ABSTRACT
Transcriptome is used to determine the induction response of Pinellia ternata (Thunb.) Breit T2 plus line (abbreviated as PT2P line) infected with Pectobacterium carotovorum. The main objective of the study was to deal with the transcriptome database of PT2P line resistance to soft rot pathogens to provide a new perspective for identifying the resistance-related genes and understanding the molecular mechanism. Results indicated that water soaking and tissue collapse started at 20 h after PT2P line was infected by P. carotovorum. A total of 1360 and 5768 differentially expressed genes (DEGs) were identified at 0 h and 20 h, respectively. After 20 h of infection, growth and development-related pathways were inhibited. Meanwhile, DEGs were promoted the colonization of P. carotovorum pathogens in specific cell wall modification processes at the early infected stage. A shift to a defensive response was triggered at 0 h. A large number of DEGs were mainly up-controlled at 20 h and were substantially used in the pathogen recognition and the introduction of signal transformation cascades, secondary metabolites biosynthesis, pathogenic proteins activation, transcription aspects and numerous transporters. Furthermore, our data provided novel insights into the transcript reprogramming of PT2P line in response to P. carotovorum infestation.
1. Introduction
Pinellia ternata (Thunb.) Breit (abbreviated as PT) is a traditional Chinese herbal medicine used in large quantities and has thousands of histories. South Asia is the main producing area. It is mainly used to treat lipid-lowering and has anti-blocking, anti-tumor, anti-early pregnancy, anti-emetic and other functions. Due to its high medicinal value, the reduction of wild PT resources has caused tension in the Chinese herbal medicine market, which requires large-scale planting. Pinellia ternata (Thunb.) Breit T2 Plus line [Citation1], hereinafter referred to as PT2P line, is a unique chromosome-doubled excellent variety in our laboratory. During the large-scale planting process, numerous diseases will reduce production of PT2P line.
Soft rot disease causes PT2P line tuber rot or even failure to harvest. Pectobacterium carotovorum, hereinafter referred to as P. carotovorum, is a strong pathogen that induces water-soaking lesions, soft rot and total collapse of the whole PT [Citation2]. The disease generally occurs in summer and spreads widely. In order to control soft rot disease, traditional measures are commonly applied, such as crop rotation, use of certified tuber seed, sterile tissue culture vaccine, proper disposal of infected plant debris, application of copper or other bactericides [Citation3]. However, no strategy is currently available to completely protect plants from damage. Therefore, an alternative approach is needed to preserve PT from soft rot disease. Hence, it is necessary for us to understand the molecular mechanism of PT in response to P. carotovorum infection and provide the necessary basis for the prevention and control of PT soft rot disease.
The existence of R genes determines the disease resistance of plants. They enable plants to recognize pathogens and actively trigger the immune system to resist microorganisms [Citation4]. Many resistant genes have been identified in the process of resisting soil-borne pathogens in a variety of plants [Citation5]. However, a global study on PT disease resistance, especially soft rot disease, is still in the developmental stage.
R genes codes cytoplasmic proteins have contained negative-triphosphate binding site (NBS) and leucine-rich repeat (LRR) domains [Citation6]. It is common knowledge that plants are exposed to various pathogens, such as fungi [Citation7], bacteria [Citation8], viruses [Citation9] and parasitic soil worms [Citation10]. These pathogenic microorganisms invade plant cells seeking nutrients to promote their proliferation, and thus, damage host cells [Citation11,Citation12]. The defense mechanism of plants depends on recognizing invasive pathogens by pattern recognition receptors (PRRs) on the surface of cytoplasm [Citation13]. It reflects the incidence of apoplastic establishment of plant contaminating pathogens through the molecules which alienated into membrane related receptor-like kinases (RLKs) or receptor-like proteins (RLPs) [Citation14]. The defense-related genes are induced by pathogen related molecular patterns (PAMPs) and/or damage related to molecular patterns (DAMPs) to participate in intracellular signaling pathways and initiate PTI reaction [Citation15]. PTI is the first layer of the defense mechanism in plants [Citation16]. Under the action of PRRs, plant cells can recognize pathogenic microbial proteins, trigger intracellular signal transduction and activate an adaptive response [Citation17]. Plant PTI is involved in the role of R gene in microbial elicitor recognition, cell signal transduction and hypersensitivity [Citation18]. Especially, PTI is initiated by the transcription of a large number of genes to cascade signal amplification through calcium-dependent protein kinases (CDPKs) [Citation19] and mitogen activated protein kinases (MAPKs) [Citation20,Citation21].
Pathogenic microorganisms can tear up the cell wall by secreting the cell wall degrading enzyme (CWDE) [Citation22] to create an invasion point, cross the plant cell wall, and obtain water and nutrients from plant protoplasts. Damaged cell wall fragments, such as galacto-oligosaccharide uronic acid, can lead to the expression of defense genes and the production of reactive oxygen species (ROS). At this time, PAMP conducts signal transduction to monitor the integrity of the cell wall, and microbial invasion is further prevented [Citation23]. Jasmonic acid (JA), salicylic acid (SA), ethylene (ET) and other plant hormones are the key regulators of anti-necrosis factor defense response, which play an important role in regulating the transduction pathway and downstream activation of defense response [Citation24].
Some members of PAMPS response genes and transcription factors (TFs) family are considered as important participants in transcriptional reprogramming and affect host susceptibility and immune defense through various ways [Citation25]. Pathogenic microorganisms have evolved a variety of strategies to inhibit host PTI, such as secret toxins [Citation26], triggering type III effectors [Citation27], synthesizing host hormone analogues or elicitors to affect host hormone levels [Citation28]. Transcriptomics can explore the molecular mechanism of plant transcription reprogramming under pathogen infection [Citation25,Citation29,Citation30]. At the same time, it helps discover new resistance genes, which provides the background for the development of plant-resistant varieties.
The first transcriptome database of PT was built by Huang [Citation31] to research the whole transcriptome information, but he has not built PT resistance to soft rot pathogens transcriptome database to reveal the molecular mechanism to prevent the soft rot disease. PT2P line is a unique chromosome-doubling strain in the laboratory, with many excellent characteristics, such as high volume, disease resistance, and high photosynthetic efficiency. This is completely different from the experimental materials of PT by Huang et al. Although Lu et.al [Citation1] have established a conventional transcriptome database of PT2P line to study its growth characteristics, they have not established transcriptome database of PT2P line resistance to soft rot pathogens. Therefore, it is necessary to establish a transcriptome database of PT2P line resistance to soft rot pathogens. In this research, it was the first time to establish the transcriptome database of PT2P line resistance to the soft rot pathogens to provide a new perspective for identifying the resistance-related genes and understanding the molecular mechanism.
2 Materials and methods
2.1. Plant material and pathogen inoculum.
Sterile PT2P line was harvested during their petiole extension period from an experimental tissue culture room at Zunyi Medical University in Guizhou province, China. First, P. carotovorum was cultivated with LB liquid medium to OD value of 0.6, which was measured at a wavelength of 600 nm. Then, PT2P line materials were inoculated with the same batch for research, set 0 h group and 20 h group, and set up a control group at the corresponding time point. The test group was co-cultivated by a PT2P line with 5 ml of P. carotovorum solution with an OD value of 0.6, and each group was repeated three times. The control group was co-cultured on the PT2P line with 5 ml of LB medium, and three repetitions were performed in each group. The co-cultivation conditions were maintained at 2000 Lx light intensity, 12 h/d illumination time and 25 ± 1°C of culture temperature.
2.2 Total RNA isolation and Illumina sequencing
Sprout samples of PT2P line were implanted in liquid nitrogen and stored at −80°C prior to RNA release. A composite sample was made by mixing an equal number of shoots. The total RNA was separated from the composite sample using the OMEGA RNA Kit in accordance with the manufacturer’s guidelines. RNA-seq library eminence control is based on the Agilent 2100 Bioanalyzer and Real-Time PCR System. Subsequently, the sequences were performed using the Illumina HiSeq ™ 2000 platform.
2.3 De novo assembly and transcriptome analysis
In the present study, the raw material in the FASTQ format was made qualitatively available using FastQC and analyzed by removing the readings of sequential adapters, anonymous nucleotides (Nradio >10%) and low quality (>5 quality scores). The enduring high-quality reading was a combined denovo via the Trinity application bundle [Citation32]. After that, the demolition sequence was removed using the CORSET software package and continued to be cut to very long unigenes. The output level of this text was standardized to allow FPKM to use RSEM and Bowtie2 with default settings [Citation33,Citation34]. Analysis of diverse gene expression (DEG) was accomplished via the DESeq2v 1.26.0 package. DEGs have | log2 (radio) | ≥1 and P value | log2 ≤ 0.05 considered important. The topGO package (v 2.37.0) in Bioconductor v 3.10 and KOBAS 3.0.0 software were used to analyze GO and KEGG enrichment of DEGs, respectively. For an overview of the full functionality, the generation was interpreted from five sources, including the eggNOG v5.0 (Evolutionary Genealogy of Genes: Non-supervised Historical Groups), GO (Gene Ontology) and KEGG release 92.0 (Kyoto Encyclopedia of Genes and Genetics) and Swiss Prot 2019_11 release. Diamond BLASTX v0.9.31 software with E-value <1 × 10–10 was used to crack against the data details of KEGG genetic annotations, eggNOG and Swiss-Prot. HMMER v3.2.1 software has been used to resist the Pfam release of the 32.0 gene annotation database. GO categories were obtained using the Blast2GO v5.2 software.
2.4 Statistical analysis
The SPSS22.0 software was used for statistical analysis. Results were shown as mean ± standard deviation (M± SD), and intergroup comparison was carried out using one-way analysis of variance (ANOVA) or Student’s t-tests. Significant difference was set to P < 0.05.
3. Results
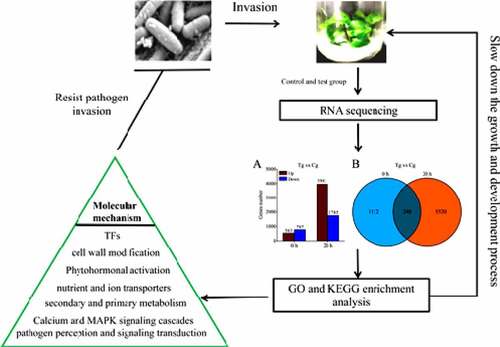
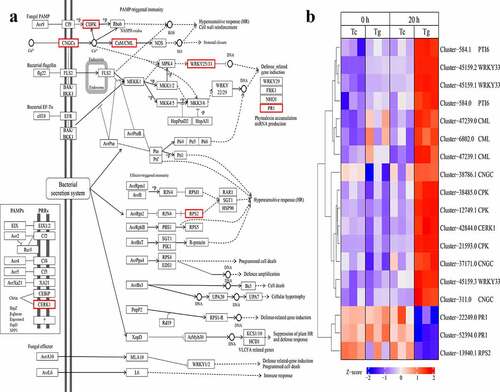
Soft rot is very harmful to PT. In order to explore the molecular mechanism of PT resistance to pathogens of soft rot infection, PT2P line and P. carotovorum were used as experimental materials. It was speculated that PT2P line would be reprogrammed transcription in the process of infection by this pathogen. Therefore, after determining the precise onset time of PT2P line being treated by the pathogen, we used RNA sequencing to analyze the DEGs of 0 h and 20 h in the process of pathogen invasion. After GO and KEGG analysis, we found that PT2P line response to the infection of P. carotovorum though regulates DEGs involves in cell wall modification, pathogen perception and signaling transduction, transcription factors, pathogenesis related and defense proteins, secondary and primary metabolisms, nutrient and ion transporters, calcium and MAPK signaling cascades, and phytohormonal activation pathway.
3.1. P. carotovorum infect PT2P line and symptoms
Through research, it was found that P. carotovorum has a strong infectivity. After 20 h of infection, PT2P line began to appear water-soaked, and some tissues became soft and collapsed ().
3.2 De novo assembly and transcriptome analysis
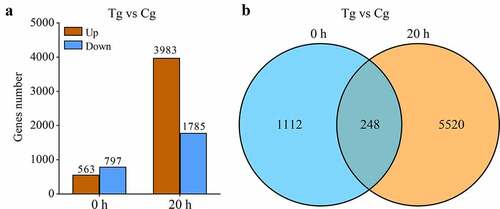
Total 575, 440, 450 pure reads were separated from 618, 196, 180 raw reads, corresponding to clean bases 86.3 Gb. The Q20/Q30 fraction, N proportion, and GC percentages were 98.54%/95.24%, 0.00% and 54.48%, respectively. Using Trinity, the clean sequences were further denovo assembled and clustered into 75, 51 unigenes. Transcript abundance findings revealed that 18, slowly countenance transcripts 612 (FPKM <1) and 19, transcripts had substantial appearance of 657 (FPKM >10). The DEGs among inoculated versus mock-inoculated trials were allocated with an edge of log2fold change ≥1. Besides, 1112 and 5520 DEGs were specifically expressed at 0 h and 20 h, respectively ( and Table S1). A total of 248 DEGs were constitutively expressed at both time points ().
3.3 DEGs functional annotations and classifications
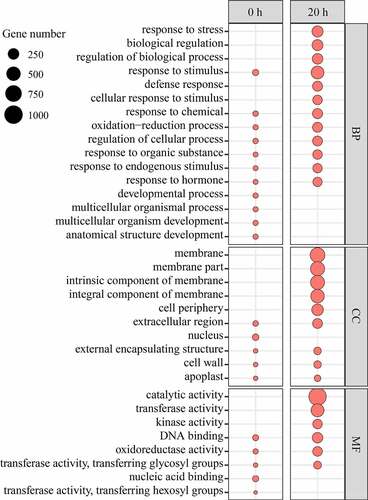
To study the potential function of these DEGs, GO enrichment analysis was carried out, and according to the results, they were divided into molecular function (MF), cell composition (CC) and biological process (BP). From the GO term analysis corresponding to BP, it was found that some BP entries only appear at 0 h, such as ‘anatomical structure development’, ‘multicellular organism development’, ‘multicellular organismal process’, ‘developmental process’. The unique BP items at 20 h include ‘response to stress’, ‘biological regulation’, ‘regulation of biological process’, ‘defense response’, ‘cellular response to stimulus’. Some BP entries appear at both time points, such as ‘response to the stimulus’, ‘response to hormone’, and ‘response to the endogenous stimulus’, ‘response to organic substance’, ‘regulation of cellular process’, ‘Oxidation-reduction process’, ‘response to chemical’ and a large number of differential genes in these items increased significantly from 0 h to 20 h.
From the GO term analysis corresponding to CC, it was found that only one CC entries ‘nucleus’ appeared at 0 h. The unique BP items at 20 h include ‘membrane’, ‘membrane part’, ‘intrinsic component of membrane’, ‘integral component of membrane’, and ‘cell periphery’. Some CC entries were appeared at both points in time, such as ‘extracellular region’, ‘external encapsulating structure’, ‘cell wall’ and ‘apoplast’. The DEGs were enriched with these items to increase significantly from 0 h to 20 h.
From the GO term analysis corresponding to MF, the unique items belonging to 0 h included ‘nucleic acid binding’, ‘transferase activity’, ‘transferring hexosyl groups’. The unique items belonging to 20 h include ‘catalytic activity’, ‘transferase activity’, ‘and ‘kinase activity’. Some CC entries appeared at both time points, such as ‘DNA binding’, ‘oxidoreductase activity’, ‘transferase activity’, ‘transferring glycosyl groups’ and the DEGs enriching these items increased significantly from 0 h to 20 h (Table S2).
3.4 DEGs involved in cell wall modification
The DEGs genes encoding glucosidase two subunit beta, xyloglucan endotransglucosylase/hydrolase protein 23/30/33 (XTH23/30/33) were up-regulated at 0 h and 20 h. However, a large number of DEGs involved in the cell wall modification process such as gene encoding the 3-ketoacyl-CoA synthase (KCS), pectinesterase (PE), cellulose synthase-Like (CesAL), xyloglucan endotransglucosylase/hydrolase (XTH), polygalacturonase inhibitor (PGI), glucosidase 2 subunit beta (GAS2), pectinesterase/pectinesterase inhibitor (PMEI) and glucan endo 1,3-beta-glucosidase were up-regulated at 20 h. In contrast, co-operative DEGs encoding diligent protein, pectate lyase (PL) and expansin (EXP) were down-regulated (Table S3).
3.5. DEGs involved in pathogen insight and signaling transduction
The DEGs receptors contain lectin domains (LecRKs) such as the G-type and L-type LecRKs at 0 h. A large number of DEGs encoding receptor kinases (RKs) were up-regulated at 20 h, such as the G-type LecRKs, L-type LecRKs, LRR receptor-like serine/threonine-protein kinase (LRRRLKs), calcium-dependent protein kinases (CDPKs), cysteine-rich receptor-like kinases (CRKs), calmodulin-binding receptor-like cytoplasmic kinase (CRCKs), mitogen-activated protein kinases (MAPKs), mitogen-activated protein kinases kinases kinases (MAPKKKs) and lysin motifs (LysMs). Meanwhile, receptors such as CBL-interacting protein kinases (CIPKs) were down-regulated (Table S4).
3.6. DEGs encoding TFs
The TFs analysis found that 20 TFs were up-regulated at 0 h and annotated as NAC, bHLH, LBD, WRKY, ERF, and G2-like. The down-regulated TFs were GeBP and C2H2. Several TFs such as WRKYs, MYBs, NACs, ZFPs, Trihelix, GeBP, B3, G2-like and C3H belonging to different families were up-regulated at 20 h. The down-regulated TFs were ZF-HD, NF-YC, GATA9, GRAS, TCP, ARF, GRF at 20 h. It was easy to notice that the pathogen-related TFs such as NACs, WRKYs and G2-like were up-regulated at 0 h and 20 h. MYB, ZFP, Trihelix and C3H were specifically up-regulated at 20 h. LBD was specifically up-regulated at 0 h (Table S5).
3.7. DEGs encoding pathogenesis and defense proteins
The DEGs analysis of pathogen-related defense proteins found that only nonspecific lipid-transfer protein 2-like (nsLTP2L) was up-regulated. The thaumatin-like protein (TLP) was down-regulated at 0 h. But after 20 h, the up-regulated genes included nsLTPs, BON1-associated protein 2 (BAP2), cationic peroxidase 1(PNC1), chitinase 1/10 L (CHI 1/10 L), endochitinase EP3-like, major allergen Pruar 1-like, major pollen allergen Bet v1-D/H-like, NDR1/HIN1-like protein 1/3 (NHL1/3), endoglucanase 12, peroxidase 12/25/51 L/N1/PX2, probable glutathione peroxidase 2, thaumatin-like protein 1, endoglucanase 12. The up-regulated defense protein does not appear at 0 h, except for nsLTPs which appear at both time points (Table S6).
3.8. DEGs involved in primary and secondary metabolisms
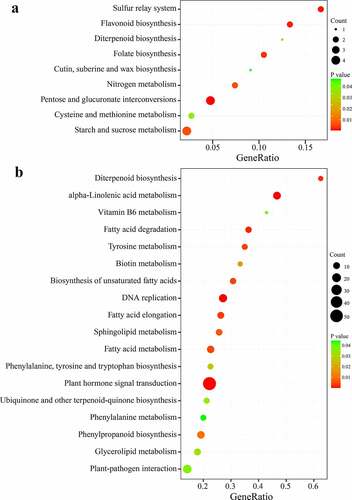
The DEGs were allocated to enriched categories to identify the pathways of PT2P line transcriptome. Among them, a large number of DEGs were related to important pathways (), such as ‘Pentose and glucuronate interconversions’, ‘Sulfur relay system’, ‘Flavonoid biosynthesis’, ‘Diterpenoid biosynthesis’, ‘Folate biosynthesis’, ‘Cutin’, ‘suberine and wax biosynthesis’, ‘Nitrogen metabolism’, ‘Cysteine and methionine metabolism’, ‘Starch and sucrose metabolism’, at 0 h. However, several defense-related pathways were a significantly enriched category at 20 h ( B), which include ‘Plant hormone signal transduction’, ‘Plant−pathogen interaction’, ‘alpha−Linolenic acid metabolism’, ‘Diterpenoid biosynthesis’, ‘Sphingolipid metabolism’, ‘Phenylalanine, tyrosine and tryptophan biosynthesis’, ‘Ubiquinone and other terpenoid−quinone biosynthesis’, ‘Phenylalanine metabolism’, ‘Phenylpropanoid biosynthesis’, ‘Glycerolipid metabolism’. Moreover, the ‘Diterpenoid biosynthesis’ pathway was constitutively enriched at two-time points (Table S7).
3.9. DEGs encoding nutrient and ion transporters
Transporter ABC transporter G family member 10-like, Aluminum-activated malate transporter 12, nucleobase-ascorbate transporter 3, AAA-ATPase, putative ABC transporter B family member 8, V-type proton ATPase subunit G. Al-activated malate transporter 12, nucleobase ascorbate transporter 3, AAA-ATPase, these three transporters also remained up-regulated at 20 h. Besides, various calcium and phospholipid along with bidirectional sugar, Cation-transporting P-type ATPase, copper, Ammonium, lysine/histidine, phosphate, sulfate, zinc, Sugar/inositol, cadmium/zinc, amino acid, GABA, nitrate, metal, organic cation/carnitine, transporters were correspondingly induced (Table S8).
3.10 Calcium and MAPK signaling pathways during P. carotovorum interactions
The data analysis found 18 DEGs were related to plant-pathogen interaction pathways at 20 h (Table S9). Among them, 15 DEGs were up-regulated and 3 DEGs were down-regulated. Cytoplasmic Ca2+ was precipitously aggregated during the perception of PAMPs. We revealed that CDPK family had been updated, regulated and phosphorylated to regulate HR. Cyclicnucleotide-gated channels (CNGCs) and CAM/CML were regulated by HR and statistical closure. Identified WRKY transport-factors (WRKY 25/33) were up-regulated and PR1 was down regulated after infection. Thus, the results suggest that the infection of P. carotovorum effects PTI pathways, and P. carotovorum bacteria can induce down-regulation of defense-related genes. Furthermore, RPS2 was down-regulated, and weakened the hypersensitivity response. Meanwhile, we found that the chitin elicitor receptor kinase 1 (CERK1) was up-regulated in response to PAMPs of P. carotovorum such as constitution ().
3.11 Phytohormonal activation in P. carotovorum infection
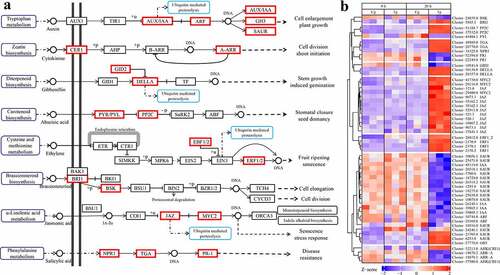
The results showed that almost no DEGs were enriched in the plant hormone signal transduction pathway at 0 h, but 55 DEGs were enriched in the plant hormone signal transduction pathway at 20 h. Among the 55 DEGs, 30 DEGs were up-regulated and 25 DEGs were down-regulated during infected by P. carotovorum (Table S10). The results showed that the expression of hormone-related genes was significantly reversed at 0 h and 20 h. Four kinds of DEGs involve the Auxin pathway which affects cell enlargement and plant growth. IAA, SAUR was down-regulated in test samples, at 20 h. A-ARR, a gene related to cytokinin synthesis, was up-regulated at 0 h but down-regulated at 20 h. Two kinds of DEGs involved the gibberellin affected stem growth and induced germination, GID2 was down-regulation and DELLA was up-regulation at 20 h. Two kinds of DEGs involve the abscisic acid which affect stomatal closure seed dormancy. PYL was down-regulated and PP2C was up-regulated at 20 h. Two kinds of DEGs were involved in the ethylene pathway which affects fruit ripening senescence. EBF1/2 and ERF1 were up-regulated at 20 h. Two DEGs were involved in the Brassinosteroid which affects cell elongation and cell division. BR11 was up-regulated and BSK was down-regulated at 20 h. Some DEGs were involved in the jasmonic acid pathway which affects ubiquitin mediated proteolysis, monoterpenoid biosynthesis, indole alkaloid biosynthesis, and senescence stress response at 20 h. NPR1 and TGA were up-regulated, but PR-1 was down-regulated ().
4. Discussion
By comparing the test group with the blank control group, we found that 563 DEGs were up-regulated and 797 DEGs were down-regulated in an experimental group at 0 h. This hints that when PT2P line was exposed to P. carotovorum, the immune response started. A large number of DEGs were changed dramatically, at 20 h. There were 3,983 DEGs that were up-regulated and 1785 DEGs that were down-regulated. It is meant that PT2P line launched a large-scale defensive response. At the same time, it was found 1112 genes specifically expressed at 0 h, and 5520 DEGs specifically expressed at 20 h. There were 248 DEGs expressed in both time points. This means that PT2P line resistance with P. carotovorum infection is more diverse and intense at 20 h.
The analysis of MF showed that only a small amount of DEGs were enriched in DNA binding, oxidoreductase activity, transfer glycosyl, nucleic acid binding, transferase activity, and transfer hexosyl function, at 0 h. The MF with abundant DEGs at 20 h was catalytic activity, transferase activity, kinase activity, DNA binding, oxidoreductase activity, transfer of glycosyl transferase activity, transferring glycosyl groups. However, the entries, which appear at 0 h, such as ‘nucleic acid binding’ and ‘transferase activity, transferring hexosyl groups’ were not detected at 20 h. This implies that the PT2P line has made tremendous adjustments to the MF in response to the pathogen invasion.
The analysis of CC showed that a small amount of DEGs were enriched in the cytoplasm, cell wall, external encapsulation structure, nucleus and extracellular region. However, there are almost no DEGs enriched in the components related to the cell membrane at 0 h. Unsurprisingly, a large number of DEGs were significantly enriched in cellular components, such as cell walls, exosomes, membrane components, membrane intrinsic components, at 20 h. PT2P line is still mainly in the stage of growth and development at 0 h. At this time, the defense response to pathogens is still in the preparatory stage. The pathogen begins to destroy the cell wall and cell membrane, causing cell component repair. Furthermore, large-scale cell component repair occurs on the cell membrane and cell wall, at 20 h.
The analysis of BP showed that a small number of DEGs were detected at 0 h. Those DEGs were begun to be enriched in ‘response to stimulus’, ‘endogenous stimuli’, ‘hormones’, and regulation of cellular processes, which are associated with the plant pathogen response, but more DEGs were involved in the development of organ structures. A large number of DEGs were significantly enriched to ‘response to stimulation’, ‘defense response’, ‘hormone response’, ‘stress response’, ‘biological regulation’, ‘regulation of cell procedures’, at 20 h. What was interesting was that there was almost no DEGs enrichment about tissue, organ structure and other developmental processes. We also observed that some GO terms shared with 0 h and 20 h, such as ‘response to stimulus’, ‘response to the endogenous stimulus’, ‘response to organic substance’, ‘regulation of cellular process’, ‘oxidation−reduction process’, ‘response to the chemical’, ‘response to hormone’. The number of DEGs enriched in those GO terms at 20 h was significantly higher than that at 0 h. The defense response at 20 h was stronger than that at 0 h. We were surprised to find that almost no DEGs were significantly enriched in development-related BP term at 20 h. Hence, it was found that PT2P line began to change growth and development state to defensive state, at 0 h. Furthermore, large-scale defense responses were caused, and the development process was slowed down, at 20 h.
The KEGG analysis found that the transcriptome was dynamic and time-dependent. Large numbers of DEGs were significantly enriched in important defense response pathways such as ‘plant endogenous hormones’, ‘plant-pathogen interactions’, ‘immune signal transduction’, ‘biosynthesis of secondary metabolites’, at 20 h.
The KEGG analysis showed that many DEGs were tangled in cell wall alteration at two times, such as KCS and XTH23/30/33 were up-regulated to restrict pathogen invasion at 0 h. The up-regulation of XTHs might be related to cell wall thickening. This suggests that a small-scale defense response was initiated even at the beginning of PT2P line exposure to pathogens. The tendency of KCS [Citation35] and CesA L [Citation36] to up-regulate expression during pathogen infection has been reported in other species. What is more, KCS involved in the biosynthesis of very long-chain fatty acids and functional wax [Citation37] improve the resistance of plants [Citation38]. PG was one of the CWDEs to resist infection. We found that PG was down-regulated and PGIP and PMEI were up-regulated in 20 h, indicating that PGIP and PMEI control PG activity [Citation39,Citation40]. The glycosidase-coding genes (beta-glucosidase (bgl44/44 l) and glucanendo 1, 3-beta-glucosidase can degrade cellulose and hemicellulose. The up-regulation of these genes proves that the cell wall structure was damaged. KCS, PE, XTH, GAS2, continuous up-regulation at 0 h and 20 h proved that the war on the cell wall at two-time points did not stop. The down-regulated genes encode diligent protein involved in the cytotoxic lignin biosynthesis [Citation41], implied that this gene is not a key gene to resist P. carotovorum infection. IAA can induce EXP, which relaxes the cell wall. As everyone knows, relaxing the cell wall was the key to plant growth, but it also makes plants vulnerable to biological invaders. It was found that IAA and EXP were down-regulated at 20 h.
Moreover, there was no differential expression of transcription factors LBD at 20 h compared with the up-regulated expression of 0 h. LBD promoted plant cells no longer relax to promote development and transform the stage for the defense pathogen invasion [Citation42,Citation43]. In our research, the secondary metabolite biosynthesis was activated on a large scale at 20 h and this metabolic process was selectively and progressively induced. The initiation of the defense response was observed at 20 h because the number of DEGs related to ‘response to stress’ in GO analysis increased from 46 at 0 h to 457 at 20 h. Simultaneously, ‘phenylalanine’, ‘tyrosine and tryptophan biosynthesis’, ‘plant-pathogen interaction’ and ‘plant hormone signal transduction’ pathways were activated. In our research, DEGs enrichment associated with the biosynthesis of terpenes-quinones, such as phenylpropane and quinone, mainly induced at 20 h. The phenylalanine biosynthetic pathway was essential to induce the biosynthesis of p-phenol and initiated the basic defense response. The ‘Flavonoid biosynthesis’, might be associated with limited the growth of pathogens, significantly induced at 0 h, but not significant enrichment at 20 h. This means that the flavonoid metabolism has begun to induce resistance at the initial stage of exposure to pathogenic bacteria, and when the disease occurs, the related resistance may have failed and switched to other more effective methods. The ‘Diterpenoid biosynthesis’ was significantly enriched at 20 h. It might contribute to the defense [Citation44] of P. carotovorum.
In our research, the data show that the PRRs were significantly induced at 20 h. The up-regulation was also reflected in the significant enrichment of the ‘kinase activity’ at 20 h. The up-regulation of WRKY may involve PAMP or effector protein directly activated by PTI pathway [Citation45]. RLK has been shown to be induced by necrotrophs immune responses, whereas Lys M-RLK can be treated as a defense activator [Citation46]. It is worth mentioning that G types of LecRKs were involved in plant defense, while RLKs might interact directly with the conduction of downstream reactions [Citation47]. P. carotovorum may induce the expression of genes related to planting hypersensitivity to promote infection. Therefore, the up-regulation of RLKs involved in this hypersensitivity reaction may promote the susceptibility of PT2P line to a certain extent.
Our data show that at 20 h, PTI activation induces various branch kinases (STKs, CRCKs, CRKs) involved in the immune-signaling pathway [Citation48,Citation49]. Our research data show that the SRK and CDPK were mainly up-regulated at 20 h, which might cause widespread plant resistance to pathogens. The TFs related to the development of the disease, such as NAC, bHLH, WRKY, ERF, G2-like were up-regulated at 0 h and these TFs were still up-regulated at 20 h. Some of the TFs with significant functions were particularly noteworthy. For example, bHLH can promote water absorption in tomato leaves and make the leaves become water soaking. Some pathogens can enter the apoplast with the diffusion of water and further infect plants [Citation50]. Our data show that bHLH were up-regulated and PT2P line showed water soaking symptoms. Therefore, we speculate that the infection of P. carotovorum on PT2P line may be similar to that of X. gardneri infecting tomatoes. The up-regulation of NAC may regulate the accumulation of sugar and amino acids in the ABA pathway and exert its anti-disease function to resist bacteria [Citation51]. In other plants, WRKYs can interact directly with PAMP or effector proteins to inhibit PTI and ETI [Citation52]. WRKYs were regulated by MAPKs and could trigger ROS outbreak [Citation53] and regulated hormone signaling pathway [Citation45]. WRKYs adjust the small RNA to promote plant immune through epigenetic mechanisms of histone methylation, proteasome-mediated degradation and intracellular retrograde signaling [Citation52]. ERF had a new function in coordinating wound defense response, and repair might be involved in cell wall cellulose biosynthesis [Citation54]. ERF up-regulated at 20 h, might play an important role in wound repair of PT2P line. Simultaneously, the specific transcription factors induced at 20 h and also aroused our interest, such as ZFPs could regulate plant resistance to pathogens [Citation55], and it is also the most widely induced transcription factor at 20 h. ERF was an integrator of the hormone pathway, directly responsible for the transcriptional regulation of several jasmonate (JA)/ethylene (ET) responsive defense genes [Citation56]. JA and ET play important roles in regulating defense responses to resist bacteria [Citation57]. It was surprising to find that all the TFs associated with growth and development such as ZF-HD [Citation58], NF-YC [Citation59], GRAS [Citation60], TCP [Citation61], ARF [Citation62], GRF [Citation63] were down-regulated at 20 h. This again confirms our hypothesis that PT2P line slowed down the growth and development process in response to P. carotovorum infection.
Our data show that in the process of abscisic acid hormone signaling, the down-regulation of PYL and up-regulation of PP2C inhibited stomatal closure and seed dormancy and increase susceptibility of PT2P line. At 20 h, a large number of DEGs were enriched in α- linoleic acid metabolic pathways which might increase jasmonic acid production. The up-regulation of JAZ and MYC2 promoted the senescence and stress response, monoterpene biosynthesis and indole alkaloid biosynthesis. Besides, we also observed that a large number of DEGs are significantly enriched in the phenylalanine metabolic pathway at 20 h, accompanying PAL up-regulation, which can also synthesize salicylic acid [Citation64]. NRP1 and TGA were up-regulated at 20 h and PR1 down-regulated, while PR1 was up-regulated at 0 h. This suggests that down-regulated PR1 weakens disease resistance and PT2P line might be prepared to strengthen resistance through DNA transcription to PR1 by regulating the salicylic acid pathway because we have found that NPR1 and TGA were up-regulated at 20 h (). This implies that the response of PT2P line to P. carotovorum infection was a tug of war in the salicylic acid pathway. It was found that a number of DEGs were significantly enriched in diterpene biosynthesis; this process can synthesize gibberellins, with GID2 down-regulation and DELLA up-regulation promoting ubiquitin mediated proteolysis. TFs were not found to be involved in this pathway, meaning that stem growth and induced germination cannot be promoted. Down-regulation of A-ARR genes inhibits cell division and seems to slow the developmental process. The down-regulation of AUX/IAA and ARF caused most SAUR and AUX/IAA down-regulation to inhibit cell enlargement and plant growth. P. carotovorum may use another virulence factor to manipulate host responses and promote colonization, which is a product of ROS, leading to oxidative bursts that regulate HR to promote susceptibility. We found that the number of DEGs enriched at ‘oxidoreductase activity’ was significantly increased from 0 h to 20 h.
In another word, this study provides a perspective to understand the molecular mechanism of PT2P line resistance to soft rot pathogens. At the same time, it will lay a theoretical foundation for controlling the soft rot caused by this pathogen in many ways, such as controlling bHLH, EXP and RLKs in an appropriate way to reduce the susceptibility of PT2P line. The ERF and other genes were regulated to speed up the repair of the cell wall, block or strengthen the specific process to control the soft rot caused by this pathogen, and even for screening resistant varieties of PT2P line.
5. Conclusion
PT2P line exhibited symptoms after P. carotovorum bacterial infection at 20 h. PT2P line down-regulated the expression of growth and development-related genes and started to reprogram disease resistance-related genes, during infection. The molecular mechanism of PT2P line resistance to pathogens is regulated by the expression of genes relative to ‘pathogen perception and signaling transduction’, ‘cell wall modification’, ‘pathogenesis-related and defense proteins’, ‘secondary and primary metabolism’, ‘nutrient and ion transporters’, ‘Calcium and MAPK signaling cascades’, ‘Phytohormonal activation’.
Supplemental Material
Download ()Acknowledgements
This work has been supported by the Zunyi Medical University Master’s Startup Funding Project (F909), Guizhou Province Department of Education Project (KY[2017]058), National Natural Science Foundation of China Project (31560079), Distinguished High-Level Talents Research Grant from a Guizhou Science and Technology Corporation Platform Talents Fund (Grant No.: [2017]5733-001 & CK-1130-002) and Zunyi City Company Science and Technology Project of Guizhou Tobacco Company (201903).
Disclosure statement
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Supplemental material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lu J, Liu JN, Sarsaiya S, et al. Phenotypic and Transcriptomic analysis of two Pinellia ternata varieties T2 line and T2Plus line. Sci Rep. 2020;10:46141.
- Hu X, Fang Q, Li S, et al. Isolation and characterization of endophytic and rhizosphere bacterial antagonists of soft rot pathogen from Pinellia ternata. FEMS Microbiol Lett. 2009;295(1):10–16.
- Somarathna T, Fernando W, Ranaweera K, et al. Antimicrobial activity and phytochemical screening of Alpinia malaccensis (Ran-kiriya) resist food-borne bacteria. J Appl Microbiol. 2018;125(5):1276–1285.
- Neupane S, Ma Q, Mathew FM, et al. Evolutionary divergence of TNL disease-resistant proteins in soybean (Glycine max) and common bean (Phaseolus vulgaris). Biochem Genet. 2018;56(4):397–422.
- Niu L, Zhong X, Zhang Y, et al. Enhanced tolerance to Phytophthora root and stem rot by over-expression of the plant antimicrobial peptide CaAMP1 gene in soybean. BMC Genet. 2020;21(1):68.
- Joshi RK, Kar B, Nayak S. Survey and characterization of NBS-LRR (R) genes in Curcuma longa transcriptome. Bioinformation. 2011;6(9):360–363.
- Doehlemann G, Okmen B, Zhu W, et al. Plant pathogenic fungi. MicrobiologySpectrum. 2017;5(1). DOI:10.1128/microbiolspec.FUNK-0023-2016
- Remus-Emsermann M, Schlechter RO. Phyllosphere microbiology: at the interface between microbial individuals and the plant host. New Phytol. 2018;218(4):1327–1333.
- Wang C, Wang C, Zou J, et al. Epigenetics in the plant-virus interaction. Plant Cell Rep. 2019 Sep;38(9):1031–1038. .
- Amoah ID, Adegoke AA, Stenstrom TA. Soil-transmitted helminth infections associated with wastewater and sludge reuse: a review of current evidence. Trop Med Int Health. 2018;23(7):692–703.
- Ohkama-Ohtsu N, Wasaki J. Recent progress in plant nutrition research: cross-talk between nutrients, plant physiology and soil microorganisms. Plant Cell Physiol. 2010;51(8):1255–1264.
- Giraldo MC, Dagdas YF, Gupta YK, et al. Two distinct secretion systems facilitate tissue invasion by the rice blast fungus Magnaporthe oryzae. Nat Commun. 2013;4(1):1996.
- Macho AP, Zipfel C. Plant PRRs and the activation of innate immune signaling. Mol Cells. 2014;54(2):263–272.
- Couto D, Zipfel C. Regulation of pattern recognition receptor signalling in plants. Nat Rev Immunol. 2016;16(9):537–552.
- Xiong J, Zhu H, Bai Y, et al. RNA sequencing-based transcriptome analysis of mature strawberry fruit infected by necrotrophic fungal pathogen Botrytis cinerea. Physiolog Mol Plant Pathol. 2018;104:77–85.
- Segonzac C, Zipfel C. Activation of plant pattern-recognition receptors by bacteria. Current opinion in biotechnology. 2011;14(1):54–61.
- Park CJ, Caddell DF, Ronald PC. Protein phosphorylation in plant immunity: insights into the regulation of pattern recognition receptor-mediated signaling. Front Plant Sci. 2012;3:177.
- Saijo Y, Loo EP, Yasuda S. Pattern recognition receptors and signaling in plant-microbe interactions. Plant J. 2018;93(4):592–613.
- Boudsocq M, Sheen J. CDPKs in immune and stress signaling. Trends Plant Sci. 2013;18(1):30–40.
- Meng X, Zhang S. MAPK cascades in plant disease resistance signaling. Annu Rev Phytopathol. 2013;51(1):245–266.
- Engelsdorf T, Kjaer L, Gigli-Bisceglia N, et al. Correction to: functional characterization of genes mediating cell wall metabolism and responses to plant cell wall integrity impairment. BMC Mol Biol. 2019;19(1):385.
- Gardner JG, Keating DH. Genetic and functional genomic approaches for the study of plant cell wall degradation in Cellvibrio japonicus. Methods Enzymol. 2012;510:331–347.
- Hematy K, Cherk C, Somerville S. Host-pathogen warfare at the plant cell wall. Curr Opin Plant Biol. 2009;12(4):406–413.
- Li N, Han X, Feng D, et al. Signaling crosstalk between salicylic acid and ethylene/jasmonate in plant defense: do we understand what they are whispering? Int J Mol Sci. 2019;20(3). DOI:10.3390/ijms20030671
- Tsuda K, Somssich IE. Transcriptional networks in plant immunity. New Phytol. 2015;206(3):932–947.
- Boller T, He SY. Innate immunity in plants: an arms race between pattern recognition receptors in plants and effectors in microbial pathogens. Science. 2009;324(5928):742–744.
- Lee JH, Kim H, Chae WB, et al. Pattern recognition receptors and their interactions with bacterial type III effectors in plants. Genes Genomics. 2019;41(5):499–506.
- Pieterse CM, Van Der Does D, Zamioudis C, et al. Hormonal modulation of plant immunity. Ann Rev Cell Develop Biol. 2012;28(1):489–521.
- Janda M, Lamparova L, Zubikova A, et al. Temporary heat stress suppresses PAMP-triggered immunity and resistance to bacteria in Arabidopsis thaliana. Mol Plant Pathol. 2019;20(7):1005–1012.
- Wise RP, Moscou MJ, Bogdanove AJ, et al. Transcript profiling in host-pathogen interactions. Annu Rev Phytopathol. 2007;45(1):329–369.
- Huang X, Jing Y, Liu DJ, et al. Whole-transcriptome sequencing of Pinellia ternata using the Illumina platform. Genet Mol Res. 2016;15(2). DOI:10.4238/gmr.15028062
- Grabherr MG, Haas BJ, Yassour M, et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat Biotechnol. 2011;29(7):644–652.
- Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9(4):357–359.
- Li B, Dewey CN. RSEM: accurate transcript quantification from RNA-Seq data with or without a reference genome. BMC Bioinformatics. 2011;12(1):323.
- Wang X, Zhi P, Fan Q, et al. Wheat CHD3 protein TaCHR729 regulates the cuticular wax biosynthesis required for stimulating germination of Blumeria graminis f.sp. tritici. J Exp Bot. 2019;7,70(2):701–713.
- Douchkov D, Lueck S, Hensel G, et al. The barley (Hordeum vulgare) cellulose synthase-like D2 gene (HvCslD2) mediates penetration resistance to host-adapted and nonhost isolates of the powdery mildew fungus. New Phytol. 2016;212(2):421–433.
- Yang T, Li Y, Liu Y, et al. The 3-ketoacyl-CoA synthase WFL is involved in lateral organ development and cuticular wax synthesis in Medicago truncatula. Plant Mol Biol. 2021;105(1–2):193–204.
- Zhang YL, Zhang CL, Wang GL, et al. The R2R3 MYB transcription factor MdMYB30 modulates plant resistance resist pathogens by regulating cuticular wax biosynthesis. BMC Mol Biol. 2019;19(1):362.
- Rathinam M, Rao U, Sreevathsa R. Novel biotechnological strategies to combat biotic stresses: polygalacturonase inhibitor (PGIP) proteins as a promising comprehensive option. Appl Microbiol Biotechnol. 2020;104(6):2333–2342.
- Tundo S, Kalunke R, Janni M, et al. Pyramiding PvPGIP2 and TAXI-III but not PvPGIP2 and PMEI enhances resistance resist Fusarium graminearum. Mol Plant-Microbe Interact. 2016;29(8):629–639.
- Sanchez-Elordi E, Sterling RM, Santiago R, et al. Increase in cytotoxic lignans production after smut infection in sugar cane plants. J Plant Physiol. 2020;244:153087.
- Lee HW, Kim MJ, Kim NY, et al. LBD18 acts as a transcriptional activator that directly binds to the EXPANSIN14 promoter in promoting lateral root emergence of Arabidopsis. Plant J. 2013;73(2):212–224.
- Lionetti V, Fabri E, De Caroli M, et al. Three Pectin Methylesterase inhibitors protect cell wall integrity for Arabidopsis immunity to Botrytis. Plant Physiol. 2017;173(3):1844–1863.
- Christie N, Myburg AA, Joubert F, et al. Systems genetics reveals a transcriptional network associated with susceptibility in the maize-grey leaf spot pathosystem. Plant J. 2017;89(4):746–763.
- Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16(2):319–331.
- Buendia L, Girardin A, Wang T, et al. LysM receptor-like kinase and LysM receptor-like protein families: an update on Phylogeny and functional characterization. Front Plant Sci. 2018;9:1531.
- Lannoo N, Van Damme EJ. Lectin domains at the frontiers of plant defense. Front Plant Sci. 2014;5:397.
- Tang D, Wang G, Zhou JM. Receptor kinases in plant-pathogen interactions: more than pattern recognition. Plant Cell. 2017;29(4):618–637.
- Wang JP, Xu YP, Munyampundu JP, et al. Calcium-dependent protein kinase (CDPK) and CDPK-related kinase (CRK) gene families in tomato: genome-wide identification and functional analyses in disease resistance. Mol Gene Genomics. 2016;291(2):661–676.
- Schwartz AR, Morbitzer R, Lahaye T, et al. TALE-induced bHLH transcription factors that activate a pectate lyase contribute to water soaking in bacterial spot of tomato. Proc Natl Acad Sci U S A. 2017;114(5):E897–E903.
- Liu Q, Yan S, Huang W, et al. NAC transcription factor ONAC066 positively regulates disease resistance by suppressing the ABA signaling pathway in rice. Plant Mol Biol. 2018;98(4–5):289–302.
- Bakshi M, Oelmuller R. WRKY transcription factors: jack of many trades in plants. Plant Signal Behav. 2014;9(2):e27700.
- Adachi H, Nakano T, Miyagawa N, et al. WRKY transcription factors phosphorylated by MAPK regulate a plant immune NADPH oxidase in nicotiana benthamiana. Plant Cell. 2015;27(9):2645–2663.
- Heyman J, Canher B, Bisht A, et al. Emerging role of the plant ERF transcription factors in coordinating wound defense responses and repair. J Cell Sci. 2018;131(2):jcs208215.
- Noman A, Aqeel M, Khalid N, et al. Zinc finger protein transcription factors: integrated line of action for plant antimicrobial activity. Microb Pathogen. 2019;132:141–149.
- Huang PY, Catinot J, Zimmerli L. Ethylene response factors in Arabidopsis immunity. J Exp Bot. 2016;67(5):1231–1241.
- Dinolfo MI, Castanares E, Stenglein SA. Resistance of Fusarium poae in Arabidopsis leaves requires mainly functional JA and ET signaling pathways. Fungal Biol. 2017;121(10):841–848.
- Liu M, Wang X, Sun W, et al. Genome-wide investigation of the ZF-HD gene family in Tartary buckwheat (Fagopyrum tataricum). Bmc Plant Biol. 2019;19(1):248.
- Palmeros-Suarez PA, Massange-Sanchez JA, Martinez-Gallardo NA, et al. The overexpression of an Amaranthus hypochondriacus NF-YC gene modifies growth and confers water deficit stress resistance in Arabidopsis. Plant Sci. 2015;240:25–40.
- Grimplet J, Agudelo-Romero P, Teixeira RT, et al. Structural and functional analysis of the GRAS gene family in grapevine indicates a role of GRAS proteins in the control of development and stress responses. Front Plant Sci. 2016;7:353.
- Chai W, Jiang P, Huang G, et al. Identification and expression profiling analysis of TCP family genes involved in growth and development in maize. Physiol Mol Biol Plants. 2017;23(4):779–791.
- Liscum E, Reed JW. Genetics of Aux/IAA and ARF action in plant growth and development. Plant Mol Biol. 2002;49(3–4):387–400.
- Wu ZJ, Wang WL, Zhuang J. Developmental processes and responses to hormonal stimuli in tea plant (Camellia sinensis) leaves are controlled by GRF and GIF gene families. Funct Integrat Genomics. 2017;17(5):503–512.
- Shine MB, Yang JW, El-Habbak M, et al. Cooperative functioning between phenylalanine ammonia lyase and isochorismate synthase activities contributes to salicylic acid biosynthesis in soybean. New Phytol. 2016;212(3):627–636.