sequences were then clustered into operational taxonomic units (OTUs) at 97% similarity and identified on the website of NCBI.
3. Results and discussion
3.1. Fermentation performance
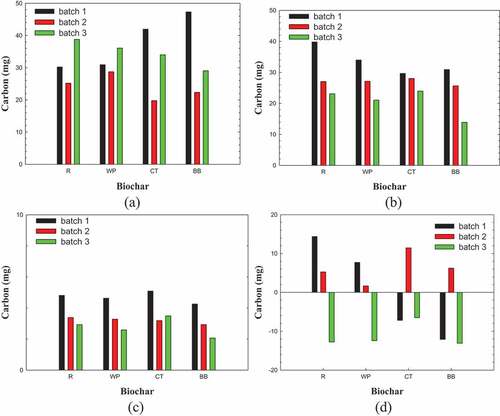
displays the flows of carbon when glucose was fed into the fermenting broths and those that arose from the production of VFA and CO2. The total VFA production rates and CO2 production rates varied with the species of biochar and declined as the number of fermentation batch increased. ) shows the flow of carbon from the biofilm. For biochars R and WP, the fed biomass firstly released carbon (batch 1); then the formed biofilm released a few carbons in batch 2; in batch 3, the biofilm grew by adsorbing a large amount of carbon. For biochars CT and BB, in batch 1, the fed biomass adsorbed carbon; then in batch 2, the biofilm released carbon, and in batch 3, the biofilm again adsorbed carbon in fermentation. The carbon flows in batch 4 were not analyzed in this study.
3.2. Identified dominant strains
lists the genera of the most dominant strains, including Sporolactobacillus spathodeae, a gram-positive, facultatively lactate producer that can yield pyruvate from glucose [Citation13]; Clostridium neunse, a gram-positive, obligate anaerobe for H2/CO2/acetate/butyrate production [Citation14]; and Clostridium paraputrificum, a gram-positive, anaerobe that can produce acetate/propionate from glucose [Citation15]. These genera were identified in abundance in almost all studied samples. The second most dominant strains in some tested samples included Tissierella creatinine, an obligately anaerobic creatinine consumer [Citation16]; Psychrobacter maritimus, gram-negative heterotrophic degrader [Citation17]; Pseudomonas vernoii, a gram-negative, aerobic degrader, or anaerobic denitrifier [Citation18]; and Sphingomonas echinoides, a gram-negative, heterotrophic organics degrader [Citation19].
Table 1. Abundances of the top 15 dominant strains identified from the consortium samples
3.3. Role of biochar amendments on fermentation performance
3.3.1. As a biased cell carrier
shows the proportions of the genera of the most abundant OTU sequences. The original consortium (C1) was enriched with Tissierella sp. and Psychrobacter sp., both of which are degraders of organic compounds. The original consortium is therefore an efficient degraded of organics. After batch 1 was tested, the consortium without added carbon (C2) was dominated by Sporolactobacillus spathodeae, Clostridium sensu stricto 11 sp., Clostridium sensu stricto 12 sp., Clostridium sensu stricto 1 sp., and Clostridium sensu stricto 5 sp., of which the first four are acid producers. Restated, the microbial community had become an effective fermenting broth that could convert glucose to VFAs.
Figure 2. Abundance proportions of dominant OTUs in genus level for the samples. C1: original consortium; C2: consortium without carbon amendment after one batch. CT1: CT biochar+biomass after batch 1; BB1: BB biochar+biomass after batch 1; WP1: WP biochar+biomass after batch 1; CT3: detached biomass from CT biochar after batch 3; BB3: detached biomass from BB biochar after batch 3; R3: detached biomass from R biochar after batch 3; CT4: attached biomass from CT biochar after batch 3; BB4: attached biomass from BB biochar after batch 3; R4: attached biomass from R biochar after batch 3; CT5: feed from CT-amended test to batch 4; R5: feed from R-amended test to batch 4
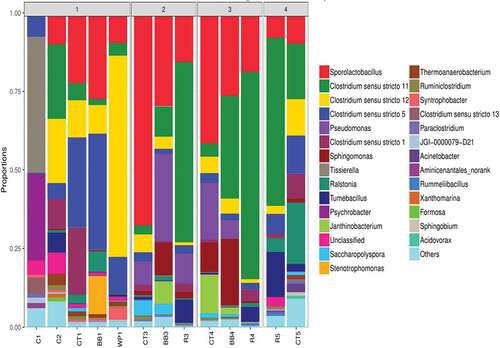
Comparing microbial communities in C2 and those in added carbon (CT1, BB1, WP1 in and ) showed that biochar consortium CT1 exhibited increased abundances of Sporolactobacillus spathodeae, Clostridium sensu stricto 5 sp., and Clostridium sensu stricto 1 sp.; biochar BB1 was enriched with Sporolactobacillus spathodeae and Clostridium sensu stricto 5 sp.; and the biofilm on biochar WP1 was enriched with Clostridium sensu stricto 5 sp. and Clostridium sensu stricto 12 sp. The addition of biochar had made the microbial consortia CT1, BB1, and WP1 fit well the fermenting environment after batch 1 (C2).
After batch 3, the biofilm on biochar CT3 had the dominant species Sporolactobacillus spathodeae, Pseudo-monas sp., Saccharopolyspora sp., and Clostridium sensu stricto 12 sp.; the biofilm on biochar BB3 was dominated by Sporolactobacillus spathodeae, Pseudomonas sp., Sphingomonas sp., and Janthinobacterium sp.; the biofilm on biochar R3 was enriched with Clostridium sensu stricto 11 sp., Sporolactobacillus spathodeae, Pseudomonas sp., and Tumebacillus sp. The biofilm that detached from biochar CT4 was enriched with Sporolactobacillus spathodeae, Pseudomonas sp., Janthinobacterium sp., and Sphingomonas sp.; the biofilm that detached from biochar BB4 contained Clostridium sensu stricto 11 sp., Sporolactobacillus spathodeae, Sphingomonas sp., and Pseudomonas sp. The supernatants from batch 3 (R5) had the dominant species Clostridium sensu stricto 11 sp., Tumebacillus sp., Sporolactobacillus spathodeae, and Clostridium sensu stricto 5 sp.; the supernatant from batch 3 (CT5) was dominated by Ralstonia sp., Clostridium sensu stricto 11 sp., Clostridium sensu stricto 5 sp., Clostridium sensu stricto 12 sp., and Sporolactobacillus spathodeae.
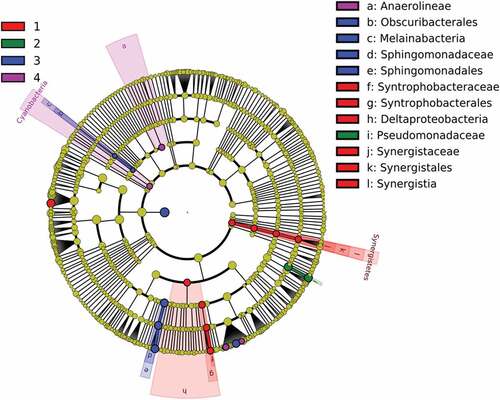
The cladogram in reveals that the species Syntrophobacteraceae and Synergistetes were enriched in samples C1, C2, BB1, CT1, WP1. Pseudomonas sp. appeared in the biomass samples after three fermentation batches (CT3, BB3, R3). The dominant strains in R4, BB4, CT4 were Sphingomanadacae, Melainabacteria, and Obscuribacterales – totally other than those species that were from biomass on biomass. The dominant strain in R5, CT5, BB5 was Anaerolinae, indicating that the suspended cells comprised very different community structures from those with the fixed cells. Restated, the biochars were ‘biased’ cell carriers.
3.3.2. Microbial community shift
presents a similarity tree that is based on a beta diversity distance matrix and a heatmap of the tested samples. The original consortium (C1) was the farthest from all other consortia, suggesting a significant shift in the microbial communities in the bacterial consortium as a result of fermentation. After batch 1, the consortium without added carbon (C2) was very different from those with added carbon (BB1, CT1, WP1), revealing the effect of added biochar on the microbial communities in the fermentation reactions. The microbial communities in BB1, CT1, and WP1 were similar, so the species of biochar did not substantially affect the relative amounts of the strains after batch 1. The high similarities between CT3 and CT4, BB3 and BB4, and R3 and R4 suggest that the main microbial communities in the fermentation broths were attached on biochar surface as biofilm.
Figure 4. Similarity analysis for the tested samples at genus levels. (a) Similarity tree; (b) heatmap. C1, C2, CT1, BB1, WP1, CT3, BB3, R3, CT4, BB4, R4, CT5, R5 are the same as defined in
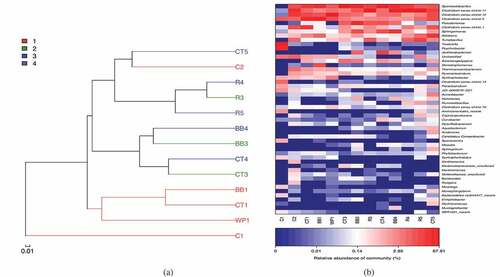
Comparing the microbial communities of CT3, CT4, BB3, and BB4 with those of CT1 and BB1 demonstrates that Sporolactobacillus spathodeae was more abundant, but Clostridium sensu stricto 5 sp. and Clostridium sensu stricto 1 sp. were less abundant. In particular, the abundances of Pseudomonas sp. and Spingomonas sp. in the latter two consortia were significantly increased. These observations indicated that, after batch 1‒3, the acid producers became less competitive whereas the heterotrophic bacteria had gradually gained competitive advantages. Comparing the microbial communities in CT5 and R5 with those in CT4 and R4 indicated that the Sporolactobacillus spathodeae and Pseudomonas sp. preferentially remained with the biofilm on the biochar surface, whereas Clostridium sensu stricto 11 sp. was distributed roughly evenly in suspended and attached forms, and Clostridium sensu stricto 5 sp. slightly favored growth in suspension. Restated, Clostridium sensu stricto 5 sp. on biochar surfaces was out-competed by heterotrophic bacteria in the biochar biofilm, consistent with the noted diminishing of homoacetogenesis throughout sequential fermentation tests [Citation12].
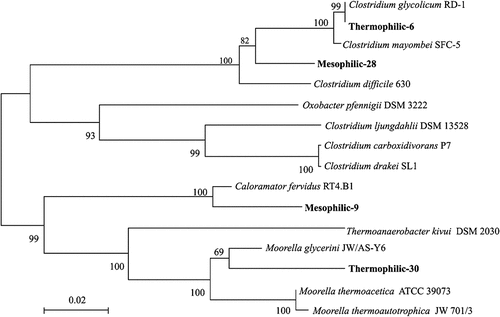
presents phylogenetic tree, based on clone libraries, that demonstrates that the two identified strains, Clostridium carboxidivorans and Clostridium drakei, which belong to Clostridium sensu stricto 5 sp., are homoacetogens. Lu et al. [Citation11] noted that in their biochar-amended fermentation test, the microbial consortium preferentially yielded acetate over butyrate at the expense of the production of H2 and CO2. These authors proposed direct interspecies electron transfer (DIET) between acid producers and the homoacetogens. The homoacetogens produced acetate from C1 compounds as the sole energy source and the co-existence of acid producers (Clostridium sensu stricto 1 sp., Clostridium sensu stricto 11 sp., Clostridium sensu stricto 12 sp.) and homoacetogens (Clostridium sensu stricto 5 sp.) on biochar CT1, BB1, and WP1 supported the formation of acetate by the reaction 4H2 + 2CO2 → CH3COOH + 2H2O. The specified homoacetogens in the consortia of interest partly supported the proposal of Lu et al. [Citation11] that homoacetogenesis led to the preferential production of acetate over butyrate in biochar-amended fermentation processes. The above observations suggest that the acid-producers dominated the batch 1, while homoacetogenesis became significant in batch 2. In batch 3, heterotrophic bacteria gradually dominated the biofilms on biochars, reducing the fermentation activities of the biofilm consortia.
3.3.3. Application prospects
The use of biochar to enhance process performance has high application prospects. The economy of fermentation processes counts on the overall efficiency from feedstocks to end products. The biochars are the products from other waste (solid waste, for instance) which can be supplied to fermenting reactors at low costs [Citation26]. The application of biochars to field fermenting reactors is straightforward with only adding a side input stream with grinded biochar appended to the existing full-scale reactor will do. Owing to the low-cost nature, the applications of biochars to fermenting broths should be economically feasible to lower income countries and regimes.
The used biochars should not be regenerated and re-used. The used biochars could be discharged as part of the effluent that enters the next handling units, such as anaerobic digester [Citation27] as electron conductors for DIET between different groups of bacteria or wastewater treatment reactor [Citation28] as biofilm carriers with retained biomass. The biochars discharged with the final waste biomass can function as settling enhancer for biomass sedimentation [Citation29], bulking agents for composting [Citation30], or simply part of fertilizers to land applications [Citation31].
4. Conclusions
Effects of adding four biochars on the microbial community in fermentation broths from glucose were examined. The biochars behaved as biofilm carriers with Sporolactobacillus spathodeae, Clostridium sensu stricto 11 sp., Clostridium sensu stricto 12 sp., Clostridium sensu stricto 1 sp., and Clostridium sensu stricto 5 sp. as the dominant genera. Fermentation considerably enriched the biofilm with acid-producers. The homoacetogens, Clostridium carboxidivorans and Clostridium drakei, were identified in a biofilm in batch 1 and batch 2 of fermentation, likely because the biochars acted as electron conductors between acid-producers and homoacetogens. In batch 3, the biofilm was enriched with heterotrophic bacteria, which outcompeted the homoacetogens in converting organic matters.
Article highlights
Four biochars shift microbial community in fermentation broths from glucose.
Dominant genera in biofilms attached on the biochars were identified.
Homoacetogens, C. carboxidivorans and C. drakei, were identified.
Heterotrophic bacteria overcompeted homoacetogens in the later fermentation stage.
Disclosure statement
No potential conflict of interest was reported by the authors.
Additional information
Funding
References
- Wainaina S, Lukitawesa AMK, Taherzadeh MJ. Bioengineering of anaerobic digestion for volatile fatty acids, hydrogen or methane production: a critical review. Bioengineered. 2019;10:437‒458.
- Preethi Usman TMM, Banu JR, Gunasekaran M, et al. Biohydrogen production from industrial wastewater: an overview. Bioresour Technol Rep. 2019;7:100287.
- Mohan SV, Amulya SDK, Katakojwala R, et al. Can circular bioeconomy by fueled by waste biorefineries – a closer look. Bioresour Technol Rep. 2019;7:100277.
- Meena RAA, Kannah YR, Sindhu J, et al. Trends and resource recovery in biological wastewater treatment system. Bioresour Technol Rep. 2019;7:100235.
- Liu X. Effects of proteases on L-glutamic acid fermentation. Bioengineered. 2019;10:646‒658.
- Xu Q, Bai F, Chen N, et al. Utilization of acid hydrolysate of recovered bacterial cell as a novel organic nitrogen source for L-tryptophan fermentation. Bioengineered. 2019;10:23‒32.
- Pan J, Ma J, Zhai L, et al. Achievements of biochar application for enhanced anaerobic digestion: a review. Bioresour Technol. 2019;292:122058.
- Duan X, Chen YZ, Yan YY, et al. New method for algae comprehensive utilization: algae-derived biochar enhances algae anaerobic fermentation for short-chain fatty acids production. Bioresour Technol. 2019;289:121637.
- Sharma P, Melkania U. Biochar-enhanced hydrogen production from organic fraction of municipal solid waste using co-culture of Enterobacter aerogenes and E. coli. Int J Hydrogen Energy. 2017;42:18865‒18874.
- Yang G, Wang JL. Synergistic enhancement of biohydrogen production from grass fermentation using biochar combined with zero-valent iron nanoparticles. Fuel. 2019;251:420‒427.
- Lu JH, Chen C, Huang C, et al. Dark fermentation production of volatile fatty acids from glucose with biochar amended biological consortium. Bioresour Technol. 2020;303:122921.
- Lu JH, Chen C, Leu SY, et al. Glucose fermentation with biochar amended consortium: sequential fermentations. Bioresour Technol. In press. DOI:10.1016/j.biortech.2020.122933
- Thamacharoensuk T, Kitahara M, Ohkuma M, et al. Sporolactobacillus shoreae sp. nov. and Sporolactobacillus spathodeae sp. nov., two spore-forming lactic acid bacteria isolated from tree barks in Thailand. Int J Syst Evol Microbiol. 2015;65:1220‒1226.
- Zhao X, Li D, Xu S, et al. Clostridium guangxiense sp. nov. and Clostridium neuense sp. nov., two phylogenetically closely related hydrogen-producing species isolated from lake sediment. Int J Syst Evol Microbiol. 2017;67:710‒715.
- Evvyernie D, Yamazaki S, Morimoto K, et al. Identification and characterization of Clostridium paraputrificum M-21, a chitinolytic, mesophilic and hydrogen-producing bacterium. J Biosci Bioeng. 2000;89:596‒601.
- Farrow JAE, Lawson PA, Hippe H, et al. Phylogenetic evidence that the gram-negative nonsporulating bacterium Tissierella (Bacteroides) praeacuta is a member of the Clostridium subphylum of the gram-positive bacteria and description of Tissierella creatinini sp. nov. Int J Syst Evol Microbiol. 1995;45:436‒440.
- Romanenko LA, Lysenko AM, Rohde M, et al. Psychrobacter maritimus sp. nov. and Psychrobacter arenosus sp. nov., isolated from coastal sea ice and sediments of the sea of Japan. Int J Syst Evol Microbiol. 2004;54:1741‒1745.
- Elomari M, Coroler L, Hoste B, et al. DNA relatedness among Pseudomonas strains isolated from natural mineral waters and proposal of Pseudomonas veronii sp. nov. Int J Syst Evol Microbiol. 1996;46:1138‒1144.
- Denner EBM, Kämpfer P, Busse HJ, et al. Note: reclassification of Pseudomonas echinoide Heumann 1962, 343AL, in the genus Sphingomonas as Sphingomonas echinoide comb. nov. Int J Syst Evol Microbiol. 1999;49:1103‒1109.
- Liou JSC, Balkwill DL, Drake GR, et al. Clostridium carboxidivorans sp. nov., a solvent-producing clostridium isolated from an agricultural settling lagoon, and reclassification of the acetogen Clostridium scatologenes strain SL1 as Clostridium drakei sp. nov. Int J Syst Evol Microbiol. 2005;55:2085‒2091.
- Keis S, Shaheen R, Jones DT. Emended descriptions of Clostridium acetobutylicum and Clostridium beijerinckii, and descriptions of Clostridium saccharoperbutylacetonicum sp. nov. and Clostridium saccharobutylicum sp. nov. Int J Syst Evol Microbiol. 2001;51:2095‒2103.
- Baek SH, Cui Y, Kim SC, et al. Tumebacillus ginsengisoli sp. nov., isolated from soil of a ginseng field. Int J Syst Evol Microbiol. 2011;61:1715‒1719.
- Valdes N, Soto P, Cottet L, et al. Draft genome sequence of Janthinobacterium lividum strain MTR reveals its mechanism of capnophilic behavior. Stand Genomic Sci. 2015;10:110.
- Duangmal K, Mingma R, Thamchaipenet A, et al. Saccharopolyspora phatthalungensis sp. nov., isolated from rhizosphere soil of Hevea brasiliensis. Int J Syst Evol Microbiol. 2010;60:1904‒1908.
- Daligault HE, Davenport KW, Minogue TD, et al. Draft genome assembly of Ralstonia pickettii type strain K-288 (ATCC 27853). Genome Announc. 2014;2:e00973–14.
- Lee DJ, Cheng YL, Wong RJ, et al. Adsorption removal of natural matters in water using biochar. Bioresour Technol. 2019;260:413‒416.
- Lu JS, Chang JS, Lee DJ. Adding carbon-based materials on anaerobic digestion performance: a mini-review. Bioresour Technol. 2020;300:122696.
- Yaashiaa PR, Kumar PS, Varjani SJ, et al. Advances in production and application of biochar from lignocellulosic feedstocks for remediation of environmental pollutants. Bioresour Technol. 2019;292:122030.
- Guo J, Jiang S, Pang Y. Rice straw biochar modified by aluminum chloride enhances the dewatering of the sludge from municipal sewage treatment plant. Sci Total Environ. 2019;654:338–344.
- Wang J, Wang S. Preparation, modification and environmental application of biochar: a review. J Clean Prod. 2019;227:1002–1022.
- Chojnacka K, Moustakas K, WItek-Krowiak A. Bio-based fertilizers: a practical approach towards circular economy. Bioresour Technol. 2020;295:122223.