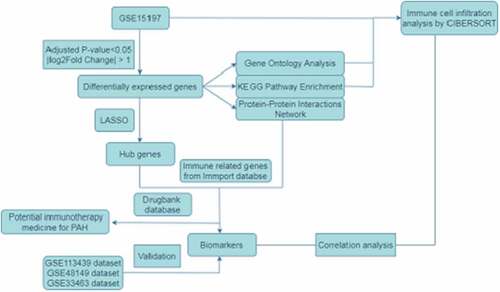
ABSTRACT
This study aimed to explore the molecular mechanisms related to immune and hub genes related to pulmonary arterial hypertension (PAH). The differentially expressed genes (DEGs) of GSE15197 were identified as filters with adjusted P value <0.05, and |Log2 fold change|> 1. Biofunctional and pathway enrichment annotation of DEGs indicated that immunity and inflammation may play an important role in the molecular mechanism of PAH. The CIBERSORT algorithm further analyzed the immune cell infiltration characteristics of the PAH and control samples. Subsequently, 16 hub genes were identified from DEGs using the least absolute shrinkage and selection operator (LASSO) algorithm. An immune related gene CX3CR1 was further selected from the intersection results of the 16 hub genes and the top 20 genes with the most adjacent nodes in the protein-protein interaction (PPI) network. GSE113439, GSE48149, and GSE33463 datasets were used to validate and proved CX3CR1 with a remarkable score of AUC to distinguish PAH samples caused by various reasons from the control group.
1. Introduction
Pulmonary arterial hypertension (PAH) is a clinical and pathophysiological syndrome caused by various heterogeneous diseases (etiology) and pathogeneses associated with pulmonary vascular structure, or functional changes that result in the development of pulmonary vascular resistance and increase in pulmonary artery pressure, subsequently triggering right heart failure or even death. It should be noted that a lowered minimum mean pulmonary artery pressure for diagnosis from 25 to 20 mmHg at rest has been described by the latest revised definition of PAH [Citation1]. Although several medicines and lung transplants had been applied to treatment PAH in clinical during the past decades, the five-year survival rate remains only 50% [Citation2,Citation3].
PAH is considered as an autoimmune inflammatory disease because of the complex changes in cytokines, chemokines, and inflammatory mediators [Citation4]. Marsh LM et al. described the different distributions of 21 types of inflammatory cells in idiopathic pulmonary arterial hypertension (IPAH) patients and normal controls in detail; their study suggested that immune imbalance played an important role in the pathogenesis of IPAH [Citation5]. Since 2015, more than 170 clinical trials focusing on PAH treatment have been registered in the National Library of Medicine clinical trials registry, and approximately 20% of the trials are related to the application of immunotherapy for treating pulmonary hypertension [Citation6]. Additionally, mesenchymal stem cell therapy has also shown potential clinical value in preclinical studies [Citation7,Citation8].
The signaling pathways and regulatory networks underlying the pathogenesis of PAH remain unknown due to the complicated etiologies. In this study, the microarray dataset of GSE15197 was adopted to screen differentially expressed genes (DEGs), exploring the gene ontology (GO), Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment, protein-protein interaction (PPI) network and further identify the immune related biomarker. We also applied the CIBERSORT algorithm [Citation9], a method to provide an estimation of the abundances of member cell types in a mixed cell population with gene expression data, to analyze the immune cell infiltration characteristics of 22 types of immune cells in PAH and normal samples. The selected biomarker was verified by GSE33463, GSE48149, GSE113439 datasets.
This study aimed to analyze the transcriptional profiles of patients with PAH based on the assessment of immune-related genes and immune cell infiltration characteristics, to explore the mechanism of immune response elicited in the pathogenesis of PAH, and to discover potential diagnostic biomarkers and therapeutic drugs to provide insights for an early diagnosis and for the development of immune-targeted therapy of PAH.
2. Materials and methods
2.1 Data source and DEGs analyses
The microarray datasets GSE15197 [Citation10], GSE33463 [Citation11], GSE48149 [Citation12] and GSE113439 [Citation13] were sourced from the GEO database. Detailed information about the datasets were provided in . All data were statistically analyzed and visualized using R software 4.0.3 and relevant packages. DEGs in the GSE15197 dataset were screened with adjusted P-value < 0.05 and |Log2 fold change|> 1 as the empirical analysis cutoff by ‘limma’ package [Citation14].
Table 1. Information of microarray dataset from GEO database
The ‘ClusterProfiler’[Citation15] and ‘org.Hs.eg.db’[Citation16] packages were used to explore the gene ontology (GO) and KEGG pathway enrichment of DEGs with a filter condition as P-value < 0.05. The results were visualized by ‘ggplot2’[Citation17] package. The protein-protein interaction (PPI) network was constructed using STRING (https://string-db.org/,Version: 11.0) [Citation18], an online web tool used to explore functional protein association networks, a filter condition with high confidence, and a minimum required interaction score of more than 0.7.
2.2 Exploring the abundance of immune cells in samples
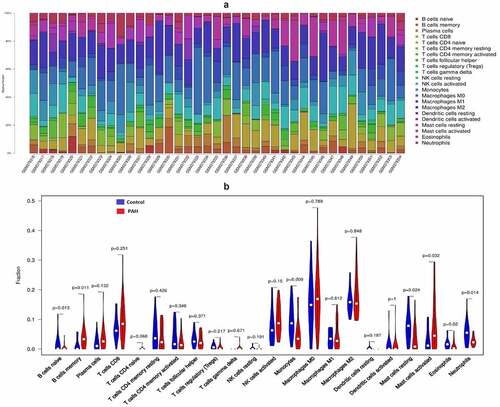
CIBERSORT, a method of exploring 22 types of tissue immune cells based on their gene expression profiles, was adopted to analyze the normalized GSE15197 expression data. A matrix of 22 types of immune cells was obtained, and all samples presented with a P-value < 0.05. The percentage of each immune cell type in the samples was calculated and displayed using a bar plot. The ‘vioplot’ package was used to compare the levels of 22 types of immune cells between the two groups. [Citation19]
2.3 Identification of hub-genes and potential immunotherapy medicine prediction
DEGs were screened using the least absolute shrinkage and selection operator (LASSO) algorithm to confirm the hub genes. We were especially interested in detecting potential immunotherapy targets and medicines for PAH, because immunotherapy is a promising field that has not been explored extensively. A dataset with 1793 unique immune-related genes was downloaded from the IMMPORT database[Citation20] (https://www.immport.org/shared/) and used to matched the immune-related hub genes. DrugBank [Citation21] (https://go.drugbank.com/) is a comprehensive pharmaceutical knowledge database, which researchers can use to study drugs which are in trial and/or have been applied clinically. We used this database to screen potential medicines for PAH and direct future studies.
2.4 Identification and validation of biomarkers
Potential biomarker of PAH was detected based on the intersections of hub genes with genes that have obvious connections with adjacent nodes in the PPI network. The discriminatory ability of the biomarker was validated using the GSE33463, GSE48149 and GSE113439 datasets and assessed using receiver operator characteristic (ROC) curves.
2.5 Correlation analysis between immune cells and biomarkers
Spearman’s rank correlation analysis was used to analyze the relationship between 22 types of immune cells and the biomarker by ‘ggstatsplot’[Citation22] and ‘ggplot2’ package. P-value < 0.05 was considered statistically significant.
3. Results
Our study indicated that, according to GO and KEGG enrichment analysis, inflammation and immune response played important roles in the pathogenesis of pulmonary hypertension. Through further analysis of immune cell infiltration, mast cells, B cells, monocytes, and neutrophils were with statistical difference (P value <0.05) and should be explored in detail. As an immune-related hub gene, CX3CR1 presents with a promising ability to be used to distinguish between patients with pulmonary hypertension caused by various reasons and those in the control group and can be considered a potential biomarker of pulmonary hypertension.
3.1 DEGs between PAH and control samples and the biofunctional analysis
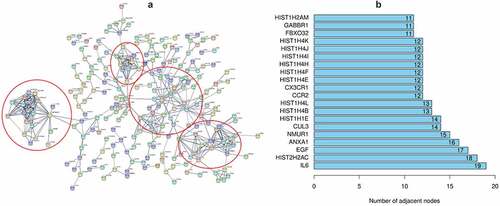
A volcano plot () was used to visualize the results of 587 DEGs, of which 321 were upregulated and 266 were downregulated. The results of KEGG pathway enrichment and gene ontology (GO) analysis are shown in . It is obvious that inflammation and immune-related pathways and biological regulatory functions play a dominant role in the enrichment results of the DEGs of PAH. The PPI network of DEGs and the top 20 genes with the most adjacent nodes are shown in .
Figure 1. DEGs and biofunctional analysis
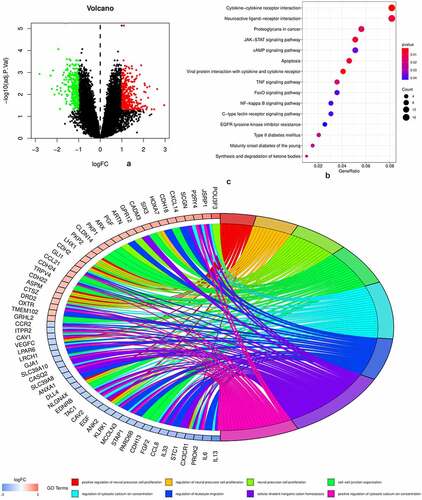
Figure 2. The PPI network of DEGs and the top 20 genes with most adjacent nodes
3.2 Immune cell infiltration of the samples
The result of CIBERSORT arithmetic analysis show the difference in abundance of 22 immune cells in the PAH and control samples. B cell naïve, monocytes, resting mast cells, eosinophils, and neutrophils were expressed at higher levels in control samples, while B cell memory and mast cell activation were significantly higher in the PAH group (P < 0.05). The matrix of result was listed in supplement file1.
3.3 Hub genes identification and potential immunotherapy medicine prediction
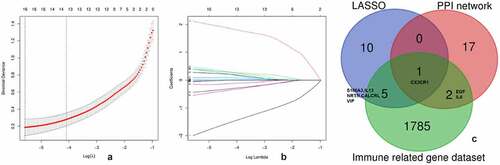
The LASSO algorithm was used to screen hub genes from the DEGs; as a result, 16 hub genes (SIAE, AKR7A2P1, NAT14, NRTN, SLC45A4, ADAD2, S100A3, LOC283481, CALCRL, IL13, VIP, GRHL2, SCG5, ABI3BP, CX3CR1, IFI44L) were identified ( and ). Three genes among the top 20 genes with most adjacent nodes in PPI network (CX3CR1, IL6, EGF) and six genes (S100A3, IL13, NRTN, CALCRL, VIP) among the hub genes by LASSO were matched with the immune related gene original from IMMPORT database (). The nine immune related genes were further investigated in the DrugBank database to predict potential medicines for PAH treatment, and the results are listed in .
Table 2. The potential medicine of immune related genes predicted in drugBank
The displays the optimal lambda (0.003) selection in the LASSO regression with 16 hub genes. The displays the coefficient profiles of the 16 hub genes. The displays the 3 immune related gene in PPI network and 6 immune related gene among the 16 hub genes selected by LASSO.
3.4 Identification and validation of PAH biomarker
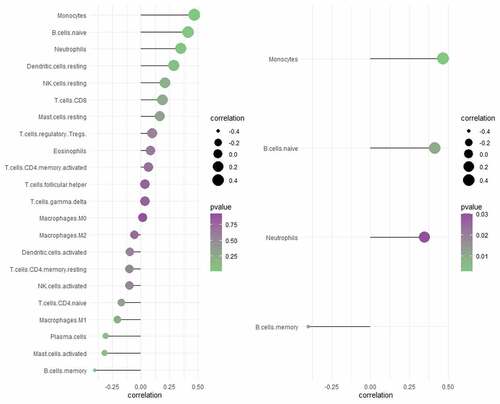
CX3CR1 was the only immune related gene considered as potential biomarker for intersecting with 16 hub genes and the top 20 genes with the most adjacent nodes in the PPI network. The discriminatory ability of CX3CR1 was validated using the GSE33463, GSE48149 and GSE113439 datasets. As shown in , CX3CR1 showed promising discriminatory ability of PAH in GSE15197 (AUC = 0.827), with a slight but acceptable decrease in GSE113439 (, AUC = 0.737), GSE48149 (, AUC = 0.773) and GSE33463 (, AUC = 0.751). Correlation analysis showed that CX3CR1 was mainly positively correlated with monocytes, B cells and neutrophils () with P-value < 0.05.
Figure 5. Identification and validation the biomarkers of PAH
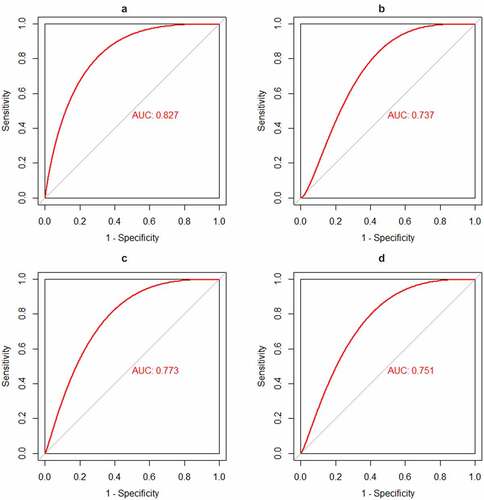
Figure 6. Correlation analysis between CX3CR1 with 22 immune cells
4. Discussion
PAH is a rare but life-threatening disease that can be categorized into five types according to the different dominant etiologies [Citation23]. Unfortunately, the treatment of PAH remains unsatisfactory, because of these multifactorial etiologies that result in a complex and uncertain pathophysiological state. In the current literature, genetic and/or gene mutations, inflammation, immunity, endothelial dysfunction, and abnormal vascular wall cell proliferation are closely related to the development of PAH [Citation24,Citation25]. We systematically analyzed the transcriptional map of PAH and explored the potential treatment medicine using bioinformatics to reveal the underlying molecular mechanisms and pathological processes, which may contribute to the prevention and treatment of the disease.
GO and KEGG pathway enrichment analyses revealed a strong involvement of inflammation and immune related process and pathways in PAH, indicating that they may be essential to the pathological processes of PAH. Based on these instructions, we explored the characteristics of 22 types of immune cell infiltration between PAH and normal samples, using the CIBERSORT algorithm. Naïve B cells, monocytes, resting mast cells, and neutrophils were expressed at higher levels in control samples (P-value < 0.05), while memory B cell and mast cell activation were significantly higher in the PAH group (P value < 0.05). Thus, the immune cells mentioned above, especially mast cells and B cells, may be closely related to the progression of PAH, a hypothesis which is supported by abundant evidences [Citation26,Citation27].
Mast cells were observed to be enriched in the pulmonary vascular tissue of PAH, and even appeared in the adjacent tissue of pulmonary perivascular cytopathy in the early stage. In a left heart failure-related pulmonary hypertension rat model treated with ketotifen (mast cell stabilizer) and a WS/WS rat model with mast cell deficiency (due to the mutation of mast cell growth factor receptor c-kit), the symptoms and vascular remodeling of pulmonary hypertension were significantly reduced. Both strategies were also effective and achieved similar results in monocrotaline-induced pulmonary arterial hypertension, suggesting that the role of mast cells extends to PAH caused by different pathogenesis [Citation28–30].
Similar to mast cells, B cells have also been directly observed infiltrating pulmonary perivascular tissue and show strong activation in the peripheral blood of PAH patients [Citation31].
Ulrich et al. reported that B cells have remarkably upregulated gene expression in the peripheral blood of PAH patients compared to healthy controls. The differentially expressed genes are mainly involved in vasomotor regulation, angiogenesis, and cell proliferation [Citation32].
Antibodies originate from B cells, which is the basis of the humoral immune response. Antibodies against pulmonary endothelial cells and fibroblasts have been detected in PAH tissue, and pathogenic auto-antibodies targeting endothelial cells are capable of inducing vascular endothelial apoptosis, and may initiate the development of PAH, which suggests that B cells may promote the pathological process of PAH [Citation33–36].
Immune cells and immune related genes are the focus of this study. According to the data set of 1793 immune related genes provided by IMMPORT database, we further selected 9 immune related genes to explore potential therapeutic drugs by Drugbank database.
Lebrikizumab is a humanized monoclonal antibody against interleukin-13 (IL-13) which has been used in trials studying the treatment of allergic asthma, atopic dermatitis, and idiopathic pulmonary fibrosis (IPF) by inhibiting the IL-13 driven Th2 inflammatory response [Citation37–39]. Erlotinib and dacomitinib are inhibitors of the epidermal growth factor receptor (EGFR) tyrosine kinase and are used to treat non-small cell lung cancer, pancreatic cancer, and several other cancer types. Accumulating evidence suggests that EGFR is closely associated with the molecular mechanism of PAH, and epidermal growth factor inhibitors such as erlotinib and dacomitinib have been used to treat PAH in animal models [Citation40–42].Interleukin-6 (IL-6) is a pleiotropic cytokine with a wide range of biological activities such as immune regulation, hematopoiesis, inflammation, and oncogenesis. By forming a high-affinity complex with human interleukin-6 (IL-6) to prevent IL-6 from binding to soluble and membrane-bound IL-6 receptors, stuximab is used to treat Castleman’s disease, a type of lymphoproliferative disease [Citation43]. Recent studies have verified that it can improve pulmonary ventilation function and reduce mortality in patients with severe COVID-19 [Citation44,Citation45]. It should be noted that the IL-6 receptor antagonist, tocilizumab, has been used in clinical trials to treat pulmonary arterial hypertension [Citation46].
Through drug prediction of hub immune-related genes, it can be inferred that immune-targeted drugs for different primary diseases demonstrate extensive research applications in the treatment of PAH. This is because there is an important immune mechanism underlying PAH. Exploration of the common immune pathway of PAH caused by different causes is of certain significance for the treatment of PAH.
CX3CR1 was considered as potential biomarker for the intersection result of the 16 hub genes and the top 20 genes with the most adjacent nodes in the PPI network. The discrimination ability of CX3CR1 was promising in both lung tissue and peripheral blood testing. CX3CR1 is the receptor for fractalkine and is regarded as a mediator of vascular and tissue damage in PAH. Jonathan Florentin et al. demonstrated that CX3CR1 played an important mediating role in the mobilization of myeloid cells from the bone marrow to blood and then to the lungs in patients with PAH [Citation47]. An increase in chemokine levels in lung tissues and the expression of chemokine receptors in blood monocytes initiate the migration of inflammatory monocytes to lung tissues. These monocytes differentiate into perivascular stromal macrophages, secrete proinflammatory cytokines, and promote vascular remodeling. Similarly, CX3CR1 also regulates neutrophil migration [Citation48]. From the perspective of CX3CR1 deficiency, some studies have confirmed that hypoxia-induced PAH can be prevented by reducing monocyte recruitment, macrophage polarization, and pulmonary artery smooth muscle cell proliferation [Citation49]. These studies indicate that CX3CR1 is an important regulatory target in the mechanism of inflammatory cell migration to lung tissues in patients with PAH. The results of the correlation analysis showed that CX3CR1 had the medium positive correlation with monocytes and neutrophils among 22 types of immune cells, which is consistent with the known studies. Currently, there is no medicine targeting CX3CR1, and a deep research about CX3CR1 and its ligand may be useful to direct the development of medicine development.
5. Conclusions and limitations
In this study, we systematically analyzed the transcriptional profiles of PAH, explored the role of immune cells in the pathogenesis of PAH, identified and verified a promising biomarker, CX3CR1. However, further in-depth studies on inflammation and immunity and validation in a larger population are essential to fully reveal the pathological processes and molecular mechanisms of PAH and to develop target medicines.
Highlights:
1. Inflammation and immune response play important roles in the pathogenesis of pulmonary arterial hypertension.
2. CX3CR1 is an important regulatory target and potential biomarker in pulmonary arterial hypertension.
3. Immune related targeted drugs may be effective in the treatment of pulmonary arterial hypertension.
Notes on contributors
Ruina Huang and Xifeng Zheng were involved in the conception and design of the study. Ruina Huang was responsible for article writing. Xifeng Zheng was responsible for visualization and scientific supervision. Junxian Wang provided effective scientific suggestions for this study. All authors reviewed and approved the final manuscript.
Supplemental Material
Download ()Acknowledgements
We would like to thank the Gene Expression Omnibus (GEO) database for the precious data used for free in scientific research. The patients involved in the database have obtained ethical approval. Our study is based on open source data, so there are no ethical issues and other conflicts of interest.
Disclosure statement
The authors declared that they have no conflicts of interests to this work. We declare that we do not have any commercial or associative interests that represents a conflict of interests in connection with the work submitted.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the Gene Expression Omnibus (GEO) database.
Supplementary material
Supplemental data for this article can be accessed here.
References
- Simonneau G, Montani D, Celermajer DS, et al. Haemodynamic definitions and updated clinical classification of pulmonary hypertension. Eur Respir J. 2019:53(1).
- Thenappan T, Ormiston ML, Ryan JJ, et al. Pulmonary arterial hypertension: pathogenesis and clinical management. BMJ. 2018;360:j5492.
- Hoeper MM, Apitz C, Grunig E, et al. Targeted therapy of pulmonary arterial hypertension: updated recommendations from the cologne consensus conference 2018. Int J Cardiol. 2018;272S:37–45.
- Rabinovitch M, Guignabert C, Humbert M, et al. Inflammation and immunity in the pathogenesis of pulmonary arterial hypertension. Circ Res. 2014;115(1):165–175.
- Marsh LM, Jandl K, Grünig G, et al. The inflammatory cell landscape in the lungs of patients with idiopathic pulmonary arterial hypertension. Eur Respir J. 2018;51(1):1701214.
- Goldenberg NM, Rabinovitch M, Steinberg BE. Inflammatory basis of pulmonary arterial hypertension: implications for perioperative and critical care medicine. Anesthesiology. 2019;131(4):898–907.
- Oh S, Jang AY, Chae S, et al. Comparative analysis on the anti-inflammatory/immune effect of mesenchymal stem cell therapy for the treatment of pulmonary arterial hypertension. Sci Rep. 2021;11(1):1–15.
- De Mendonca L, Felix NS, Blanco NG, et al. Mesenchymal stromal cell therapy reduces lung inflammation and vascular remodeling and improves hemodynamics in experimental pulmonary arterial hypertension. Stem Cell Res Ther. 2017;8(1):1–15.
- Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457.
- Rajkumar R, Konishi K, Richards TJ, et al. Genomewide rna expression profiling in lung identifies distinct signatures in idiopathic pulmonary arterial hypertension and secondary pulmonary hypertension. Am J Physiol Heart Circ Physiol. 2010;298(4):H1235–H1248.
- Mura M, Cecchini MJ, Joseph M, et al. Osteopontin lung gene expression is a marker of disease severity in pulmonary arterial hypertension. Respirology. 2019;24(11):1104–1110.
- Hsu E, Shi H, Jordan RM, et al. Lung tissues in patients with systemic sclerosis have gene expression patterns unique to pulmonary fibrosis and pulmonary hypertension. Arthritis Rheumatism. 2011;63(3):783–794.
- Poh LNF, Cheadle C, Berger AE, et al. Erythroid-specific transcriptional changes in pbmcs from pulmonary hypertension patients. PLoS ONE. 2012;7(4):e34951.
- Ritchie ME, Phipson B, Wu D, et al. Limma powers differential expression analyses for rna-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47.
- Yu G, Wang LG, Han Y, et al. Clusterprofiler: an r package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287.
- Carlson M, Falcon S, Pages H, et al. org. Hs. eg. db: Genome wide annotation for Human. R package version, 2017, 3(1)
- Ginestet C. Ggplot2: elegant graphics for data analysis. Journal of the Royal Statistical Society: Series A (Statistics in Society). 2011;174(1):245–246.
- Mering C. String: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261.
- Hu K. Become competent within one day in generating boxplots and violin plots for a novice without prior r experience. Methods and Protocols. 2020;3(4):64.
- Bhattacharya S, Andorf S, Gomes L, et al. Immport: disseminating data to the public for the future of immunology. Immunol Res. 2014;58(2–3):234–239.
- Wishart DS, Feunang YD, Guo AC, et al. Drugbank 5.0: a major update to the drugbank database for 2018. Nucleic Acids Res. 2018;46:D1074–D1082.
- Patil I. Ggstatsplot: ‘Ggplot2ʹ based plots with statistical details. 2018.
- Galiè N, Humbert M, Vachiery J-L, et al. 2015 esc/ers guidelines for the diagnosis and treatment of pulmonary hypertension. Eur Heart J. 2016;37(1):67–119.
- Guignabert C, Dorfmuller P. Pathology and pathobiology of pulmonary hypertension. Semin Respir Crit Care Med. 2013;34(5):551–559.
- Morrell NW, Aldred MA, Chung WK, et al. Genetics and genomics of pulmonary arterial hypertension. Eur Respir J. 2019;53(1).
- Breitling S, Hui Z, Zabini D, et al. The mast cell–b cell axis in lung vascular remodeling and pulmonary hypertension. Am J Physiol Lung Cell Mol Physiol. 2017;312(5):L710–L721.
- Klouda T, Yuan K. Inflammation in pulmonary arterial hypertension. Lung Inflammation in Health and Disease, Volume I. 2021;1303:351.
- Koudstaal T, Boomars KA, Kool M. Pulmonary arterial hypertension and chronic thromboembolic pulmonary hypertension: an immunological perspective. J Clin Med. 2020;9(2):9.
- Savai R, Pullamsetti SS, Kolbe J, et al. Immune and inflammatory cell involvement in the pathology of idiopathic pulmonary arterial hypertension. Am J Respir Crit Care Med. 2012;186(9):897–908.
- Hoffmann J, Yin J, Kukucka M, et al. Mast cells promote lung vascular remodelling in pulmonary hypertension. Eur Respir J. 2012;40:515–517.
- Tuder RM, Groves BM, Badesch DB, et al. Exuberant endothelial cell growth and elements of inflammation are present in plexiform lesions of pulmonary hypertension. Am J Pathol. 1994;144:275–285.
- Ulrich S, Taraseviciene-Stewart L, Huber LC, et al. Peripheral blood b lymphocytes derived from patients with idiopathic pulmonary arterial hypertension express a different rna pattern compared with healthy controls: a cross sectional study. Respir Res. 2008;9:9.
- Li C, Liu P, Song R, et al. Immune cells and autoantibodies in pulmonary arterial hypertension. Acta Biochim Biophys Sin (Shanghai). 2017;49(12):1047–1057.
- Liu XD, Guo SY, Yang LL, et al. Anti-endothelial cell antibodies in connective tissue diseases associated with pulmonary arterial hypertension. J Thorac Dis. 2014;6(5):497–502.
- Tamby MC. Antibodies to fibroblasts in idiopathic and scleroderma-associated pulmonary hypertension. Eur Respir J. 2006;28(4):799–807.
- Nicolls MR. Autoimmunity and pulmonary hypertension: a perspective. Eur Respir J. 2005;26(6):1110–1118.
- Loh TY, Hsiao JL, Shi VY. Therapeutic potential of lebrikizumab in the treatment of atopic dermatitis. J Asthma Allergy. 2020;13:109–114.
- Corren J, Lemanske RF Jr, Hanania NA, et al. Lebrikizumab treatment in adults with asthma. N Engl J Med. 2011;365(12):1088–1098.
- Maher TM, Costabel U, Glassberg MK, et al. Phase 2 trial to assess lebrikizumab in patients with idiopathic pulmonary fibrosis. Eur Respir J. 2021;57(2).
- Dahal BK, Cornitescu T, Tretyn A, et al. Role of epidermal growth factor inhibition in experimental pulmonary hypertension. Am J Respir Crit Care Med. 2009;181(2):158–167.
- Sagar D, Vandana P. Development of anti-angiogenic erlotinib liposomal formulation for pulmonary hypertension: a qbd approach. Drug Delivery & Translational Research. 2019;9(5):980–996.
- Yu X, Zhao X, Zhang J, et al. Dacomitinib, a new pan-egfr inhibitor, is effective in attenuating pulmonary vascular remodeling and pulmonary hypertension. Eur J Pharmacol. 2019;850:97–108.
- Gliga S, Orth HM, Lübke N, et al. Multicentric castleman’s disease in hiv patients: a single-center cohort diagnosed from 2008 to 2018. Infection. 2021;1–7.
- Gritti G, Raimondi F, Bottazzi B, et al. Siltuximab downregulates interleukin-8 and pentraxin 3 to improve ventilatory status and survival in severe covid-19. Leukemia. 2021;35(1):1–5.
- Albiach L, Agüero D, Ambrosioni J, et al. Tocilizumab reduces the risk of icu admission and mortality in patients with sars-cov-2 infection.
- Hernandez-Sanchez J, Harlow L, Church C, et al. Express: clinical trial protocol for transform-uk: a therapeutic open label study of tocilizumab in the treatment of pulmonary arterial hypertension. Pulm Circ. 2017;8(1):204589321773582.
- Florentin J, Coppin E, Vasamsetti SB, et al. Inflammatory macrophage expansion in pulmonary hypertension depends upon mobilization of blood-borne monocytes. J Immunol. 2018;200(10):3612–3625.
- Florentin J, Zhao J, Tai -Y-Y, et al. Interleukin-6 mediates neutrophil mobilization from bone marrow in pulmonary hypertension. Cell Mol Immunol. 2021;18(2):374–384.
- Amsellem V, Abid S, Poupel L, et al. Roles for the cx3cl1/cx3cr1 and ccl2/ccr2 chemokine systems in hypoxic pulmonary hypertension. Am J Respir Cell Mol Biol. 2017;56(5):597.