ABSTRACT
Xuebijing Injection have been found to improve the clinical symptoms of COVID-19 and alleviate disease severity, but the mechanisms are currently unclear. This study aimed to investigate the potential molecular targets and mechanisms of the Xuebijing injection in treating COVID-19 via network pharmacology and molecular docking analysis. The main active ingredients and therapeutic targets of the Xuebijing injection, and the pathogenic targets of COVID-19 were screened using the TCMSP, UniProt, and GeneCard databases. According to the ‘Drug-Ingredients-Targets-Disease’ network built by STRING and Cytoscape, AKT1 was identified as the core target, and baicalein, luteolin, and quercetin were identified as the active ingredients of the Xuebijing injection in connection with AKT1. R language was used for enrichment analysis that predict the mechanisms by which the Xuebijing injection may inhibit lipopolysaccharide-mediated inflammatory response, modulate NOS activity, and regulate the TNF signal pathway by affecting the role of AKT1. Based on the results of network pharmacology, a molecular docking was performed with AKT1 and the three active ingredients, the results indicated that all three active ingredients could stably bind with AKT1. These findings identify potential molecular mechanisms by which Xuebijing Injection inhibit COVID-19 by acting on AKT1.
1. Introduction
Corona Virus Disease 2019 (COVID-19) is an infectious disease caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2). It was first identified in December 2019 in Wuhan, Hubei, China, and has since spread around the world. The WHO has declared that the COVID-19 outbreak constitutes a Public Health Emergency of International Concern (PHEIC). This disease can be clinically classified as mild, severe, or critical. Fever, dry cough, and fatigue are the main manifestations, and patients classified as severe can rapidly progress to acute respiratory distress syndrome (ARDS) and multiple organ failure (MOF), amongst other conditions. Unfortunately, at present, there is no cure officially approved for this disease, creating a formidable challenge in its treatment, prognosis, and control. Traditional Chinese medicines (TCM), that are characterized as being anti-viral and affecting multiple pathways and targets, have been proven to be significantly effective in treating COVID-19.
The Xuebijing (XBJ) injection is a traditional Chinese medicine that is based on the XueFuZhuYu Decoction (XFZYD). Its main herbal components are Carthami Flos, Paeoniae Radix Rubra, Chuanxiong Rhizoma, Salviae Miltiorrhizae Radix et Rhizoma, and Angelicae Sinensis Radix, and it can have several effects, such as anti-inflammation, anti-oxidation, immune regulation, improving blood stasis, anti-endotoxin, and anti-shock. Previously, it has been used to treat patients with diseases like severe pneumonia and sepsis. In 2019, the National Drug Catalog for Basic Medical Insurance, Work-related Injury Insurance and Maternity Insurance stated that XBJ injection should be used in the first aid treatment of critically ill patients. Furthermore, the Diagnosis and Treatment Protocol for COVID-19 (Trial Versions 4, 5, 6, 7, 8) states that it should be used to treat severe and critically ill patients with COVID-19. However, its mechanisms in the treatment of COVID-19 have not yet been elucidated.
Highly pathogenic viruses should be handled under strict laboratory conditions and in compliance with specific biosafety procedures, this study has attempted to investigate the possible molecular mechanisms of XBJ injection when treating COVID-19 via computer simulation based on network pharmacology and molecular docking analyses; thereby avoiding potential laboratory biosafety hazards and improving the efficiency of research into infectious disease outbreaks and modern Chinese medicines. Recently, network pharmacology-based drug repositioning has become increasingly important for the research and development of drugs. Network pharmacology is an application tool based on systems biology theory and network analysis statistics, that conducts network analysis for specific biological systems, and designs multi-target drug molecules by selecting specific signal nodes and comparing signal pathways. Zhou et al. [Citation1] identified 16 candidate drugs and 3 potential drug combinations for the treatment of human coronaviruses (HCoVs) through network predictions using over 2000 FDA-approved drugs and systems pharmacology and network pharmacology analyses. These results provide guidance for drug repositioning in the prevention and control of COVID-19. Molecular docking is an important computer-aided drug design method used to study receptor-ligand interactions such as the conformational space, binding energy, and chemical environment via stoichiometry. It has been widely used to elucidate and predict the therapeutic effects, as well as the underlying mechanisms of drugs. In recent years, molecular docking has developed into a common and preferred technique when reseraching Chinese medicines and has exhibited proven efficiency when screening for active ingredients [Citation2,Citation3].
This study has improved upon the network pharmacology and molecular docking anlaysis methods, and determined how to identify the core target in the gene network using the maximum Degree value, which indicates the role of the nodes in network pharmacology. The cluster Profiler package in R language was then used to screen the latest Gene Ontology and KEGG pathways for the core target. The network pharmacology results were verified using protein-small molecule docking to simulate the binding between active ingredients and the core target. These analyses were used to predict the active ingredients, targets, and signaling pathways, provide a molecular basis to investigate the mechanisms by which the COVID-19 inflammatory stress response is inhibited by the XBJ injection, as well as enhance the available theoretical support for the wider application of this drug in the treatment of COVID-19.
2. Materials and methods
2.1 Main active ingredients, potential targets, and network building for the XBJ injection
2.1.1 Screening of the main active ingredients and potential targets for the XBJ injection
The main active ingredients were screened from the 5 main herbal components of the XBJ injection, including Carthami Flos, Paeoniae Radix Rubra, Chuanxiong Rhizoma, Salviae Miltiorrhizae Radix et Rhizoma and Angelicae Sinensis Radix using the Traditional Chinese Medicine Systems Pharmacology Database (TCMSP [Citation4],http://tcmspw.com/tcmsp.php) according to oral bioavailability (OB) ≥ 30% and drug likeness (DL) ≥ 0.18. The drug targets of the main active ingredients were then screened through the Targets information of TCMSP. Finally, gene symbol conversion was performed for the targets screened from the TCMSP according to UniProt [Citation5] (https://www.uniprot.org/). Additional information was added according to the literature, and 206 targets were obtained.
2.1.2 Construction of the ingredient-target network for the XBJ injection
Cytoscape 3.7.2 [Citation6] was used to import the main active ingredients and targets of the XBJ injection, and the active ingredients were numbered to build the ingredient-target network.
2.2 Screening of COVID-19 targets
In Genecards (https://www.genecards.org/) [Citation7], coronavirus disease 2019, coronavirus pneumonia, coronavirus, and novel coronavirus 2019 were used as the keywords for retrieval respectively, and the results were exported in Excel. Then 277 potential targets for COVID-19 were screened for score ≥1 and the information was added according to the literature.
2.3 Analysis of the XBJ targets when treating COVID-19
2.3.1 Construction of the protein-protein interaction (PPI) network
Junction targets for the XBJ injection and COVID-19 were imported into the STRING database (https://string-db.org/cgi/input.pl) [Citation8], and the relevant parameters were set as follows:
Basic Settings: ① Network type: full network (the edges indicating both functional and physical protein associations); ② meaning of network edges: evidence (line color indicates the type of interaction evidence); ③ active interaction sources: Textmining, Experiments, Databases, Co-expression, Neighborhood, Gene Fusion, and Co-occurrence; ④ minimum required interaction score: medium confidence (0.400); ⑤ max number of interactors to show: 1st shell: none/query proteins only, 2nd shell: none.
Advanced Settings: ① network display mode: interactive svg; ② network display options: hide disconnected nodes in the network.
The PPI network for the junction targets for the XBJ injection and COVID-19 was constructed using the STRING database, and the results were exported to a TSV(Tab-separated values) file for further analysis using Cytoscape 3.7.2 (① for analysis, click Tools > Network Analyzer > Network Analysis > Analyze Network > Treat the network as undirected; ② for mapping, click Tools > Network Analyzer > Network Analysis > Generate Style from Statistics) to optimize the PPI network and screen for the core target with the maximum Degree value.
2.3.2 GO enrichment analysis and KEGG pathway analysis for the core target for the XBJ injection and COVID-19
Gene Ontology (GO) includes three aspects: biological process (BP), cellular component (CC) and molecular function (MF). GO enrichment analysis and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analysis were conducted for the core target for the XBJ injection and COVID-19 using R (version 3.6.1) [Citation9] and the clusterProfiler [Citation10] package, respectively. In addition, Cytoscape 3.7.2 was used to plot the results.
2.4 Verification of the molecular docking for the results of network pharmacology
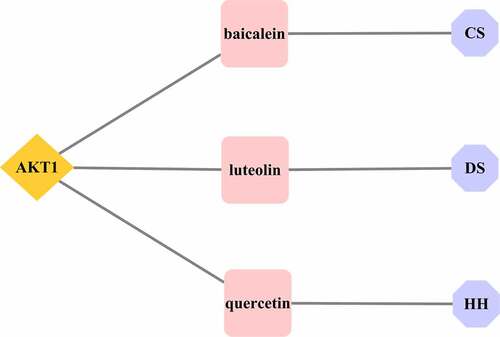
According to the network pharmacology results, AKT1 was identified as the core target of the XBJ injection when treating COVID-19, and the active ingredients of the XBJ injection, baicalein, luteolin, and quercetin were all connected with the core target AKT1. Consequently, further molecular docking verification was carried out based on these results.
RCSB PDB databases were used to download the 3D structure PDB file of the core target AKT1 (PDB ID: 1UNP), and it was then imported into the Discovery Studio 2019 Client (DS) to remove solvent molecules and ligands. Solvents were removed from the structure, while polar hydrogen atoms were added to the 1UNP by clicking ‘Add Polar’, and a forcefield was applied to the 1UNP by selecting ‘Apply Forcefield’. The PubChem database was used to obtain the SDF files of the active ingredients for the core target AKT1 as small molecules, which were imported into the DS. Apply Forcefield was used to give the force field to the small molecules. Finally, the CDOCKER (Dock Ligands) was used for protein-small molecule docking, by adjusting the Top Hits-Pose Cluster Radius to 0.5, and the other parameters were their default values. CDOKER [Citation11] is a high-accuracy molecular docking method based on the CHARMm force field. With this method, high-temperature kinetics are used to search for the flexible conformation of the ligand molecules. The lower the CDOCKER interaction energy is, the more stable the conformation of the ligand binding to the receptor is thought to be. Furthermore, ≤−5.0 kcal·mol−1 indicates that the protein and small molecule can bind, and ≤−7.0 kcal·mol−1 indicates a strong binding ability.
3. Results
This study aimed to investigate the molecular targets and mechanisms of the Xuebijing injection when used to treat COVID-19 using network pharmacology and molecular docking analyses. According to the ‘Drug-Ingredients-Targets-Disease’ network, AKT1 was identified as the core target, and baicalein, luteolin, and quercetin were identified as the active ingredients of the Xuebijing injection that were associated with AKT1. The results of the GO enrichment analysis showed that response to lipopolysaccharide was the most significant biological process in which AKT1 was involved, the cellular component was spindle, and its molecular function involoved nitric-oxide synthase regulator activity. The results of the KEGG pathway analysis showed that AKT1 was involved in the treatment of COVID-19, mainly by regulating TNF and other signaling pathways. Based on the results of the network pharmacology, molecular docking analysis was performed with AKT1 and the three active ingredients, and the results indicated that all three active ingredients could bind stably with AKT1.
3.1 Main active ingredients, potential targets and network building for the XBJ injection
3.1.1 Screening of the main active ingredients and potential targets
The 5 herbal components were retrieved using TCMSP. According to the screening requirements of an OB ≥ 30% and DL ≥ 0.18, we identified 44 active ingredients, including 9 from Carthami Flos, 8 from Paeoniae Radix Rubra, 6 from Chuanxiong Rhizoma, 19 from Salviae Miltiorrhizae Radix et Rhizoma, and 2 from Angelicae Sinensis Radix. MOL000449-Stigmasterol and MOL000358-beta-sitosterol were the common active ingredients mapping to Carthami Flos, Paeoniae Radix Rubra, and Angelicae Sinensis Radix. MOL002714- baicalein was the common active ingredient mapping by Carthami Flos and Paeoniae Radix Rubra, while MOL000006-luteolin was the common active ingredient mapping to Carthami Flos and Salviae Miltiorrhizae Radix et Rhizoma. Following the removal of the duplicates, a total of 38 non-repeated active ingredients were identified. In addition, 206 non-repeated drug targets were obtained from the TCMSP, according to the active ingredients mentioned above.
3.1.2 Construction of the ingredient-target network
The ingredient-target network for the XBJ injection was constructed using Cytoscape 3.7.2 and included 38 non-repeated active ingredients and 206 non-repeated potential targets (see and ). The core targets in the ingredient-target network, including PTGS1, PTGS2, NCOA2, KCNH2, SCN5A, ADRB2, RXRA, PGR, NR3C2, NCOA1, and PDE3A, were screened according to Degree ≥ 5.
Figure 1. Ingredient-target network for the XBJ injection. HH denotes the active ingredient of Carthami Flos; CS denotes the active ingredient of Paeoniae Radix Rubra; CX denotes the active ingredient of Chuanxiong Rhizoma; DS denotes the active ingredient of Salviae Miltiorrhizae Radix et Rhizoma; G1 and G2 denote the active ingredients of Carthami Flos, Paeoniae Radix Rubra and Angelicae Sinensis Radix; C1 denotes the common active ingredient of Carthami Flos and Paeoniae Radix Rubra; D1 denotes the common active ingredient of Carthami Flos and Salviae Miltiorrhizae Radix et Rhizoma; blue diamonds represent the targets
Table 1. XBJ injection Herbal Components- Active Ingredients- ID
3.2 Screening of COVID-19 targets
GeneCards was used to identify 277 targets of COVID-19, and STRING and Cytoscape were used to screen out the free targets. These targets, such as TNF, IL6, GAPDH, TP53, and IL10, were then arranged in order of degree (see ).
Table 2. Targets of COVID-19
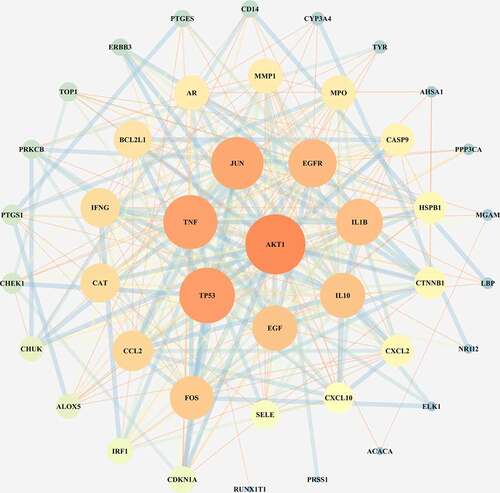
3.3 PPI network of targets for XBJ injection in treating COVID-19
The 46 junction targets of the Xuebijing injection and COVID-19 were imported into the STRING database to construct a PPI network (average node degree was 13.1, PPI enrichment p-value < 1.0 × 10−16, avg. local clustering coefficient was 0.675). The PPI network was then optimized using the Cytoscape software (see ), and the core target AKT1 was identified according to Degree ≥ 34. Secondary targets were also screened according to 24 ≤ Degree < 34, and included TP53, TNF, JUN, EGFR, IL1B, IL10, and EGF.
3.4 Analysis of the core target AKT1 from the XBJ injection when treating COVID-19
3.4.1 GO enrichment analysis
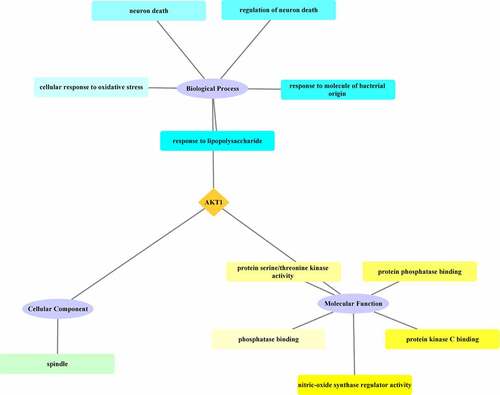
GO enrichment analysis of the core target AKT1 from the junction targets mapped for the XBJ injection and COVID-19 was conducted using the clusterProfiler package and Cytoscape 3.7.2 was used to plot the results. According to the results of the GO enrichment analysis, the main biological processes in which AKT1 was involved included response to lipopolysaccharide, response to molecule of bacterial origin, regulation of neuron death, neuron death, and cellular response to oxidative stress. The cellular component was spindle. The molecular functions in which AKT1 was found to be involved included nitric-oxide synthase regulator activity, protein kinase C binding, protein phosphatase binding, protein serine/threonine kinase activity, and phosphatase binding. See .
3.4.2 KEGG pathway analysis
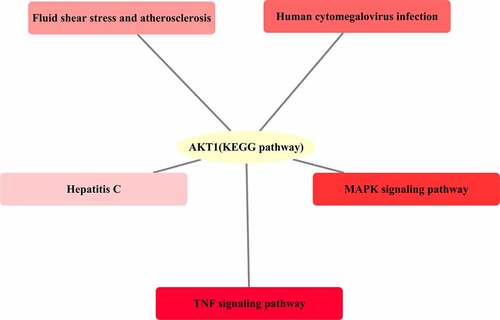
The KEGG pathway analysis of the 46 junction targets mapped to the XBJ injection and COVID-19 was conducted using the clusterProfiler package and Cytoscape 3.7.2 was used to plot the results. The results showed that the KEGG pathways in which AKT1 was involved included the TNF signaling pathway, MAPK signaling pathway, human cytomegalovirus infection, fluid shear stress and atherosclerosis, and hepatitis C (see ).
3.4.3 Active ingredients that are connected with the core target AKT1
The active ingredients of the XBJ injection, included baicalein (Paeoniae Radix Rubra), luteolin (Salviae Miltiorrhizae Radix et Rhizoma), and quercetin (Carthami Flos), and they were all connected with AKT1, suggesting that these three active ingredients may be effective for XBJ injection in the prevention and treatment of COVID-19 (see ).
3.5 Verification of molecular docking for the XBJ injection when treating COVID-19 by the Results of Network Pharmacology
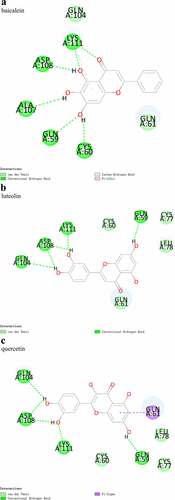
The activity of the three active ingredients obtained through the network pharmacology analysis were validated using protein-small molecule docking.The results showed stable binding between the three active ingredients (baicalein, luteolin, and quercetin) and AKT1, with binding energies of −25.5854 kcal·mol−1, −31.5575 kcal·mol−1, and −31.53 kcal·mol−1, respectively. The binding models are shown in −6c.
4. Discussion
According to the Diagnosis and Treatment Protocol for COVID-19 (Trial Version 8), critically ill patients are more likely to develop dyspnea and/or hypoxemia within a week, and severe patients, like those with SARS and MERS, may suffer from a cytokine storm (CS), which can rapidly develop into acute respiratory distress syndrome, septic shock, and multiple organ failure. The XBJ injection has been shown in previous studies to be effective at improving end-point outcomes for severe community-acquired pneumonia (CAP) and reduced the 28-day mortality in septic patients [Citation12].
In this study, 38 non-repeated active ingredients and 206 non-repeated potential targets of the XBJ injection, 277 potential targets of COVID-19, and 46 junction targets were screened using network pharmacology analysis, and the core target from the junction targets was identified to be AKT1. AKT1 is a member of the AKT kinase family that regulates metabolism, proliferation, cell survival, growth, and angiogenesis through a series of downstream substrates. AKT regulates the metabolism of fat, amino acids, and other substances by activating AS160 and PFKFB2, and also induces the TSC1/TSC2 complex and mTORC signal transduction after the stimulation of inflammatory factors, so as to regulate the growth of the endothelium, restore the endothelial barrier function, and promote proliferation of the lung fibroblasts [Citation13–15]. A previous study has shown that overexpressed constitutively active AKT1 can promote viral protein synthesis [Citation16]. Furthermore, activation of the PI3K/AKT pathway is indispensable for coxsackievirus B3 infection. Dominant negative mutant of AKT1 can significantly dampen viral RNA expression and further reduce viral capsid protein expression and viral release [Citation17]. The replication of another coronavirus, Middle East respiratory syndrome coronavirus, was remarkably inhibited when kinase inhibitors that target PI3K/AKT were adminstered [Citation18]. Collectively, AKT1 could be an ideal target with a broad-spectrum antiviral effect. These results suggest that the XBJ injection is an effective control and treatment for COVID-19, probably due to its regulation of AKT1.
According to the results of the GO enrichment analysis, the response to lipopolysaccharide (LPS) was the most significant biological process that AKT1, the core target of junction targets to the XBJ injection and COVID-19, was involved. LPS is a major component in the cell walls of Gram-negative bacteria. It has been reported that LPS can damage alveolar epithelial cells and capillary endothelium cells, resulting in changes to the intercellular space and permeability [Citation19,Citation20]. AKT could regulate the NF-κB signal transduction through phosphorylated IKKα and Tpl2 in order to ameliorate lipopolysaccharide-induced alveolar blood stasis and abnormal fibrinolysis in mice [Citation21]. It has been suggested that XBJ injection may inhibit response to lipopolysaccharide by regulating AKT1 expression, thus alleviating lung injury caused by LPS. The results of this study showed that the most significant molecular function of AKT1 is the nitric-oxide synthase regulator activity. Nitric-oxide synthase (NOS) is an isozyme that is reportedly associated with pneumonia and pulmonary fibrosis [Citation22]. These results suggest that XBJ injection may regulate NOS activity through AKT1, which may help to prevent and treat COVID-19.
According to the results of the KEGG pathway analysis, the TNF signaling pathway is the most significant pathway in which the core target AKT1, that is the core target of junction targets to XBJ injection and COVID-19, was involved. Tumor necrosis factors (TNF) include TNF-α and TNF-β which can promote the expression of proinflammatory factors and participate in systemic inflammatory responses, while it can also produce an anti-infection effect and prevent early viral proteins from being synthesized to inhibit viral replication and kill infected cells. Therefore, the XBJ injection can regulate the TNF signal pathway through AKT1, so that it has anti-inflammation and anti-virual effects, and thus the potential to reduce the inflammation caused by COVID-19.
The protein-small molecule docking analysis revealed that all three active ingredients (baicalein, luteolin, and quercetin) had perfect CDOCKER Interaction Energy with AKT1 (1UNP), with the highest found in luteolin. This inferred that luteolin was the most important active ingredient contributing to the therapeutic effects of the XBJ injection on COVID-19. The analysis of the binding model showed that there were conventional hydrogen bonds between the luteolin and Gln104, Asp108, Lys111, and Gln59 of the AKT1 and van der Waals between luteolin and Cys60, Gln61, Cys77, and Leu78 of AKT1. It has been reported that luteolin can mitigate airway inflammation by regulating the body’s antioxidative stress response and the COX-2 signaling pathway. Luteolin is also shown to relieve myocardial ischemic injury by modulating the mitogen-activated protein kinase (MAPK)/NF-κB pathway and suppressing enterovirus type 71 infections by inhibiting cell apoptosis and inflammatory factor secretion, demonstrating its antiviral properties. Additionally, luteolin and quercetin are found to induce cell apoptosis by inhibiting the phosphorylation and activation of AKT [Citation23–25]. Despite all these findings, it has not been reported whether these three active ingredients relieve COVID-19 symptoms through the direct regulation of AKT1. In this study, molecular docking was implemented to further explore the mechanisms of action of the three active ingredients in treating COVID-19. Furthermore, considering that baicalein, luteolin, and quercetin are found in many herbal components, these active ingredients may not be unique to the XBJ injection, indicating the possibility of discovering antiviral TCM combinations in other ancient prescriptions containing baicalein, luteolin, and quercetin.
In this study, potential targets were screened, a PPI network was constructed, and enrichment analysis was conducted to find potential pathways. The XBJ injection was shown to produce synergistic therapeutic effects on COVID-19 with ‘multi- active ingredients, multi-target and multi-pathway’, and molecular docking was performed to verify such results. However, the present study has some limitations: 1) The study results are based on existing data available in the given database, and differences in the results may arise if different databases were used. Therefore, future studies should focus on multidatabase analysis and databases that are updated regularly. 2) The biological network in this study was constructed using qualitative data, but TCM prescriptions usually have a complex composition, as well as intricate interactions, making it extremely difficult to perform an analysis based on the quantitative data. Consequently, the reliability of the conclusions from this study need to be further verified. Therefore, in vivo and in vitro experiments should be conducted to further examine the molecular mechanisms of the XBJ injection for the treatment of COVID-19.
5. Conclusion
In conclusion, according to the network pharmacology and molecular docking analyses, the predicted active ingredients in the XBJ injection include baicalein, luteolin, and quercetin. The XBJ injection may inhibit the lipopolysaccharide-mediated inflammatory response, modulate NOS activity, and regulate the TNF signaling pathway by affecting the function of the AKT1, thereby ultimately suppressing the excessive inflammatory response associated with COVID-19. Due to the limitations of the computational methods for chemistry and biology, the results need to be verified with follow-up experiments to provide a basis for the treatment of COVID-19 with TCM.
Abbreviations
Disclosure of potential conflicts of interest
No potential conflict of interest was reported by the author(s).
Highlight
(1) AKT1 was identified as the core target of Xuebijing injection used to treat COVID-19.
(2) Xuebijing injection may inhibit lipopolysaccharide-mediated inflammatory response.
(3) Xuebijing injection may regulate nitric-oxide synthase activity and the TNF signal pathway.
(4) Baicalein, luteolin, and quercetin may be the active ingredients of Xuebijing injection connected with AKT1.
Data availability statement
Publicly available datasets were analyzed in this study. This data can be found here: [GeneCards] at [https://www.genecards.org/], [Traditional Chinese Medicine Systems Pharmacology Database (TCMSP)] at [http://tcmspw.com/tcmsp.php], [UniProt] at [https://www.uniprot.org/], [STRING] at [https://STRING-db.org/cgi/input.pl], [Pubchem] at [https://pubchem.ncbi.nlm.nih.gov/], [RCSB Protein Data Bank] at [http://www.rcsb.org/].
Correction Statement
This article has been republished with minor changes. These changes do not impact the academic content of the article.
Additional information
Funding
References
- Zhou Y, Hou Y, Shen J, et al. Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Published Cell Discov. 2020;6(1:14.
- Moslemi D, Nokhandani AM, Otaghsaraei MT, et al. Management of chemo/radiation-induced oral mucositis in patients with head and neck cancer: a review of the current literature. Radiother Oncol. 2016;120(1):13–20.
- Barbosa SCM, Pereira VBM, Wong DVT, et al. Amifostine reduces inflammation and protects against 5-fluorouracil-induced oral mucositis and hyposalivation. Braz J Med Biol Res. 2019;52(3):e8251. Published.
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6:13. Published.
- The UniProt Consortium.UniProt: the universal protein KnowledgeBase [published correction appears.Nucleic Acids Res.2017;45(D1):D158–D169. Nucleic Acids Res. 2018 Mar 16; 46(5):2699
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003 Nov;13(11):2498–2504.
- Stelzer G, Rosen N, Plaschkes I, et al. The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics. 2016;54(1):1.30.1–1.30.33. Published.
- Szklarczyk D, Gable AL, Lyon D, et al. STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res. 2019;47(D1):D607–13.
- R Core Team. R: a language and environment for statistical computing; 2019. Available from: https://www.R-project.org/. Vienna, Austria: R Foundation for Statistical Computing
- Yu G, Wang LG, Han Y, et al. clusterProfiler: an R package for comparing biological themes among gene clusters. Omics. 2012;16(5):284–287.
- Ding X, Wu Y, Wang Y, et al. Accelerated CDOCKER with GPUs, parallel simulated annealing, and fast Fourier transforms. J Chem Theory Comput. 2020;16(6):3910–3919.
- Song Y, Yao C, Yao Y, et al. Xuebijing injection versus placebo for critically ill patients with severe community-acquired pneumonia: a randomized controlled trial. Crit Care Med. 2019;47(9):e735–43.
- Zhou Y, Yang Y, Liang T, et al. The regulatory effect of microRNA-21a-3p on the promotion of telocyte angiogenesis mediated by PI3K (p110α)/AKT/mTOR in LPS induced mice ARDS. J Transl Med. 2019;17(1):427. Published.
- Xu F, Zhou F. Inhibition of microRNA-92a ameliorates lipopolysaccharide-induced endothelial barrier dysfunction by targeting ITGA5 through the PI3K/Akt signaling pathway in human pulmonary microvascular endothelial cells. Int Immunopharmacol. 2020;78:106060.
- Xie T, Xu Q, Wan H, et al. Lipopolysaccharide promotes lung fibroblast proliferation through autophagy inhibition via activation of the PI3K-Akt-mTOR pathway. Lab Invest. 2019;99(5):625–633.
- Wang X, Zhang H, Abel AM, et al. Role of phosphatidylinositol 3-kinase (PI3K) and Akt1 kinase in porcine reproductive and respiratory syndrome virus (PRRSV) replication. Arch Virol. 2014;159(8):2091–2096.
- Esfandiarei M, Luo H, Yanagawa B, et al. Protein kinase B/Akt regulates Coxsackievirus B3 replication through a mechanism which is not caspase dependent. J Virol. 2004;78(8):4289–4298.
- Kindrachuk J, Ork B, Hart BJ, et al. Antiviral potential of ERK/MAPK and PI3K/AKT/mTOR signaling modulation for Middle East respiratory syndrome coronavirus infection as identified by temporal kinome analysis. Antimicrob Agents Chemother. 2015;59(2):1088–1099.
- Zemans RL, Colgan SP, Downey GP. Transepithelial migration of neutrophils: mechanisms and implications for acute lung injury. Am J Respir Cell Mol Biol. 2009;40(5):519–535.
- Gerber A, Heimburg A, Reisenauer A, et al. Proteasome inhibitors modulate chemokine production in lung epithelial and monocytic cells. Eur Respir J. 2004;24(1):40–48.
- Liu B, Wu Y, Wang Y, et al. NF-κB p65 Knock-down inhibits TF, PAI-1 and promotes activated protein C production in lipopolysaccharide-stimulated alveolar epithelial cells type II. Exp Lung Res. 2018;44(4–5):241–251.
- El-Kashef DH. Nicorandil ameliorates pulmonary inflammation and fibrosis in a rat model of silicosis. Int Immunopharmacol. 2018;64:289–297.
- Lee WJ, Wu LF, Chen WK, et al. Inhibitory effect of luteolin on hepatocyte growth factor/scatter factor-induced HepG2 cell invasion involving both MAPK/ERKs and PI3K-Akt pathways. Chem Biol Interact. 2006;160(2):123–133.
- Im E, Yeo C, Lee EO. Luteolin induces caspase-dependent apoptosis via inhibiting the AKT/osteopontin pathway in human hepatocellular carcinoma SK-Hep-1 cells. Life Sci. 2018;209:259–266.
- Granado-Serrano AB, Martín MA, Bravo L, et al. Quercetin induces apoptosis via caspase activation, regulation of Bcl-2, and inhibition of PI-3-kinase/Akt and ERK pathways in a human hepatoma cell line (HepG2). J Nutr. 2006;136(11):2715–2721.