ABSTRACT
The main aim of this work was to evaluate differential expression and biological functions of circulating miRNA and whole peripheral blood (PB) genes in patients affected by venous thromboembolism (VTE) and in healthy subjects. Circulating miRNA sequences and PB expression profiles were obtained from GEO datasets. Ten miRNAs with the most significant differential expression rate (dif-miRNA) were subjected to miRbase to confirm their identity. Dif-miRNA targets were predicted by TargetScan and aligned with differentially expressed genes to obtain overlapping co-genes. Biological functions of co-genes were analyzed by Gene Ontology and KEGG analysis. Interaction network of dif-miRNAs, co-genes, and their downstream pathways were studied by analyzing protein–protein interaction (PPI) clusters (STRING) and determining the crucial hubs (Cytoscape).
MiR-522-3p and miR-134 dif-miRNAs are involved in protein translation and apoptosis by regulating their respective co-genes in PB. Co-genes are present in nucleolus and extracellular exosomes and are involved in oxidative phosphorylation and ribosome/poly(A)-RNA organization. The predicted PPI network covered 107 clustered genes and 220 marginal joints, where ten hub genes participating in PPIs were found. All these hub genes were down-regulated in VTE patients. Our study identifies new miRNAs as potential biological markers and therapeutic targets for VTE.
VTE, including deep vein thrombosis (DVT) and pulmonary embolism (PE), is the third most common acute and life-threatening cardiovascular condition, after myocardial infarction and stroke [Citation1]. According to the epidemiological researches, the incidence rates of PE and DVT account for 0.39–1.15‰ and 0.53–1.62‰, respectively [Citation2,Citation3]. The annual mortality caused by PE in US reaches near 200,000, and most patients ultimately die within the first 1 h of PE presentation. In recent years, the diagnosed VTE cases have grown in China, with the proportion of PE patients rapidly increasing from 0.26‰ to 1.45‰ in 10 years [Citation4,Citation5]. Thus, early diagnosis and treatment of VTE can have great clinical relevance to reduce mortality.
MicroRNAs (miRNAs or miR) are highly conserved small non-coding RNA (containing about 22 nucleotides) that regulate gene expression by binding to the 3ʹ-untranslated complementary region of target mRNAs and inducing their degradation or translation inhibition. Other than in cellular, miRNAs are also produced extracellular [Citation6]. Recent evidence revealed different biological functions of circulating miRNAs, suggesting their possible role in the progresses of stroke, cardiovascular and metabolic diseases [Citation7–10].
In order to unveil new biomarkers predisposing to VTE and potentially altered in high-risk population, the main goal of this study was to screen key circulating miRNAs related to the occurrence and progression of VTE disease and that were not identified by previous bioinformatics analyses. In this study, we analyzed new indicators of poor VTE prognosis and identified new potential therapeutic targets for this refractory disease. We compared differential whole blood gene expression in VTE patients and healthy donors, by using bioinformatic approaches and querying the Gene Expression Omnibus (GEO) datasets. We then associated biological functions to representative differentially expressed genes and enriched relevant signaling pathways by Kyoto Encyclopedia of Genes and Genomes (KEGG). We also established the protein–protein interaction networks to identify key hub genes and investigated related cellular biological process that may be regulated by circulating miRNAs. Based on the results, this study might provide new diagnostic tools for the prevention and early diagnosis of PTE.
Materials and methods
Microarray data
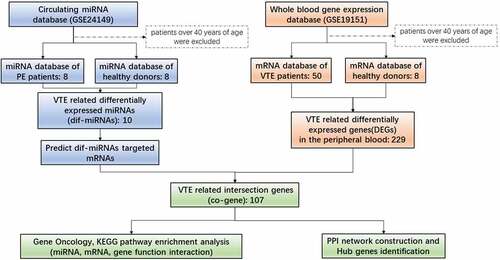
In order to evaluate the biological functions of circulating miRNAs in whole blood of VTE patients, we used expression data of circulating miRNA GSE24149 and whole blood gene expression GSE19151 from GEO database (https://www.ncbi.nlm.nih.gov/gds). GSE24149 was collected in 2010 in Shanghai, China. It contains plasma miRNA profiles from 10 PE patients and 10 healthy donors (age range: 20–82 years, no ethnic information available). GSE19151 dataset was collected in 2009 in Durham, USA. It contains gene expression profiles of plasma obtained from whole blood of 70 VTE patients and 63 control donors (age range: 18–84 years; 25 African Americans, 6 African Origin/Black, 95 Caucasians, 4 Hispanic or Latino, 1 Indian, 1 mixed, and 1 unclear ethnicity). Patients over 40 years of age were excluded from the study.
Identification of differentially expressed miRNAs and mRNAs relevant to VTE
The online bioinformatic tool GEO2 (http://www.ncbi.nlm.nih.gov/geo/geo2r/) was used to compare multiple GEO lists and recognize significant expression differences. The automatic algorithm of GEO2R is designed to calibrate p values with false discovery rate (FDR) correction when performing multiple t-test analysis. Here, we utilized GEO2R to extract the differential expression lists. The differentially expressed genes were defined with the significance of |log FC| > 1 and p < 0.05.
Acquisition of VTE-related gene list
miRbase (http://mirbase.org/index.shtml) is a primary open repository which archives published miRNA sequences and annotation and provides target gene prediction service. The latest version is 22.
TargetScan (http://www.targetscan.org/vert_72/) is an online software, widely used for prediction of miRNA binding sites on target genes. It can search conserved 8-mer, 7-mer and 6-mer sites that are compatible with miRNA seed regions. We searched for the 10 most differentially upregulated or downregulated miRNAs (dif-miRNA) into miRbase to confirm their identities, followed by target gene prediction with TargetScan. We then compared these predicted target genes with GSE19151 list to obtain an intersection list (co-gene).
GO and KEGG analysis of miRNA target genes
GO is bioinformatic analysis widely used to enrich and assign genes to pre-defined functional characteristics comprising biological process, cellular components, and molecular function. Also, KEGG database is an extensively used database, storing plentiful resources of genome, biological pathways, diseases and data related to chemical substances and drugs. In this study, we analyzed DEG with GO annotations and KEGG pathway enrichment on Database for Annotation, Visualization and Integrated Discovery (DAVID): https://david.ncifcrf.gov/. P <0.05 and gene count ≥ 5 were considered statistically significant.
PPI establishment and hub gene identification
The STRING database (http://string-db.org/) provides information on the protein–protein interaction (PPI). In order to analyze the PPI of co-genes, we mapped co-gene interaction network through STRING database and extracted PPI pairs based on a combination score >0.4. Subsequently, the PPI network was visualized through Cytoscape (www.cytoscape.org/). The tendency of PPI was evaluated by the Cytoscape plug-in CytoHubba. Gene nodes with multiple interactive connections were crucial in maintaining the stability of the entire network. In our research, the top-ten nodes with the most active connectivity were identified as Hub genes.
Results
Differentially expressed circulating miRNAs and peripheral blood whole blood genes (DEG)
In order to unveil differentially expressed circulating miRNAs and differentially expressed genes (DEG) in whole peripheral blood, we compared the expression profiles of VTE patients and control group in the two datasets analyzed.
From the analysis of GSE24149 dataset of circulating miRNA, 220 and 12 circulating miRNAs were found to be upregulated and downregulated, respectively, in PE patients, as compared to healthy control groups. Among the differentially expressed miRNAs, ten dif-miRNAs with the higher |LogFC| value were further analyzed (). In parallel, from the analysis of GSE19151 dataset of whole blood gene expression profile, among the 229 DEGs in the peripheral blood of VTE patients, 2 were up-regulated and 227 were down-regulated ().
Table 1. Top ten miRNAs with the most significant differential expressions in peripheral blood of VTE patients
Table 2. Differentially expressed genes (DEGs) in peripheral blood of VTE patients
Detection and functional enrichment analysis of co-genes in VTE patients
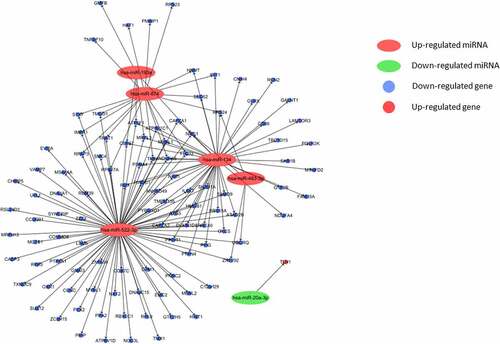
We used TargetScan to detect target mRNAs of the circulating dif-miRNAs. DEG of GSE19151 were compared with common DEGs and named co-genes (, ). We then focused on the 107 co-genes and investigated their biological function by using the DAVID online bioinformatic tool for GO enrichment and KEGG pathway analysis.
Table 3. Overlapping gene list (co-genes) of dif-miRNAs target genes and DEGs in VTE patients
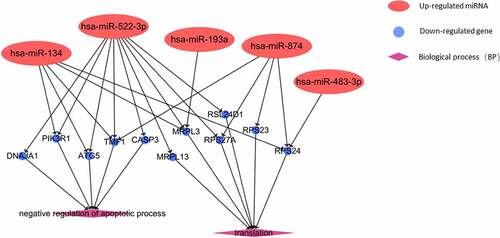
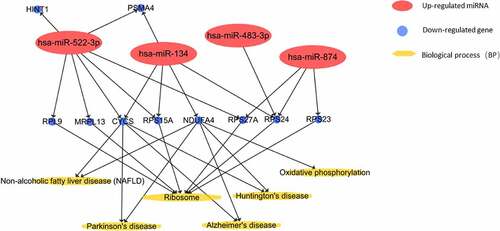
GO enrichment analysis covers three fields, including biological processes, cellular component, and molecular function. The results of GO analysis suggested that the identified co-genes were involved in translation and negative regulation of apoptosis. These processes were, next, related to dif-mRNA and co-genes by constructing a miRNA-gene-biological process interaction network (). The network map showed that 11 co-genes participated in the two biological processes and 5 dif-miRNAs played a pivotal regulatory role. Co-genes were associated with cellular components, including nucleolus and extracellular exosomes. MF analysis implied that co-genes were actively involved in structural constituents of ribosomes and poly(A) RNA binding activity (). In addition, the results of KEGG pathway analysis showed that co-genes were mainly present in pathways related to ribosomes, oxidative phosphorylation, as well as in Parkinson’s disease, nonalcoholic fatty liver disease (NAFLD), Huntington’s and Alzheimer’s disease ().
Table 4. Gene ontology (GO) function enrichment analysis of co-genes in VTE patients
Table 5. KEGG enrichment analysis of in VTE patients
PPI network construction and Hub genes identification
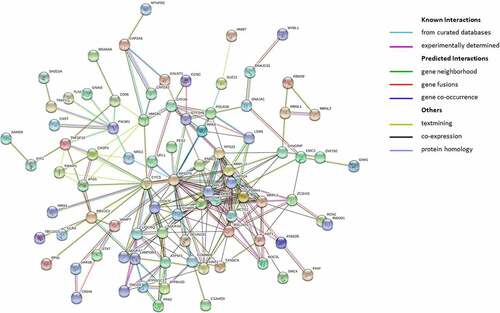
In order to clarify the interaction network between the downstream proteins of co-genes and explore the core regulatory genes, STRING was used to predict the interaction of 107 co-genes at protein level.
As shown in , a total of 107 gene nodes and 220 marginalized genes are present in PPI network. We further identified the top-ten Hub genes evaluated by connectivity in the PPI network (). Among them, we found a high connectivity for ribosomal protein S27a (RPS27A) (score = 24), together with cytochrome c, somatic (CYCS; score = 17), mitochondrial ribosomal protein L13 (MRPL13; score = 16), ribosomal protein L9, ribosomal protein S15a (RPL9, RPS15A; score = 15), ribosomal protein S24 (RPS24; score = 14), ribosomal protein S23 and proteasome 20S subunit alpha 4 (RPS23, PSMA4; score = 13), histidine triad nucleotide binding protein 1 and for NDUFA4 mitochondrial complex associated (HINT1, NDUFA4; score = 12). All these hub genes were down-regulated in VTE patients.
Table 6. Top ten co-genes genes (hub genes) with highest connectivities
Finally, we established an interaction network among dif-miRNA, Hub gene, and KEGG pathway ().
Discussion
VTE is a common peripheral vascular disease (PVD). Thrombus shedding in acute stage of VTE may cause fatal pulmonary embolism, which is one of the primary causes of sudden clinical death. Recent studies showed that PE and DVT are frequent clinical manifestations of VTE. Deep vein thrombosis and pulmonary embolism are manifestations of venous thromboembolism at different stages, especially in patients with lower extremity DVT. Current case reports showed that deep vein thrombosis as well as pulmonary embolism are the most preventable causes of death in hospital [Citation11]. Early detection, diagnosis and treatment are clinically important to reduce the VTE fatality. Although the D-dimer evaluation, color Doppler blood flow imaging, venography and other approaches have increased the sensitivity and specificity of VTE diagnosis, there is still an urgent need for better, alternative diagnostic methods, also due to nonspecific symptoms of VTE and the restriction of current detective approaches.
Although miRNAs are mainly present in cells, they can be released extracellularly through the exosome secretion pathway. Circulating miRNA are involved in intercellular communications, in biological functional regulation, as well as can promote disease progression [Citation12–15]. Moreover, research showed that circulating miRNAs regulate gene expression through its impact on mRNA transcription [Citation16,Citation17]. Our results demonstrated that several circulating miRNAs can take active part in post-transcriptional modifications and mRNA stabilization through regulating nuclear genes and participating in poly(A) RNA binding processes (). Nowadays, the measurement of circulating miRNAs as VTE diagnostic approach is drawing great attention. Compared to conventional protein biomarkers, circulating miRNA has many advantages, including: ① the stability of circulating miRNA makes the detection reproducible; ② the variabilities of circulating miRNA expression are specifically related to the disease; ③ sample collection for circulating miRNAs is less invasive, more convenient for detection and analysis; ④the increase of circulating miRNA level can be considered as an early signal of disease, compared to protein biomarkers.
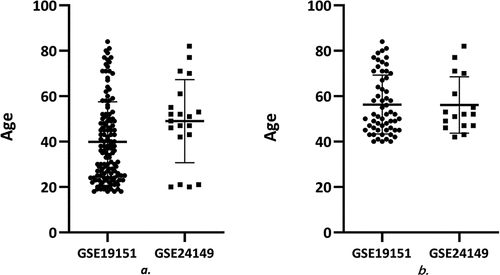
In this study, circulating miRNA and gene expression analyses were performed with the GEO database. We analyzed circulating miRNAs and genes differentially expressed in the peripheral whole blood of VTE patients and healthy donors. GSE24149 contains only data relative to 4 subjects under 40. The mean age ± standard deviation was 49 ± 18.31 years, the median was 50 years, the 25% and 75% percentiles were 42.25 and 59.5 years, respectively. However, 73 out of 133 samples of GSE19151 came from subjects under 40 and 2 from subjects with unknown age. The mean age ± standard deviation was 39.85 ± 17.66 years, the median was 36 years, the 25% and 75% percentiles were 25 and 50 years, respectively. Considering that differences in age distribution could affect analysis results, patients over 40 years were excluded from the study. Data adjustment reduced differences in age distribution of the two datasets, and further analysis of the datasets showed that the mean age distribution ± standard deviation in GSE24149 (16 samples) was 56.13 ± 12.40 years, the median was 52 years, the 25% and 75% percentiles were 47 and 67.75 years, respectively. After adjustment, the mean of age distribution ± standard deviation in GSE19151 (58 samples) was 56.33 ± 12.98 years, the median was 52.00 years, the 25% and 75% percentiles were 45 and 68.50 years, respectively (). We identified one up-regulated and 106 down-regulated co-genes and constructed a PPI network to further identify the relationships between these dif-miRNAs and co-genes. We then focused on 10 critical hub genes presumably targeted by dif-miRNAs. Our study on VTE-related miRNA regulation proposes a novel methodological approach for early VTE diagnosis.
Figure 5. Age distribution in each dataset before and after preprocessing (a. before preprocessing b. after preprocessing)
According to the traditional view, venous thrombosis is related to red blood cells, producing ‘red thrombus’ rich in fibrin, whereas arterial thrombus is rich in platelets, forming ‘white thrombus.’ However, current experimental studies have shown that platelet activation is also involved in venous thrombosis [Citation18]. Platelets can be used not only as a donor of miRNA but also as a recipient of circulating miRNA. In recent years, studies have found that miRNAs can be combined with the mRNA of multiple key factors in the coagulation-anticoagulation and fibrinolysis system and regulate the process of thrombosis and dissolution at gene level. For example, miR-223, miR-96, miR-200b, etc. can inhibit platelet protein expression through miRNA-mRNA pathway, thereby affecting platelet function. miR-494, miR-27a, miR-27b can be combined with the 3ʹUTR of tissue factor pathway inhibitor (TFPI) mRNA and promote thrombosis by down-regulating the expression of TFPIα [Citation19]. Studies have shown that there is a certain amount of ribosomal protein in platelets [Citation20]. For example, ribosomal protein S6K1 and SLFN14 are present in circulating platelets and exert their unique effects in the process of hemostasis and thrombosis [Citation21,Citation22]. The polymorphism of ribosomal protein MRPL37 predisposes to recurrent venous thromboembolism [Citation23]. In our study, the Hub genes RPS27A, MRPL13, RPL9, RPS23, and RPS24 belong to the ribosomal protein family. We observed circulating hsa-miR-522-3p, -134, -874, -483 – through KEGG pathway analysis. miR-522-3p participates in the metabolic process of ribosomal protein family by regulating the aforementioned Hub genes. At present, few reports on the relationship between these miRNA-ribosomal protein family genes and venous thrombosis are available. Therefore, we speculate that the identified miRNA-genes may be closely related to platelet activation and may be new potential markers.
Vascular endothelial cells play an important role in the process of thrombosis. After endothelial cells are destroyed, collagen and intravascular tissue factors are exposed to the bloodstream, with thrombus beginning to form. The exposed collagen triggers the activation and accumulation of platelets, whereas tissue factors trigger the production of thrombin that converts fibrinogen into fibrin and activates platelets. Therefore, endothelial dysfunction can be considered as a risk factor for thrombosis. In recent years, a number of studies have suggested that miR-31, miR-20a, and miR-29b can regulate the expression level of TNFSF15–TNFRSF25 through related cell pathways [Citation24–26], and the pathological up-regulation of TNFSF15–TNFRSF25 can lead to the occurrence and progression of primary venous thromboembolism by exerting its pro-apoptotic and anti-proliferative activities on endothelial cells [Citation27]. In vitro studies have also demonstrated that miR-134 can aggravate glucose-induced endothelial cell dysfunction, whereas miR-874-3p can antagonize the damage of high glucose to endothelial cells [Citation28,Citation29]. Our research shows that circulating hsa-miR-134, -522-3p and -874 are involved in the inhibition of apoptotic processes during the progression of VTE by regulating CASP3, ATG5, DNAJA1, PIK3R1, and TMF1 genes. The latter can in turn regulate the apoptosis of vascular endothelial cells in VTE.
Exosomes can promote the exchange of peptides, miRNAs, mRNA and mitochondrial DNA between cells and tissues. They play an important role in many physiological and pathological processes and can be used as diagnostic markers for various cardiovascular diseases [Citation30]. Vascular endothelial cells and platelets can produce a large number of exosomes. The apoptotic bodies, MVs and exosomes released by caspase-3-activated VECs were analyzed by large-scale proteomics. Protein expression profiles of the body showed that the 20S proteasome activity in circulating exosome-like vesicles increased after vascular injury in mice [Citation31]. Platelet-derived exosomes are rich in a variety of miRNAs with regulatory functions. Among them, the levels of miR-223, miR-339 and miR-21 are related to platelet activation, with miR-223 being the most abundant miRNA in platelets and contributing to platelet activation, reactive secretion, adhesion and aggregation [Citation32]. These exosomal miRNAs can be used as biomarkers to predict thrombosis [Citation33]. Antiplatelet therapy can significantly reduce the levels of these miRNAs. Similarly, we suggest that circulating miR-20a- 3p, -134, -522-3p, and -874 can regulate the expression of genes encoding components of exosomes, including TLN1, LAMTOR3, STX7, and IMPA1, thus controlling the involvement of exosomes in the pathophysiological activities associated to VTE.
Although we found new targets with potential research value, our research has some limitations. The analyzed datasets are all taken from the GEO library. Thus, considering the differences in datasets on the aspects of experimental conditions and candidates’ ethnicity and living environment, there are limitations in further clarifying the association between miRNA-gene and disease. Future experiments verifying the relevance of these miRNA-mRNA pairs in specific populations, as well as tests in animal models are needed. Secondly, in addition to peripheral whole blood, miRNAs may also be enriched in other potential target tissues, such as endothelium of diseased blood vessels and thrombus components.
Conclusion
In this study, we showed a comprehensive analysis of the entire genome of VTE patients performed by analyzing their circulating miRNA and gene interactions. We also described the relationship between several miRNA-based circulating biomarkers and key genes in peripheral whole blood. Our findings explained the biological functions of these blood biomarkers in VTE patients, thus providing new targets with strong research value.
Highlights
1. Some new miRNAs and hub genes as potential biological markers and therapeutic targets for VTE were identified.
2. We identify new gene regulation mechanisms involved in the progression of VTE.
3. We provide new useful tools to identify those individuals potentially subjected to VTE.
Disclosure of conflict of interest
The authors declared that they do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Additional information
Funding
References
- Raskob GE, Angchaisuksiri P, Blanco AN, et al. Thrombosis: a major contributor to global disease burden[J]. Arterioscler Thromb Vasc Biol. 2014;34(11):2363–2371.
- Wendelboe AM, Raskob GE. Global burden of thrombosis: epidemiologic aspects[J]. Circ Res. 2016;118(9):1340–1347.
- Keller K, Hobohm L, Ebner M, et al. Trends in thrombolytic treatment and outcomes of acute pulmonary embolism in Germany. Eur Heart J. 2020;41(4):522–529.
- Fesmire FM, Brown MD, Espinosa JA, et al. Critical issues in the evaluation and management of adult patients presenting to the emergency department with suspected pulmonary embolism. Ann Emerg Med. 2011;57(6):628–652.
- Xu XF, Yang YH, Zhai ZG, et al. Prevalence and incidence of deep venous thrombosis among patients in medical intensive care unit. Zhonghua Liu Xing Bing Xue Za Zhi. 2008;29(10):1034–1037.
- Zampetaki A, Willeit P, Drozdov I, et al. Profiling of circulating microRNAs: from single biomarkers to re-wired networks. Cardiovasc Res. 2012;93(4):555–562.
- Kumar S, Williams D, Sur S, et al. Role of flow-sensitive microRNAs and long noncoding RNAs in vascular dysfunction and atherosclerosis. Vasc Pharmacol. 2019;114:76–92.
- Ultimo S, Zauli G, Martelli AM, et al. Cardiovascular disease-related miRNAs expression: potential role as biomarkers and effects of training exercise. Oncotarget. 2018;9(24):17238–17254.
- Barutta F, Bellini S, Mastrocola R, et al. MicroRNA and microvascular complications of diabetes. Int J Endocrinol. 2018;2018:6890501.
- Dewdney B, Trollope A, Moxon J, et al. Circulating MicroRNAs as biomarkers for acute ischemic stroke: a systematic review. J Stroke Cerebrovasc Dis. 2018;27(3):522–530.
- Michota FA. Bridging the gap between evidence and practice in venous thromboembolism prophylaxis: the quality improvement process. J Gen Intern Med. 2007;22(12):1762–1770.
- Xu H, Du X, Xu J, et al. Pancreatic β cell microRNA-26a alleviates type 2 diabetes by improving peripheral insulin sensitivity and preserving β cell function. PLoS Biol. 2020;18(2):e3000603.
- Wang S, Min J, Yu Y, et al. Differentially expressed miRNAs in circulating exosomes between atrial fibrillation and sinus rhythm. J Thorac Dis. 2019;11(10):4337–4348.
- Patil M, Henderson J, Luong H, et al. The art of intercellular wireless communications: exosomes in heart disease and therapy. Front Cell Dev Biol. 2019;7:315.
- Sundar IK, Li D, Small RI. RNA-sequence analysis of plasma-derived extracellular vesicle miRNAs in smokers and patients with chronic obstructive pulmonary disease as circulating biomarkers. J Extracell Vesicles. 2019;8(1):1684816.
- Chen J, Tian J, Tang X, et al. MiR-346 regulates CD4⁺CXCR5⁺ T cells in the pathogenesis of Graves’ disease. Endocrine. 2015;49(3):752–760.
- Rodríguez-Muñoz A, Martínez-Hernández R, Ramos-Leví AM, et al. Circulating microvesicles regulate Treg and Th17 differentiation in human autoimmune thyroid disorders. J Clin Endocrinol Metab. 2015;100(12):E1531–E1539.
- Jerjes-Sánchez C, Villarreal-Umaña S, Ramírez-Rivera A, et al. Improving adjunctive treatment in pulmonary embolism and fibrinolytic therapy. The role of enoxaparin and weight-adjusted unfractionated heparin. J Thromb Thrombolysis. 2009;27(2):154–162.
- Ali HO, Arroyo AB, González-Conejero R, et al. The role of microRNA-27a/b and microRNA-494 in estrogen-mediated downregulation of tissue factor pathway inhibitor α. J Thromb Haemost. 2016;14(6):1226–1237.
- Mills EW, Wangen J, Green R, et al. Dynamic regulation of a ribosome rescue pathway in erythroid cells and platelets. Cell Rep. 2016;17(1):1–10.
- Fletcher SJ, Pisareva VP, Khan AO, et al. Role of the novel endoribonuclease SLFN14 and its disease-causing mutations in ribosomal degradation. RNA. 2018;24(7):939–949.
- Gao W, Wang K, Zhang L, et al. Pharmacological inhibition of S6K1 facilitates platelet activation by enhancing Akt phosphorylation. Platelets. 2019;30(2):241–250.
- Sundquist K, Ahmad A, Svensson PJ, et al. Polymorphisms in PARK2 and MRPL37 are associated with higher risk of recurrent venous thromboembolism in a sex-specific manner. J Thromb Thrombolysis. 2018;46(2):154–165.
- Deng HT, Liu HL, Zhai BB, et al. Vascular endothelial growth factor suppresses TNFSF15 production in endothelial cells by stimulating miR-31 and miR-20a expression via activation of Akt and Erk signals. FEBS Open Bio. 2016;7(1):108–117.
- Zhang K, Cai HX, Gao S, et al. TNFSF15 suppresses VEGF production in endothelial cells by stimulating miR-29b expression via activation of JNK-GATA3 signals. Oncotarget. 2016;7(43):69436–69449.
- Ergun S, Oztuzcu S. Sequence-based analysis of 5ʹUTR and coding regions of CASP3 in terms of miRSNPs and SNPs in targeting miRNAs. Comput Biol Chem. 2016;62:70–74.
- Della Bella S, Calcaterra F, Bacci M, et al. Pathologic up-regulation of TNFSF15-TNFRSF25 axis sustains endothelial dysfunction in unprovoked venous thromboembolism. Cardiovasc Res. 2020;116(3):698–707.
- Wang HW, Su SH, Wang YL, et al. MicroRNA-134 contributes to glucose-induced endothelial cell dysfunction and this effect can be reversed by far-infrared irradiation. PLoS One. 2016;11(1):e0147067.
- Lo WY, Peng CT, Wang HJ. MicroRNA-146a-5p mediates high glucose-induced endothelial inflammation via targeting interleukin-1 receptor-associated Kinase 1 expression. Front Physiol. 2017;8:551.
- Villasante A, Marturano-Kruik A, Ambati SR, et al. Recapitulating the size and cargo of tumor exosomes in a tissue-engineered model. Theranostics. 2016;6(8):1119–1130.
- Dieudé M, Bell C, Turgeon J, et al. The 20S proteasome core, active within apoptotic exosome-like vesicles, induces autoantibody production and accelerates rejection. Sci Transl Med. 2015;7(318):318ra200.
- Chyrchel B, Totoń-Żurańska J, Kruszelnicka O, et al. Association of plasma miR-223 and platelet reactivity in patients with coronary artery disease on dual antiplatelet therapy: a preliminary report. Platelets. 2015;26(6):593–597.
- Tan M, Yan HB, Li JN, et al. Thrombin stimulated platelet-derived exosomes inhibit platelet-derived growth factor receptor-beta expression in vascular smooth muscle cells. Cell Physiol Biochem. 2016;38(6):2348–2365.