ABSTRACT
Bladder cancer is one of the most common malignant tumors worldwide. Accordingly, its incidence and mortality are high. One of the characteristics of cancer is genomic instability. New studies suggest that long non-coding RNAs (lncRNAs) play an important role in maintaining genomic instability. This study aimed to identify a genomic instability-associated lncRNA signature to predict the outcome of patients with bladder cancer. We downloaded data for bladder cancer patients from The Cancer Genome Atlas database to obtain lncRNA expression profiles as well as somatic mutation profiles. Using the lncRNA computational framework, a genomic instability-related lncRNA signature (GIlncSig) was established and the prognostic value of this signature was assessed and validated. A five-lncRNA signature based on genomic instability (CFAP58-DT, MIR100HG, LINC02446, AC078880.3, and LINC01833) was obtained from 58 differentially expressed lncRNAs. Patients were divided into high-risk and low-risk groups, with the high-risk group having a substantially worse prognosis than the low-risk group. Univariate and multivariate Cox analyses indicated that GIlncSig may be an independent prognostic factor; this finding was subsequently validated. In addition, enrichment analysis indicated that GIlncSig is associated with genomic instability in bladder cancer. GIlncSig has a predictive value for the prognosis of bladder cancer patients and provides guidance for the clinical treatment of these patients.
Introduction
Bladder cancer is one of the most common malignant tumors in the world. In fact, it is the fourth most common malignant tumor in men, and its incidence in women is increasing yearly [Citation1]. At the time of diagnosis, approximately 70% – 75% of patients have non-muscle-invasive bladder cancer (NMIBC) and 25% – 30% have muscle-invasive bladder cancer (MIBC) [Citation2,Citation3]. The treatment modality differs between NMIBC and MIBC.In fact, the treatment modality of NMIBC is transurethral resection of bladder tumor and postoperative intravesical chemotherapy, whereas that of MIBC is radical cystectomy [Citation4]. The prognosis of MIBC is markedly worse than that of NMIBC [Citation5]. Therefore, new biomarkers are needed to predict the outcome of patients with bladder cancer and assess the clinical efficacy of treatment.
Genomic instability is inextricably related to the occurrence and development of tumors and is considered to be one of the characteristics of tumors [Citation6,Citation7]. Long non-coding RNAs (lncRNAs) are most commonly defined as non-coding RNAs greater than 200 nucleotides in length [Citation8]. In recent years, lncRNAs have been reported to play an important role in tumor development and metastasis [Citation9–11], as well as genomic instability. Wang [Citation12] et al. found a lncRNA GUARD, that is important for maintaining genome integrity. Further,Kirsten [Citation13] et al. suggested that mitosis-associated lncRNAs have a remarkble impact on the maintainance of breast cancer genome stability. However, exploring the role of the lncRNAs associated with genomic instability in cancer is extensively needed.
In this study, we sought to obtain relevant lncRNA data as well as mutation data for bladder cancer from The Cancer Genome Atlas (TCGA) database and construct a genomic instability-related lncRNA signature (GIlncSig) to explore its predictive role in the prognosis of bladder cancer. Our hypothesis is that GIlncSig has the ability to predict the prognosis of patients with bladder cancer. The aim of our study is to evaluate and validate the prognostic ability of GIlncSig and its independent prognostic value. Our goal is to guide the clinical use of GIlncSig in bladder cancer.
Methods
Data collection and processing
Transcriptional profiles, clinical data, and somatic mutation profiles of patients with bladder cancer were downloaded from TCGA database (https://portal.gdc.cancer.gov/). Subsequently, the expression levels of the lncRNAs and mRNAs in bladder cancer samples were extracted separately from the transcriptional profiling data. We obtained 395 samples with complete lncRNA expression profiles, mRNA expression profiles, clinical features, and somatic mutation expression profiles. Subsequently, 395 bladder cancer samples were randomly divided into training and testing sets. The training set contained 200 samples and was used to build the prognostic risk model. The prognostic risk model contained 195 samples and was used to validate the established prognostic risk model. The Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) database was searched to obtain the dataset, GSE31684, for bladder cancer, which contains survival data for validation. The clinical and pathological characteristics of the tumor samples are shown in .
Table 1. Clinical and pathological characteristics of bladder cancer samples in this study
Identifying the genomic instability-related lncRNAs
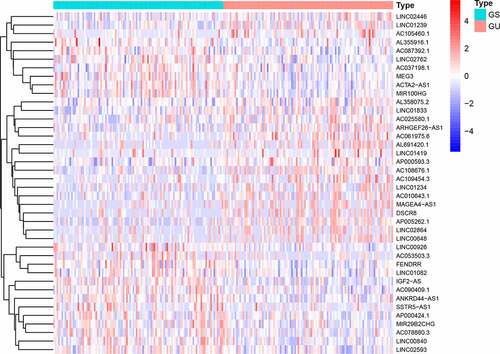
We used the computational framework designed by [Citation14] et al. to identify the lncRNAs associated with genomic instability. Briefly, we calculated the number of somatic mutations in each sample, arranged the samples in ascending order of the number of somatic mutations, assigned the first 25% to the genomic stable (GS)-groups and the last 25% to the genomic unstable (GU) like-groups, and used the ‘limma’ package of R version 4.0.2 to perform differential analysis of the lncRNA expression profiles in the two groups (false discovery rate (FDR) < 0.05, fold change (FC) > 1). The differentially expressed lncRNAs are genomic instability-related lncRNAs.
Statistical analysis
Based on the resulting genomic instability-associated lncRNAs, univariate Cox regression analysis was used to identify prognostically-relevant lncRNAs among the genomic instability-related lncRNAs. According to the prognostically-relevant lncRNAs, GIlncSig was constructed using multivariate Cox regression analysis with the formula: RiskScore = ∑ki=1 CiEi, where RiskScore represents the risk score of the patient, Ci represents the coefficient of the ith lncRNA, and Ei represents the expression level of the ith lncRNA in this patient. According to the median risk value of patients in the training set, patients with risk values greater than the median value were placed in the high-risk group, and patients with risk values lower than the median value were placed in the low-risk group.
The survival difference between the high-risk and low-risk groups was compared by the ‘survdiff’ and ‘survfit’ functions in the ‘survival’ package of R using the Kaplan–Meier (KM) method and the log-rank test (p < 0.05 indicated a significant difference). The accuracy of GIlncSig was assessed using a time-dependent receiver operating characteristic (ROC) curve. Univariate and multivariate Cox regression analyses and stratified analyses were employed for independent prognostic assessment of GIlncSig.
Enrichment analysis
We identified mRNAs that were expressed in pairs with the lncRNAs associated with genomic instability; each lncRNA selected the top ten mRNAs associated with it. Thereafter, a co-expression network was constructed. The ‘clusterProfiler’ package and ‘ggplot2ʹ package of R were used to perform Gene Ontology (GO) enrichment analysis and Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway analysis of the mRNAs co-expressed with the lncRNAs.
Chemosensitivity of GilncSig
We used the ‘pRRophetic’ package of R to calculate the half inhibitory concentration (IC50) of the commonly-used chemotherapeutic agents for bladder cancer and compare the difference in IC50 between the high- and low-risk groups of GIlncSig.
Results
In this study, we constructed a prognostic signature through the TCGA database, and we hypothesized that the signature has the ability to predict the prognosis of patients with bladder cancer, and the goal of our study is to provide a guiding role for the clinical practice of patients with bladder cancer through this signature. In the present study, we first downloaded data from bladder cancer patients from the TCGA database, picked out genomic unstable lncRNAs and established a lncRNA signature. Subsequently, the prognostic performance of the signature was evaluated and validated, and the model was combined with the clinical data of bladder cancer patients to provide a guiding role for the clinical practice of bladder cancer patients.
Identification of genomic instability-related lncRNAs
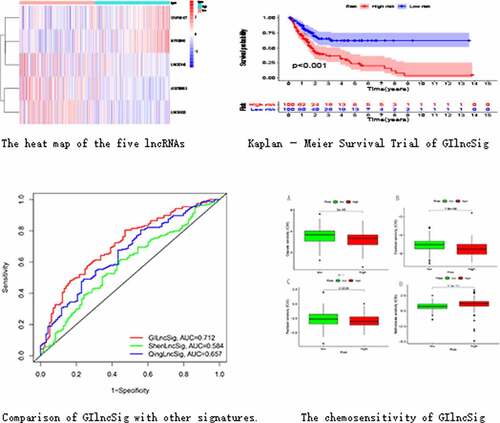
We divided the samples into GS-like and GU-like groups, according to somatic mutations, and compared the lncRNA expression between the two groups to identify differentially expressed lncRNAs. As shown in , 58 differentially expressed lncRNAs were identified.
Enrichment analysis
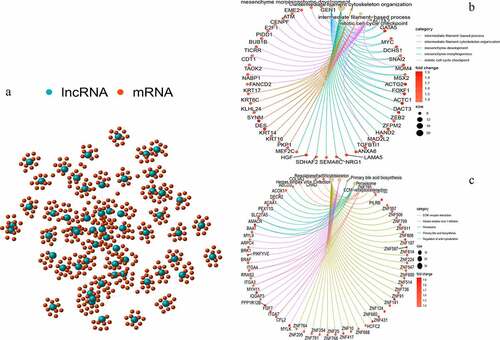
We performed a co-expression analysis of the resulting lncRNAs to construct a co-expression network (). GO and KEGG enrichment analyses of the mRNAs co-expressed by lncRNAs were also conducted. Based on GO enrichment analysis, the mRNAs in this co-expression network were mainly enriched in the intermediate filament-based process, intermediate filament organization cytoskeleton, and mitotic cell cycle (); these are important links to genomic instability. According to KEGG pathway enrichment analysis, there were two enriched pathways: peroxisome and regulation of the actin cytoskeleton, which are associated with genomic instability (). These lncRNAs were thus defined as genomic instability-related lncRNAs.
Figure 2. Co-expression network and enrichment analysis of differential lncRNAs. (a) Expression networks of differential lncRNAs and their associated mRNAs. The blue circles represent lncRNAs and red circles represent mRNAs. (b) Gene Ontology (GO) enrichment analysis and (c) Kyoto Encyclopedia of Gene and Genomes (KEGG) pathway analysis of mRNAs associated with differential lncRNAs
Identification of the GIlncSig
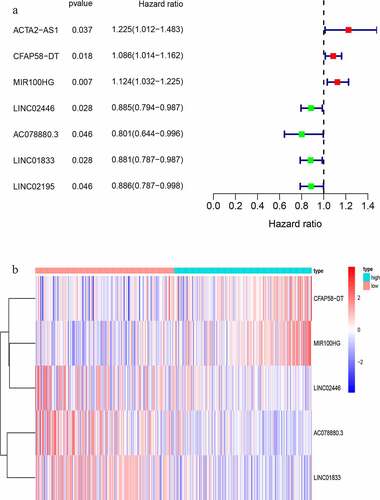
We randomly divided 396 bladder cancer samples into a training group (n = 200) and a test group (n = 196). Thereafter, the expression levels of the differential lncRNAs in the samples were combined with prognostic information using the ‘limma’ package of R. Based on univariate COX analysis, seven lncRNAs were identified to be associated with bladder cancer prognosis (). Multivariate Cox analysis was also employed to further screen the resulting seven lncRNAs to obtain a signature composed of five lncRNAs associated with bladder cancer prognosis (). A GIlncSig consisting of five lncRNAs was then constructed. The risk score of this signature was calculated as follws: expression of CFAP58-DT*0.137287+ expression of MIR100HG*0.100184+ expression of LINC02446* (−0.182782) +expression of AC078880.3* (−0.174686) +expression of LINC01833* (−0.164337). The risk value of the samples in the training group was calculated using the above formula. Thereafter, the median value was used to divide the samples into high- and low-risk groups. The expression levels of GIlncSig in the high- and low-risk groups are shown in .
Table 2. LncRNAs associated with the prognosis of bladder cancer patients obtained after multivariate COX analysis
Figure 3. Identification of genomic instability-related lncRNA signature (GIlncSig). (a) Prognostic relevant lncRNAs obtained after univariate COX analysis. (b) Expression heatmap of five lncRNAs obtained after multivariate COX analysis. The abscissa is the bladder cancer sample, with blue representing the high-risk group and red representing the low-risk group
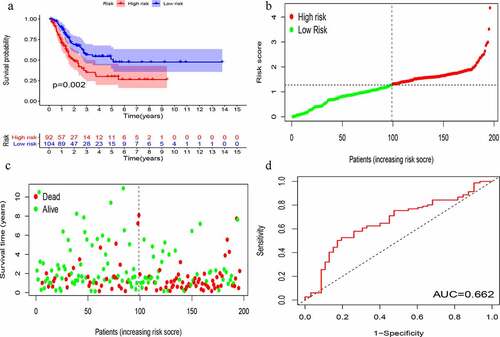
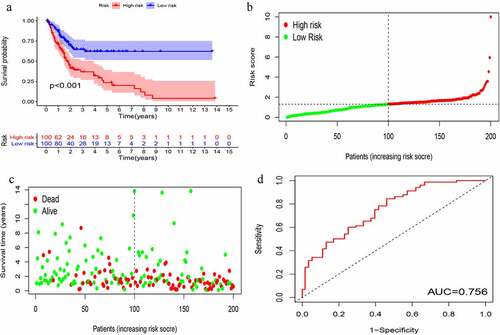
KM survival analysis was carried out on GIlncSig. As shown in , in the training group, the survival rate of patients in the low-risk group was significantly higher than that of patients in the high-risk group. Further, time-dependent ROC curve analysis showed that the area under the ROC curve (AUC) at 5 years was 0.756 (). The results of the test group were consistent with those of the training group (). These results suggest that patients in the high-risk group had a worse prognosis than those in the low-risk group.
Figure 4. Evaluation of instability-related lncRNA genomic signature (GIlncSig) in the training group. (a) Kaplan – Meier Survival Trial of GIlncSig for High and Low Risk Groups. (b) Risk score for each tumor sample. (c) Survival outcome for each tumor sample. (d) Time-dependent ROC curve at 5 years
Clinical correlation analysis of the GIlncSig
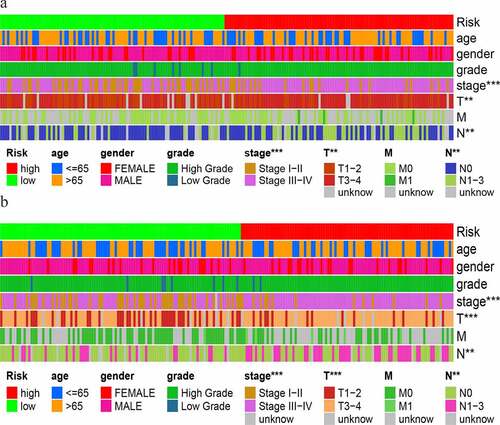
We used the Wilcoxon signed-rank test to explore the relationship between risk and the clinical characteristics. As shown in , the stages, T stage and N stage, were significantly correlated with the risk score in the training group. This result was also obtained in the test group ().
Independent prognostic analysis of GIlncSig
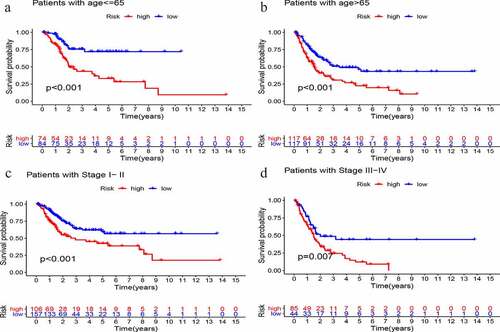
Multivariate Cox regression analysis of age, sex, grade, stage, and GIlncSig risk score was performed to determine the independent prognostic value of GIlncSig. As shown in , in addition to the risk score, age and stage also have important prognostic values. Therefore, we performed a further validation to determine whether the prognostic value of GIlncSig is independent of age and stage. Briefly, we divided all patients into young and elderly groups according to age, with 65 years as the cutoff. GIlncSig was then used to divide each group into high-risk and low-risk groups. As shown in , in the young group,the survival rate of the high-risk group was significantly lower than that of the low-risk group (P < 0.001). Similar results were obtained for elderly patients (; P < 0.001). Subsequently, we divided the patients into early (stage I–II) and advanced (stage III–IV) groups according to the pathological stage, and further divided the early group into high- and low-risk groups. As shown in , there was a significant difference in survival between the high- and low-risk groups. Moreover, the same results were obtained in the advanced stage group (; P = .007). Based on the above results, GIlncSig has an independent prognostic value in bladder cancer.
Table 3. Independence of GIlncSig by univariate and multivariate COX analysis
Comparison of the GILncSig to other LncRNA-Related signatures
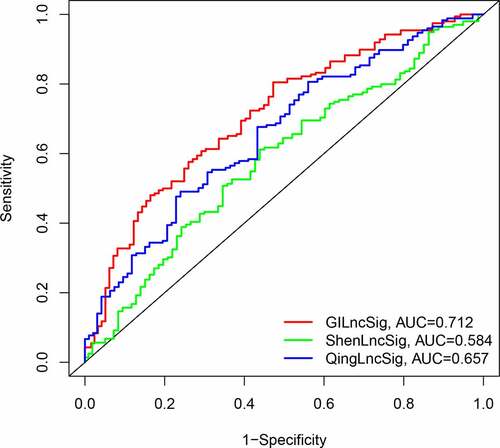
We compared the resulting GIlncSig to other published signatures related to lncRNAs; the first signature is the 8-lncRNA signature (ShenlncSig) by Shen [Citation15] et al. and the second signature is the 6-lncRNA signature (QinglncSig) by Qing [Citation16] et al. As shown in , the 5-year AUC of GIlncSig was 0.712; this value was higher than that of ShenlncSig (AUC = 0.584) and QinglncSig (AUC = 0.657). Our AUC values are based on all bladder cancer samples. There were five lncRNAs in our signature, this number is lower than that in ShenlncSig and QinglncSig (eight and six, respectively). Altogether, these findings suggest that GIlncSig has a better prognostic value than the other signatures.
Validation of the GILncSig in the GEO database
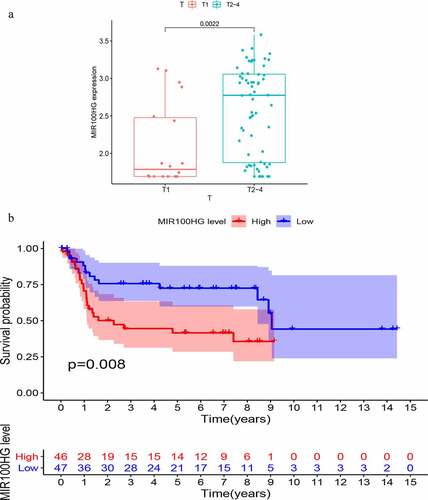
To further validate the prognostic value of GIlncSig, we found three datasets, GSE32894, GSE31684 and GSE48075, with clinical features in the GEO database. However, only one lncRNA (MIR100HG) was found in the expression matrix of GSE36184. Therefore, we proceeded to explore the association between MIR100HG and bladder cancer. This lncRNA was associated with the T stage of bladder cancer. As shown in , the T stage is positively correlated with the expression of MIR100HG (P = 0.0022). Finally, a KM survival analysis was performed for MIR100HG. As shown in , the expression of MIR100HG was negatively correlated with patient survival (P = 0.008). Such findings are similar to those obtained using TCGA database.
Chemosensitivity of the GIlncSig
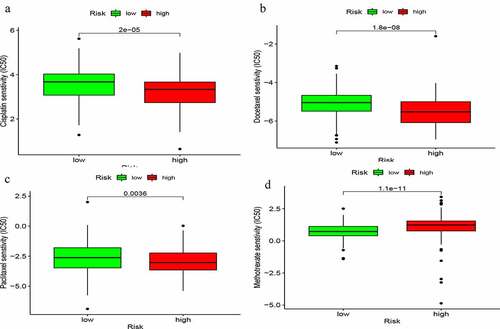
We explored the relationship between commonly-used chemotherapeutic agents for bladder cancer and the risk of the GIlncSig. As shown in , the sensitivity of cisplatin, docetaxel, and paclitaxel in the low-risk group was higher than that in the high-risk group. However, the sensitivity of methotrexate was lower in the low-risk group than the high-risk group (). These findings suggest that GIlncSig may have the ability to predict chemosensitivity in bladder cancer.
Discussion
Genomic instability plays an important role in guiding the development of tumors and can lead to heterogeneity, thereby inducing tumor occurrence [Citation17,Citation18]. Presently, the main source of genomic instability is believed to be the inactivation of DNA repair and abnormalities in gene transcription and replication [Citation19]. Genomic instability is suggested to be closely related to bladder cancer and can be used as a clinical biomarker [Citation20]. In recent years, changes in lncRNA expression have been demonstrated to promote tumor development and progression, and can be employed as a novel tumor biomarker [Citation21,Citation22]. LncRNAs have been reported to be associated with bladder cancer progression [Citation23]. Moreover, a close association has been identified between lncRNAs and genomic instability. Sungyul [Citation24] et al. suggested that the lncRNA, NORAD, plays an important role in maintaining genomic instability. However, the association between the lncRNAs associated with genomic instability and bladder cancer is still poorly studied and is in its infancy. Therefore, we constructed a GIlncSig to explore its prognostic value in bladder cancer. Most of the lncRNA signatures for the construction of bladder cancer in the past are prognostic models of immune-related genes. Our difference is that we group samples based on gene mutations and perform differential analysis to obtain differentially expressed genes. And after the multivariate COX analysis, we further stratified the analysis of other independent prognostic factors other than the risk score. We also added the correlation analysis between the risk score and chemotherapy drugs to enhance the prognostic performance of our signature. These are not available in previous studies.
First, we screened 58 lncRNAs displaying genomic instability and performed an enrichment analysis on their co-expressed genes. Accordingly, the co-expressed mRNAs were found to be mainly enriched in the intermediate filament cytoskeleton organization and mitotic cell cycle. Some studies have revealed a correlation between intermediate filaments and genomic instability [Citation25,Citation26]. The mitotic cell cycle is also associated with genomic instability, and its alteration can directly affect genome stability [Citation27,Citation28]. The GIlncSig model in this study is comprised of five lncRNAs; CFAP58-DT and MIR100HG are associated with worse prognosis, and LINC02446, AC078880.3, and LINC01833 are associated with better prognosis. Based on the results of the KM survival analysis, GIlncSig displayed a strong prognostic value, which was significantly correlated with the survival rate of patients. After univariate and multivariate Cox analyses, we proceeded to perform independent prognostic analysis. In addition to risk score, age and stage were found to have a prognostic value. By continuing the stratified analysis, GIlncSig was also found to have an independent prognostic value in bladder cancer. In fact, GIlncSig was recognized to have a better prognostic value than other previously published lncRNA signatures. We also validated this model using the GEO database. According to the results, the expression of MIR100HG was associated with poor prognosis in bladder cancer, thereby aligning with the data analysis results from TCGA database. Finally, we conducted an analysis of the correlation between commonly used chemotherapy drugs and the signature. The results showed that, except for methotrexate, the sensitivity of cisplatin, docetaxel and paclitaxel in the high-risk group was significantly lower than that in the low-risk group. Therefore, we guess that this model may have the ability to predict the sensitivity of chemotherapy drugs, and in the grouping by this signature, the chemotherapy effect of the high-risk group may be worse than that of the low-risk group.
Of the five lncRNAs in the GIlncSig, CFAP58-DT and AC078880.3 have not been reported in the literature. MIR100HG plays a role in the induction of many tumors, such as gastric cancer [Citation29], breast cancer [Citation30], and osteosarcoma [Citation31], however, its role in bladder cancer has not been reported in the literature. According to a study by Zhang [Citation32] et al., LINC02446 has an inhibitory effect on the proliferation and metastasis of bladder cancer cells. Currently, only few reports exist on LINC01833; however, this lncRNA has been demonstrated to be correlated with tumors [Citation33,Citation34]. We believe that this model not only has an important predictive role in the prognosis of bladder cancer, but may also be employed to identify new biomarkers for bladder cancer that can be further studied.
Although we identified and validated a GIlncSig that can predict the outcome of bladder cancer, this study had a few limitations. First, relatively few datasets (i.e., from TCGA and GEO databases) were collected. More independent datasets are thus needed for further validation. Second, more biological experiments are needed to validate and explore the mechanism of action of GIlncSig in terms of genomic instability. We are currently conducting clinical trials, but this is a very time-consuming process. We will clinically verify our conclusions in follow-up studies.
Conclusion
In conclusion, the GIlncSig can predict the outcome of patients with bladder cancer as well as the sensitivity of this tumor to chemotherapeutic drugs. This signature may thus have some guiding significance in clinical practice.
Author contributions
L.Z. and L.F.Z. were responsible for conceiving and designing this study, H.W. and Z.Y.Z. wrote the main manuscript, S.L.G. and was responsible for preparing the tables, C.L. and Z.Z. were responsible for preparing figures, and H.W. and X.Y.X. were responsible for re-examination. All authors approved the submission of this study.
Disclosure of potential conflicts of interest
The authors declare that they have no competing interests.
Supplemental Material
Download ()Acknowledgements
This work was supported by the Young Talent Development Plan of Changzhou Health Commission (No. CZQM2020065), Young Scientists Foundation of Changzhou No.2 People’s Hospital (2019K008), Changzhou Sci & Tech program (CJ20190100), the Second Hospital of Changzhou Discipline Funding (YJXK202013), Changzhou Innovation Team Funding (XK201803), Changzhou Top Talent Project (RC201620).
Dat availability statement
All the data sets of this study are available in the TCGA database(https://portal.gdc.cancer.gov/) and GEO database (http://www.ncbi.nlm.nih.gov/geo/).
Supplementary materials
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30.
- Seidl C. Targets for therapy of bladder cancer. Semin Nucl Med. 2020;50(2):162–170.
- Lenis A, Lec P, Chamie K, et al. Bladder cancer: a review. JAMA. 2020;324(19):1980–1991.
- Li F, Guo H, Wang Y, et al. Profiles of tumor-infiltrating immune cells and prognostic genes associated with the microenvironment of bladder cancer. Int Immunopharmacol. 2020;85:106641.
- Song Y, Jin D, Chen J, et al. Identification of an immune-related long non-coding RNA signature and nomogram as prognostic target for muscle-invasive bladder cancer. Aging (Albany NY). 2020;12(12):12051–12073.
- Negrini S, Gorgoulis VG, Halazonetis TD. Genomic instability — an evolving hallmark of cancer. Nat Rev Mol Cell Biol. 2010;11(3):220–228.
- Andor N, Maley CC, Ji HP. Genomic instability in cancer: teetering on the limit of Tolerance. Cancer Res. 2017;77(9):2179–2185.
- Ma L, Bajic V, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. 2013;10(6):925–933.
- Liu H, Wan J, Chu J. Long non-coding RNAs and endometrial cancer. Biomed Pharmacother. 2019;119:109396.
- Weidle UH, Birzele F, Kollmorgen G, et al. Long non-coding RNAs and their role in metastasis. Cancer Genomics Proteomics. 2017;14(3):143–160.
- Fang Y, Fullwood M, Roles F. Roles, functions, and mechanisms of long non-coding RNAs in cancer. Genomics Proteomics Bioinformatics. 2016;14(1):42–54.
- Hu WL, Jin L, Xu A, et al. GUARDIN is a p53-responsive long non-coding RNA that is essential for genomic stability. Nat Cell Biol. 2018;20(4):492–502.
- Tracy KM, Tye CE, Ghule PN, et al. Mitotically-Associated lncRNA (MANCR) affects genomic stability and cell division in aggressive breast cancer. Mol Cancer Res. 2018;16(4):587–598.
- Bao S, Zhao H, Yuan J, et al. Computational identification of mutator-derived lncRNA signatures of genome instability for improving the clinical outcome of cancers: a case study in breast cancer. Brief Bioinform. 2020;21(5):1742–1755.
- Shen D, Zhang Y, Zheng Q, et al. A competing endogenous RNA network and an 8-lncRNA prognostic signature identify MYO16-AS1 as an oncogenic lncRNA in bladder cancer. DNA Cell Biol. 2021;40(1):26–35.
- Qing L, Gu P, Liu M, et al. <p>Extracellular Matrix–related six-lncRNA signature as a novel prognostic biomarker for bladder cancer. Onco Targets Ther. 2020;40:12521–12538.
- Shen Z. Genomic instability and cancer: an introduction. J Mol Cell Biol. 2011;3(1):1–3.
- Duijf P, Nanayakkara D, Nones K, et al. Mechanisms of genomic instability in breast cancer. Trends Mol Med. 2019;25(7):595–611.
- Tubbs A, Nussenzweig A. Endogenous DNA damage as a source of genomic instability in cancer. Cell. 2017;168(4):644–656.
- Vacher S, Suybeng V, Girard E, et al. Genomic instability signature of palindromic non-coding somatic mutations in bladder cancer. Cancers (Basel). 2020;12(10):2882.
- Bhan A, Soleimani M, Mandal S. Long noncoding RNA and cancer: a new paradigm. Cancer Res. 2017;77(15):3965–3981.
- Wang L, Cho K, Li Y, et al. Long noncoding RNA (lncRNA)-Mediated competing endogenous RNA networks provide novel potential biomarkers and therapeutic targets for colorectal cancer. Int J Mol Sci. 2019;20(22):5758.
- Zhuang C, Ma Q, Zhuang C, et al. LncRNA GClnc1 promotes proliferation and invasion of bladder cancer through activation of MYC. The FASEB Journal. 2019;33(10):11045–11059.
- Lee S, Kopp F, Chang T, et al. Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell. 2016;164(1–2):69–80.
- Vargas J, Hatch E, Anderson D, et al. Transient nuclear envelope rupturing during interphase in human cancer cells.. Nucleus (Austin, Tex.). 2012;3(1):88–100.
- Graziano S, Kreienkamp R, Coll-Bonfill N, et al. Causes and consequences of genomic instability in laminopathies: replication stress and interferon response.. Nucleus (Austin, Tex.). 2018;9(1):258–275.
- Vitale I, Galluzzi L, Castedo M, et al. Mitotic catastrophe: a mechanism for avoiding genomic instability. Nat Rev Mol Cell Biol. 2011;12(6):385–392.
- Petsalaki E, Zachos G. DNA damage response proteins regulating mitotic cell division: double agents preserving genome stability. FEBS J. 2020;287(9):1700–1721.
- Li J, Xu Q, Wang W, et al. MIR100HG: a credible prognostic biomarker and an oncogenic lncRNA in gastric cancer. Biosci Rep. 2019;39(4).
- Wang S, Ke H, Zhang H, et al. LncRNA MIR100HG promotes cell proliferation in triple-negative breast cancer through triplex formation with p27 loci. Cell Death Dis. 2018;9(8):805.
- Su X, Teng J, Jin G, et al. ELK1-induced upregulation of long non-coding RNA MIR100HG predicts poor prognosis and promotes the progression of osteosarcoma by epigenetically silencing LATS1 and LATS2. Biomed Pharmacothe. 2019;109:788–797.
- Zhang X, Zhang J, Zhao W, et al. Long non-coding RNA LINC02446 suppresses the proliferation and metastasis of bladder cancer cells by binding with EIF3G and regulating the mTOR signalling pathway. Cancer Gene Ther. 2021. DOI:10.1038/s41417-020-00285-2.
- Zhang Y, Li W, Lin Z, et al. <p>The long noncoding RNA Linc01833 enhances lung adenocarcinoma progression via MiR-519e-3p/S100A4 axis. Cancer Manag Res. 2020;12:11157–11167.
- Jiang Y, Chen J, Ling J, et al. Construction of a Glycolysis-related long noncoding RNA signature for predicting survival in endometrial cancer. J Cancer. 2021;12(5):1431–1444.