ABSTRACT
This study aimed to identify potential pharmacological targets of triptolide regulating the tumor microenvironment (TME) of stomach adenocarcinoma (STAD) patients. A total of 343 STAD cases from The Cancer Genome Atlas (TCGA) were assigned into high- or low-score groups applying Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE). Hub genes were identified from differentially expressed genes (DEGs) shared by stromal- and immune-related components in the TME of STAD patients using R software. Cox regression analysis was used to identify genes significantly correlated with STAD patient survival. Triptolide target genes were predicted from the Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP). Top 30 genes filtered by Cytohubba from 734 DEGs were screened as hub genes. Forty-two genes were found to be at high risk for STAD prognosis. Thirty-four targets of triptolide were predicted using the TCMSP database. Importantly, C-X-C chemokine receptor type 4 (CXCR4) was identified as a potential target of triptolide associated with the TME in STAD. Analysis of survival highlighted the association between CXCR4 upregulation with STAD progression and poor prognosis. Gene Set Enrichment Analysis (GSEA) confirmed that genes in the CXCR4- upregulated group had significant enrichment in immune-linked pathways. Additionally, triptolide targets were found to be significantly enriched in CXCR4-related chemokine and cancer-related p53 signaling pathways. Molecular docking demonstrated a high affinity between triptolide and CXCR4. In conclusion, CXCR4 may be a therapeutic target of triptolide in the treatment of STAD patients by modulating the TME.
Introduction
Cancer contributes substantially to the global disease burden in terms of both incidence and mortality [Citation1]. Stomach cancer is the third-largest contributor to cancer mortality in the low-to-medium Human Development Index (HDI) nations, and one of the most fatal diseases in the above-average-high HDI nations [Citation2]. According to a report on cancer statistics in 2012, gastric cancer is frequently diagnosed (15.82%) and is a main contributor to cancer-related mortality (17.70%) in China [Citation3]. The important risk factors for gastric cancer consist of Epstein-Barr virus infection, Helicobacter pylori infection, family history, environmental factors, age, and sex [Citation4,Citation5]. Among various treatment strategies for patients with gastric cancer, surgery still represents the ideal therapeutic modality, but the prognosis remains poor [Citation6]. Therefore, postoperative adjuvant treatment regimens are urgently needed to reduce the recurrence and improve survival.
Potential postoperative options for gastric cancer patients include chemotherapy, radiotherapy, and targeted therapy [Citation7]. However, the sensitivity of patients to these therapies is limited, and adverse reactions, including nausea and vomiting, often accompany these treatment regimens. Accordingly, novel agents, including traditional Chinese medicines (TCM) or their extracts, which can not only increase the sensitivity to chemotherapy and radiation therapy but also cause minimal toxicity, are needed for patients with gastric cancer [Citation8]. The widespread use of TCM has a long history of at least 2,000 years in China. Besides acupuncture, Chinese herbal medicines have been shown to be a low-risk and effective therapeutic option in oncology patients that enhances quality of life and survival odds [Citation9,Citation10]. Triptolide, a diterpene triepoxide extracted from Tripterygium wilfordii Hook, is a pharmacologically active compound with anti-inflammatory, immunosuppressive, and antitumor activities [Citation11–13].
The tumor microenvironment (TME) is a continuously evolving and complex cellular ecosystem that surrounds the tumors, and consists of stromal cells, immune cells, fibroblasts, blood vessels, and endothelial cells [Citation14]. TME plays a key role in tumor progression and patient survival. Triptolide exerts its protective effect through modulating the tumor immune microenvironment by regulating tumor stromal components [Citation15]. More interestingly, previous studies have highlighted that triptolide reduces chemoresistance of cancers [Citation16]. However, the role of triptolide in the TME of stomach adenocarcinoma (STAD) has not been adequately examined.
In the present study, using a bioinformatics approach, we aimed to identify the potential pharmacological targets of triptolide that modulate the TME of patients with STAD. STAD cases were obtained from The Cancer Genome Atlas (TCGA) database, and RNA-seq data of these cases were analyzed using the Estimation of STromal and Immune cells in MAlignant Tumor tissues using Expression data (ESTIMATE) to identify the overlapping stromal and immune-related genes. The target genes of triptolide were also investigated using the TCMSP database. The intersection of stromal-/immune-related differentially expressed genes (DEGs) with triptolide target genes revealed C-X-C chemokine receptor type 4 (CXCR4) to be a potential therapeutic target for triptolide in modulating the TME of STAD. Finally, the potential interaction of CXCR4 with triptolide was evaluated.
Material & methods
Data sources & processing
Transcriptomic RNA-Seq dataset for STAD was collected from TCGA database by selecting adenomas and adenocarcinomas as ‘disease-type,’ transcriptome profiling as ‘data-category,’ gene expression quantification as ‘data-type,’ and HTSeq-FPKM ‘workflow-type.’ A total of 373 cases (30 normal and 343 tumor samples) were included in the gene expression dataset, which contained gene expression data for each sample. Ensembl IDs were converted to gene symbols based on the transcriptome annotation file downloaded from the Ensembl database (Homo sapiens sapiens.GRCh38.102.chr.gtf).
To obtain clinical information of the patients, including clinicopathological staging, survival time and status, we downloaded the clinical dataset for STAD from TCGA database by selecting clinical as ‘data category’ and BCR xml as data ‘format.’ To determine the purity of the tumor as well as the degree of infiltrating stromal- or immune-related cells within tumor tissues, we calculated the stromal, immune, and ESTIMATE scores for each tumor sample from STAD RNA-seq data using R estimate package based on the ESTIMATE algorithm [Citation17]. The ESTIMATE scoring result and clinical dataset of STAD were then merged using R loaded with limma package for further analysis. Ethics permission was not necessary since all data used in this study was collected from publicly available online database repositories.
Correlation analysis
Eligible patients were stratified into subgroups to assess the relationships between calculated stromal/immune scores in the TME of STAD patients and clinicopathological characteristics, including clinical staging (stages I–IV), differentiation grade (G0–G3), and TNM classification (T1–T4, N0–N3, M0–M1) through R limma and ggbubr packages. P-values were based on the Wilcoxon signed-rank test, and p < 0.05 was considered to confer statistical significance. Patients with missing data were censored, with no imputation for massing values.
Identification of differently expressed genes (DEGs)
A total of 343 tumor samples were separated into ESTIMATE low- and high-score groups as determined by the median cutoffs for stromal- and immune-related scoring, as previously described [Citation18]. By matching the gene expression data and TME scoring results for each sample, DEGs with |logFC| > 1 and false discovery rate (FDR) < 0.05 based on the Wilcoxon test were identified in stromal and immune high-score vs. low-score groups [Citation19,Citation20]. Heatmaps for the DEGs were generated using the pheatmap package (R). Intersected DEGs were identified between the high-score vs. low-score samples using the VennDiagram package of R. Symbols of all intersected DEGs were transformed to entrez IDs using the R package (org.Hs.eg.db) for enrichment analysis.
Gene Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analyses were carried out using R packages of clusterProfiler, org.Hs.eg.db, enrichplot, and ggplot2 [Citation21]. A cutoff q value < 0.05 was deemed to confer statistical significance, while the 10 top-ranking biological processes (BP), molecular functions (MF) and cellular components (CC), as well as the top 30 statistically relevant pathways were visualized.
Generation of protein–protein interaction (PPI) networks and screening of hub genes
The PPI networks of the DEGs were generated using STRING, an online public database for predicting functional associations between proteins [Citation22], and optimized by Cytoscape, an open-source software that integrates biomolecular interaction networks [Citation23]. Proteins with a combined score (minimum required interaction score) of more than 0.95, were kept. The top 30 genes filtered by Cytohubba using the MCC method were screened as hub genes.
Cox regression analysis
By merging the survival data from the clinical dataset of STAD and gene expression data, we obtained the survival time, survival status, and gene expression quantifications of each STAD sample, and those with missing values were excluded. Using the R package of survival, we identified genes related to prognosis based on the analysis of univariate Cox regression and Kaplan–Meier approach, with statistical significance set at p < 0.05.
Prediction of triptolide targets
The Traditional Chinese Medicine Systems Pharmacology Database and Analysis Platform (TCMSP) was used to identify potential human protein targets of triptolide [Citation24]. To convert names of the protein targets to corresponding gene symbols, we downloaded the annotation information from the Uniprot database filtered by reviewed records and Homo sapiens as organisms. GO and KEGG enrichment analysis of these targets was conducted using R. 2D structure of triptolide in .sdf format was obtained from the PubChem database for molecular docking.
Intersected genes between PPI network, Cox analysis, and triptolide targets
To identify the potential target of triptolide that affect the prognosis of STAD, 30 high-ranking hub genes with most adjacent nodes in the PPI network, prognosis-related genes generated by Cox regression analysis, and target genes of triptolide were intersected using Bioinformatics & Evolutionary Genomics, an online database that provides a free-to-use tool to calculate and draw Venn diagrams [Citation25]. We then compared the differences in expression of the intersected genes between normal and tumor samples of STAD, and p < 0.05 based on the Wilcoxon test was considered significant. Using R packages of survival and survminer, we analyzed possible correlations between expression levels of the intersected genes, patient survival, and clinicopathological staging.
Gene Set Enrichment Analysis (GSEA)
We further investigated the mechanisms used by the intersected genes involved in the TME and prognosis of STAD using the software GSEA v4.0.3. The 343 tumor samples downloaded from TCGA were separated into gene (CXCR4) high-and low-expression groups. GSEA for KEGG pathways was performed, and the permutation quantity was set to 1000. A False discovery rate q value (FDR q-val) < 0.05 was deemed to be statistically significant.
Molecular docking
Structure of the target protein was downloaded from the RCSB PDB database (https://www.rcsb.org), and the open-source computer program, PyMol, was used to remove water and ligands from the protein. The 2D structure of triptolide (PubChem CID: 107,985) was provided by PubChem (https://pubchem.ncbi.nlm.nih.gov/) and was imported into the software ChemOffice to optimize its structure by calculating MM2 minimized energy [Citation26]. The structure of triptolide was then saved in .sdf format and transformed to .pdb format. Open-source AutoDock Tools 1.5.6 was applied to add hydrogens to the structure of the target protein and the Gasteiger charges were calculated. Atomic charges were added, atomic types were allocated to triptolide, and flexible bonds were set as rotatable by default. The structure of triptolide in .pdbqt format was then obtained. The target protein (as receptor) and triptolide (as ligand) were imported into AutoDock Vina for molecular docking, and PyMol was used for visualization. The binding free energy range was set to 5, and the number of docking modes was set as 20. The active binding point was determined based on the coordinates of the ligands in the CXCR4 complex, 3OE6 [Citation27]. The grid box for CXCR4 was identified by setting the spacing as 0.375 and grid point numbers in the x, y, and z dimensions as 40, 40, and 40, respectively. The grid centers were 17.679, 28.847, and 73.411 for the x, y, and z centers, respectively.
Results
In the present study, the potential prognostic biomarkers in regulating the TME of STAD patients by triptolide treatment were evaluated using bioinformatics analysis. First, STAD cases were obtained from the TCGA database. Then, the overlapping stromal and immune-related genes were obtained using ESTIMATE. Triptolide target genes were obtained from the TCMSP database. Importantly, the intersection of these stromal-/immune-related DEGs with triptolide target genes revealed CXCR4 to be a potential target of triptolide in modulating the TME of patients with STAD. Finally, correlation analysis revealed that CXCR4 overexpression was linked to STAD progression and poor survival. Furthermore, molecular docking studies revealed that triptolide demonstrates high affinity for CXCR4.
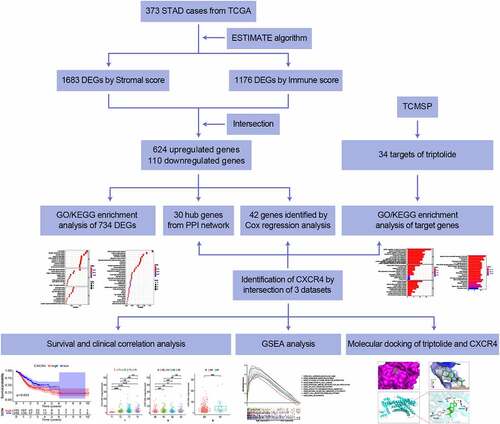
Study workflow
To analyze the effect of triptolide on the prognosis of STAD cases, initial determination for the amount of stromal- and immune-related facets within STAD TME were required, depending on transcriptome RNA-seq data of 343 STAD cases from TCGA. A PPI network was developed, followed by univariate Cox regression analysis based on DEGs shared by stromal/immune-related scoring. A total of 34 triptolide target genes were predicted by searching the TCMSP database and GO/KEGG-enrichment analysis was performed for these genes. Through intersection analysis based on the hub genes in the PPI network, prognosis-related genes from Cox regression analysis, and target genes of triptolide, we identified CXCR4 as the overlapping gene. With a focus on CXCR4, we carried out correlation analysis of survival and clinicopathological staging, GSEA, and molecular docking verification of CXCR4 and triptolide. Details of the process used for the analysis are shown in Graphical Abstract 1.
Correlation of stromal and immune components with clinicopathological features in STAD cases
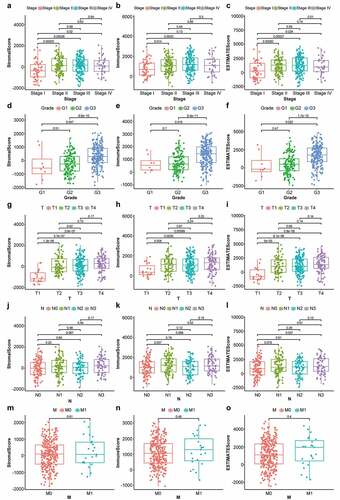
First, the changes in stromal-, immune-, and ESTIMATE-scores were evaluated in various cancer stages and differentiation grades. As shown in , the stromal scores were found to be positively correlated with STAD staging, showing a significant difference between stage I and stage II/III/IV (p < 0.05); immune scores were markedly lower in stage I than in stages II and III () (p < 0.05); ESTIMATE scores, the sum of stromal and immune scores, were markedly lower in stage I than in stages II/II/IV () (p < 0.05). For differentiation grading, significant differences were observed between G1 and G3 as well as G2 and G3 among patients with different stromal- () (p < 0.05), immune- () (p < 0.05), and ESTIMATE- scores () (p < 0.05). A positive correlation was observed between T1 and T2/T3/T4 among patients with different stromal-, immune-, and ESTIMATE- scores () (p < 0.05). Stromal and immune scores showed no significant difference among cases with various N classifications () (p > 0.05); the sum of stromal and immune scores, ESTIMATE scores, were significantly lower in N0 than in N3 () (p < 0.05). No significant differences were observed in the score distribution among patients with or without metastasis () (p > 0.05). These results suggested that the stromal and immune components played a significant role in the progression of STAD.
Figure 1. Box plots of correlation between ESTIMATE scores with clinicopathological staging characteristics. (2a, 2b, 2 c) StromalScore, ImmuneScore and ESTIMATEScore of STAD patients in each stage. (2d, 2e, 2 f) StromalScore, ImmuneScore and ESTIMATEScore of STAD patients in each grade. (2 g, 2 h, 2i) StromalScore, ImmuneScore and ESTIMATEScore of STAD patients in each tumor classification. (2 j, 2k, 2 l) StromalScore, ImmuneScore and ESTIMATEScore of STAD cases with or without lymph node metastasis. (2 m, 2 n, 2o) StromalScore, ImmuneScore and ESTIMATEScore of STAD cases with or without distant metastasis. P-values based on the Wilcoxon signed-rank test; p < 0.05 was statistically significant
Intersected DEGs between stromal and immune scores
For a detailed analysis of the differences in gene expression related to stromal and immune components of STAD TME, we used R software and performed a comparative study between high- and low-score samples. Using median cutoffs, a total of 1495 genes were found to be upregulated and 188 genes were downregulated in the stromal high-scoring group (1683 DEGs by stromal score in total, ); 866 genes were upregulated and 310 were downregulated within the immune-related high-scoring group (1176 DEGs by immune score in total, ). As shown in the Venn diagram, 624 genes were upregulated and 110 were downregulated in high-ranking cohorts for both stromal and immune-related scorings (). These 734 genes (624 upregulated +110 downregulated) were chosen for further analysis.
Figure 2. Heatmaps, Venn diagrams, and enrichment analysis of the DEGs. (2a) Heatmapped DEGs within the stromal high-scoring group versus low-scoring group (|logFC| >1; FDR < 0.05 based on Wilcoxon test); red represents higher expression and blue represents lower expression. (2b) Heatmapped DEGs in immune high-scoring group versus low-scoring group (|logFC| >1; FDR < 0.05 based on Wilcoxon test); red represents higher expression and blue indicates lower expression. (2 c) Venn diagram of 624 commonly upregulated genes (stromal/immune-linked scorings). (2d) Venn diagram of 110 commonly downregulated genes (stromal/immune-linked scorings). (2e) Ten highest-ranking GO terms for BP, CC and MF enriched using DEGs shared by stromal and immune scores (q value < 0.05). (2 f) Top 30 KEGG terms enriched by DEGs and ranked by q values from highest to lowest (q value < 0.05)
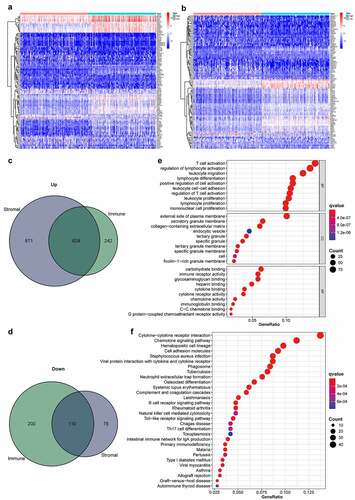
GO function and KEGG pathway enrichment assessments were conducted, focusing solely on the 734 intersected DEGs. As indicated by GO enrichment analysis, the DEGs were significantly associated with immune functions, such as biological functions of leukocyte proliferation, regulation of lymphocyte activation, T-cell activation, and molecular function of immune receptor activity (). Results from KEGG-enrichment analysis showed that cytokine–cytokine receptor interaction, B cell receptor signaling pathway, chemokine-signaling pathway, and hematopoietic cell lineage were significantly enriched by the DEGs () (q value < 0.05). Therefore, the roles of the shared DEGs correlated with immune-related activities, suggesting that the engagement of immune factors likely played a predominant role in the TME of STAD.
Identification of CXCR4 in the intersection analysis of PPI network, univariate Cox regression analysis, and triptolide targets
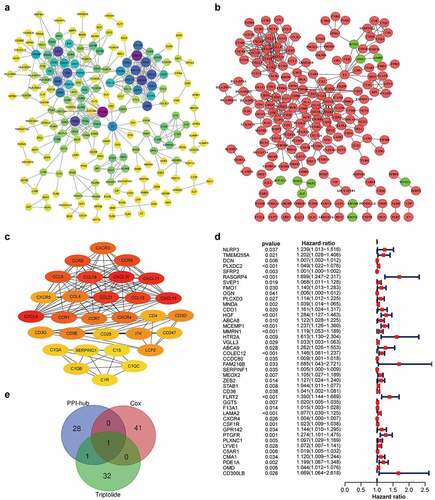
A PPI network of these 734 DEGs was created based on the STRING database and Cytoscape software (). A sub-network of 30 highest-ranking genes filtered by the Cytohubba MCC method is presented in . Univariable Cox regression analysis of the DEGs revealed the identities of 42 genes at high risk for STAD prognosis (). Thirty-four target genes were predicted using the TCMSP database (). Through intersection analysis of the hub genes within the PPI network, univariate Cox regression analysis, and target genes of triptolide, we identified CXCR4 as the sole overlapping target of the above three datasets (). CXCR4 was deemed to be a high-risk factor for STAD prognosis by Cox regression analysis (p = 0.01; hazards ratio [HR], 1.004; 95% CI [1.001 − 1.007]).
Table 1. List of 34 triptolide targets and their respective UniProt Entry IDs and gene names
Figure 3. CXCR4, a hub gene from the PPI network, correlated with STAD prognosis and was identified as a target gene of triptolide. (3a) PPI network (STRING) based on DEGs (combined score = 0.95). (3b) Interaction network constructed using Cytoscape v3.7.2 based on DEGs with a minimum required interaction score > 0.95; green and red illustrate downregulated and upregulated genes, respectively. (3 c) Sub-network of 30 hub genes filtered by Cytohubba MCC method; node score increases from light to dark color. (3d) Forest plot of Cox regression analysis with DEGs; genes with p < 0.05 and HR > 1 were identified as high-risk factors for STAD prognosis. (3e) Venn diagram of 30 hub genes from the PPI network, 42 prognosis-related genes based on Cox regression analysis, and 34 target genes of triptolide
CXCR4 expression linked to survival of patients with STAD
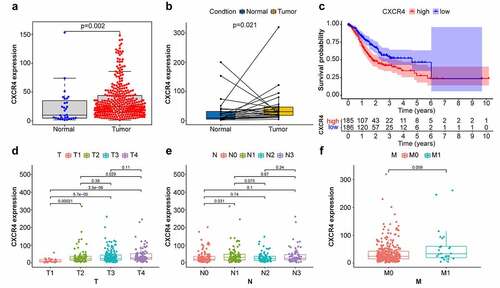
Comparison of CXCR4 expression levels in normal and STAD tumor tissues revealed that CXCR4 was significantly upregulated in tumor tissues () (p = 0.002; p = 0.021). To investigate possible links between CXCR4 expression and survival of patients with STAD, tumor samples were separated into CXCR4 high-and low-expression cohorts. As illustrated in , the 5-year survival rate was lower in the high-expression cohort compared to the low-expression cohort (p = 0.003). Correlation analysis between CXCR4 expression and TNM classification revealed that CXCR4 was significantly upregulated in T2, T3, and T4 stages relative to T1 stage, as well as T4 compared with T2 () (p < 0.05); its association with N and M stages was not significant () (p > 0.05). These results indicated that CXCR4 expression was negatively linked to the prognosis of patients with STAD.
Figure 4. Comparison between CXCR4 expression with prognosis/clinicopathological features in STAD patients. (4a, 4b) Difference analysis and paired difference analysis showed significantly higher CXCR4 expression levels in STAD tissues versus normal tissues (p < 0.05, Wilcoxon test). (4 c) Patients were categorized into CXCR4 high-/low-expressing cohorts; survival curve showed that CXCR4 was linked to poor prognosis of STAD patients (p = 0.003). (4d) CXCR4 expression levels were significantly higher in T2/3/4 versus T1 and in T4 versus T2 (p < 0.05). (4e) No correlation detected between CXCR4 expression and N classification (p > 0.05). (4 f) CXCR4 showed no difference between patients with and without metastasis (p = 0.059)
Pathways associated with CXCR4 in patients with STAD
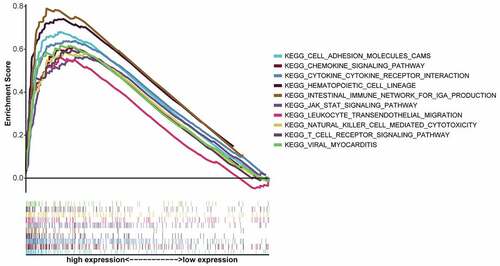
To further understand the underlying mechanism of CXCR4 expression involved in the progression of STAD, GSEA was conducted on the CXCR4-high-expression cohort for the 10 highest-ranking pathways (by FDR q value), as FDR q values were higher than 0.05 for all pathways enriched by the CXCR4 low-expression group (). As shown in , most pathways enriched in the CXCR4 high-expression cohort were immune- or tumor-related, including leukocyte trans-endothelial migration, Janus kinase 2 (JAK)-signal transducer and activator of transcription (STAT) signaling pathway, T-cell receptor signaling pathway, cytokine–cytokine receptor interaction, hematopoietic cell lineage, natural killer (NK) cell-mediated cytotoxicity, and chemokine-signaling pathway.
Table 2. Top 10 pathways enriched by genes in the CXCR4 high-expression group based on GSEA (in the order of FDR q value)
Triptolide bound to CXCR4 with high affinity
As illustrated in , the targets of triptolide were biologically active in immune cell proliferation, and their MFs were significantly enriched in chemokine receptor binding, CCR chemokine receptor binding, and cytokine receptor binding (q value < 0.05). The targets of triptolide were significantly enriched in the chemokine, nuclear factor kappa B (NF-κB), p53, and hypoxia-inducible 1 (HIF−1) signaling pathways; it is worth noting that several cancer pathways were significantly enriched by triptolide targets, including pacreatic cancer, colorectal cancer and small cell lung cancer ( and ) (q value < 0.05).
Figure 6. Enrichment analysis of triptolide target genes and molecular docking between triptolide and CXCR4. (6a) Ten highest-ranking GO terms for BP, CC, and MF enriched by triptolide targets (q value < 0.05). (6b) Top 15 KEGG pathways enriched by triptolide targets and ranked by q values from the highest to the lowest. (6 c) The optimal mode of molecular interaction between triptolide and CXCR4 is shown in its three-dimensional model and the binding point is located in the center of the active binding pocket. (6d) Hydrophobicity of molecular interaction between triptolide and CXCR4. (6e) Hydrogen bonds are formed with Tyr121, Arg188, and Tyr255
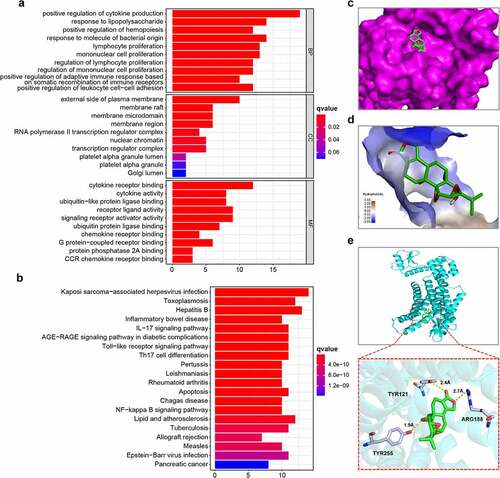
Table 3. List of 15 cancer- and CXCR4-related KEGG pathways significantly enriched by 34 targets of triptolide
Next, the potential docking of CXCR4 with triptolide was investigated. As shown in , the first mode of molecular docking between triptolide and CXCR4 was the optimal one, as it had the lowest binding free energy of −8.6 kcal/mol and its distance from the center of the active binding pocket was 0. The 3D presentation of the optimal docking mode is shown in . As illustrated in , the hydrophobic interactions between triptolide and CXCR4 were relatively weak. Hydrogen bonds were formed with Tyr121, Arg188, and Tyr255 ().
Table 4. Molecular docking parameters of 20 docking modes
Discussion
According to the 2018 statistical report on cancers, stomach cancer accounted for 5.7% of newly diagnosed cancer cases and 8.2% of cancer-related mortality, thus constituting the fifth most frequently identified tumors and third-most mortality-inducing tumors worldwide[Citation1]. As immunotherapy has shown promising prospects as a novel therapeutic modality for cancers, such as gastric and lung cancer, increasing number of studies have focused on the role of immune cells in promoting or inhibiting oncogenesis. The report that pembrolizumab, a programmed death-1 (PD-1) inhibitor, exhibited efficacy in advanced STAD cases, highlights the role of TME in STAD[Citation28]. However, the upregulation of programmed death-ligand 1 (PD-L1) appears to act as a risk factor for disease progression and survival [Citation29,Citation30]. Therefore, identification of more immune-related biomarkers is needed to predict the prognosis and evaluate the role of immune-related facets within the TME in STAD cases. To improve the efficacy of immunotherapy in the treatment of gastric cancer and to develop and discover candidate drugs for immunotherapy, it is essential to understand the complex immune microenvironment within STADs, and to identify more predictive biomarkers to achieve personalized treatment with immunotherapy.
This investigation focused on identifying TME-related genes correlated with survival/TMN classifications in STAD cases, based on TCGA data. Correlation analysis of stromal and immune components in STAD TME with clinicopathological profiles in the STAD cases revealed that immune score increased as the tumor progressed from stage I to stages II, III, and IV, from well-differentiated (G1) to poorly differentiated (G2/3), and from T1 to T2/T3/T4. This suggests that immune infiltration is likely associated with the progression of STAD, particularly from the early stage to later or advanced stages. This is consistent with previous report that STAD progression can be influenced by its innate phenotype and extrinsic tumor immune microenvironment as well [Citation31]. As a newly developed algorithm, ESTIMATE uses the unique characteristics of cancer tissue transcription profiles to infer tumor cell structure and different infiltrating normal cells[Citation32]. The algorithm estimates stromal and immune scores based on the specific gene expression signatures of stromal and immune cells to predict the level of infiltration of stromal and immune cells[Citation33]. In the present study, to further determine the relationship between triptolide and the TME of patients with STAD, we first identified 624 upregulated and 110 downregulated genes as the overlapping genes between stromal and immune scores.
CXCR4 has been found to be the most widely expressed chemokine receptor in most tumors and its expression in tumor tissues correlates with metastases in several different cancers, including gastric cancer [Citation34]. Studies have shown that CXCR4 blockade may serve as a therapeutic target for cancers by altering the tumor microenvironment and inhibiting tumor growth [Citation35]. Through a series of transcriptomic analyses of STAD samples from TCGA, we noted that upregulated CXCR4 was highly associated with STAD progression (from stage I to stages II/III/IV, T1 to T2/3/4) and poor survival. Triptolide had an oral bioavailability (OB) of 51.29%, drug likeness of 0.68, and molecular weight of 360.44, which met the filtering criteria of OB > 30% and DL > 0.18 [Citation24]. Importantly, we identified CXCR4 as a target of triptolide, a small molecule extracted from Tripterygii Radix, and validated their binding activity via molecular docking. Triptolide exhibits antitumor activity in a multi-pathway, multi-target, and broad-spectrum manner. The relatively high binding affinity between triptolide and CXCR4 indicated that triptolide represents a potential CXCR4 inhibitor candidate that may be used to improve prognosis and slow progression in STAD patients.
CXCR4, a chemokine receptor family member and a G protein-coupled receptor, has seven transmembrane regions, one extracellular N-terminal, and one intracellular C-terminal. The chemokine receptor-ligand pair of CXCR4 and CXCL12, a specific CXCR4 ligand, plays a pivotal role within the chemokine superfamily [Citation36]. The CXCR4/CXCL12 axis is implicated in STAD survival, growth, proliferation, and metastasis, and its inhibition is effective in suppressing STAD development and metastasis [Citation37]. CXCR4/CXCL12 is associated with a series of pathophysiological processes, including embryonic hematopoiesis, vasculogenesis, inflammation, and immune surveillance. In myeloid cells, CXCR4-mediated signals inhibit natural killer (NK) cell-mediated tumor immune surveillance and consequently promote tumor expansion [Citation38]. Recently, Liu et al. reported that DUXAP8, a long non-coding RNA (lncRNA), plays an active role in activating cell proliferation, migration, and invasion of papillary thyroid carcinoma through regulating CXCR4 mediated by miR-223-3p [Citation39]. In another study, Zou et al. found that CXCR4 was overexpressed in the TME of soft tissue sarcomas [Citation40]. Studies have shown that CXCR4 antagonism has antitumor effects in metastasized breast cancers in mice, when combined with inhibition of indoleamine 2,3-dioxygenase 1 (IDO1), by reducing intratumoral infiltration of regulatory T-cells (Tregs) and myeloid-derived suppressor cells (MDSCs) [Citation41], together with CXCR4 blockade by plerixafor (AMD3100), possibly resulting in tumor control and animal survival by increasing effector T-cell invasion and memory T-cell development within the TME of ovarian cancer [Citation42].
Our results showed that CXCR4 expression level increased with STAD progression, which is consistent with previous report that CXCR4/CXCL12 axis promotes STAD development through the process of angiogenesis and vascular endothelial growth factor (VEGF) expression by activating its downstream the Janus kinase 2/signal transducer and activator of transcription (JAK2/STAT) signaling pathway [Citation43]. Furthermore, CXCR4 inhibition disrupts homeostatic functions in various cancers, including gastric cancer [Citation44]. As CXCR4 is involved in tumor progression and metastasis by targeting multiple cells within the TME [Citation45], the correlation between CXCR4 expression and TME was additionally analyzed. As demonstrated by the GSEA results, there was high enrichment of immune-linked pathways in the CXCR4-upregulated cohort, including the NK cell-mediated cytotoxicity, T-cell receptor-, JAK-STAT-, and chemokine-signaling pathways. Thus, upregulation of CXCR4 along with the subsequent activation of the downstream pathways may have oncogenic roles in STAD, suggesting that CXCR4 blockade may be therapeutically beneficial in STADs.
The effects of triptolide are associated with the regulation of immune activity, including T-cell pro-apoptotic activity and Treg downregulation [Citation46]. Triptolide has also demonstrated antitumor activity in multiple solid cancers, such as non-small cell lung cancer, colon, prostate, and liver carcinomas [Citation47–50]. In this study, we found that the targets of triptolide were functionally associated with immune cell proliferation, chemokine receptor binding, and CCR chemokine receptor binding, and were significantly enriched in chemokine signaling, p53, NF-κB, and HIF−1 signaling pathways. The chemokine-signaling pathway has been identified as a key factor within the TME and directly or indirectly regulates tumor cell proliferation, cancer stem cell features, and tumor metastases [Citation51]. Cancer stem cell mutation in p53 leads to CXCR4 upregulation, as this gene can be downregulated through p53 signaling [Citation52]. NF-κB signaling pathway is positively associated with CXCR4 expression in cancer cells, and several compounds have been shown to downregulate CXCR4 activation by suppressing the NF-κB pathway [Citation53–55]. HIF-1 has been reported to be involved in upregulating CXCR4, thus enhancing its activity in STAD [Citation56].
Furthermore, we found that triptolide bound to CXCR4 with high affinity, as illustrated in the molecular docking results. Therefore, we speculate that CXCR4 is a novel drug target for triptolide against STAD in improving the prognosis of patients with STAD. Although triptolide is effective in inhibiting cancers, its toxicity must not be ignored. Novel delivery strategies and derivatives or analogs of triptolide have been developed to attenuate its systemic toxicities while improving efficacy [Citation57,Citation58].
Analysis using ESTIMATE led to the identification of TME-associated genes in the STAD TME. CXCR4 represents a prospective and novel prognostic biomarker and a potential therapeutic target of triptolide in STAD TME. Given the continuous renewal of databases, further investigations are necessary to further verify the effects of CXCR4 expression in STAD TME and to validate the efficacy and safety of triptolide in STAD TME through CXCR4-related signaling pathways.
Conclusion
In summary, using bioinformatics analyses, we identified triptolide as a potential candidate for STAD therapy by targeting CXCR4 based on DEGs identified through stromal-/immune-related facet comparative analyses within the tumor microenvironment of STAD patients. These findings may lead to the development of new drugs for STAD treatment.
Highlights
(1) Immune components of the STAD TME are closely associated with the progression of STAD
(2) Triptolide targets are enriched in chemokine- and cancer-related signaling pathways
(3) CXCR4 represents a potential target of triptolide in regulating the TME of STAD
Author contributions
Data curation: Hairong Qiu, Rui Gao, Jianglong Shi; Formal data analysis: Hairong Qiu, Han Yu, Xiaobo Zhang; Methodology: Hairong Qiu, Han Yu, Xiaobo Zhang; Writing-original draft: Hairong Qiu; Writing-review and editing: Hairong Qiu, Xiaobo Zhang, Han Yu; Conceptualization: Hairong Qiu and Tao Shen; Supervision: Tao Shen; Project administration: Tao Shen.
Acknowledgements
We thank TCGA, TCMSP, PubChem, and RCSB PDB databases for providing publicly accessible datasets. We are also thankful for the all the public domain softwares and analytical tools used in this study, including but not limited to R software supported by R Core Team, PyMol, and AutoDock Tools.
Data availability statement
The datasets analyzed in the present study are publicly available at https://portal.gdc.cancer.gov/ and https://tcmspw.com/tcmsp.php. Structures of triptolide and CXCR4 were available online at https://pubchem.ncbi.nlm.nih.gov/. and http://www1.rcsb.org/, respectively.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Bray F, Jemal A, Grey N, et al. Global cancer transitions according to the Human Development Index (2008-2030): a population-based study. Lancet Oncol. 2012;13(8):790–801.
- Chen W, Zheng R, Baade PD, et al. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132.
- Van Cutsem E, Sagaert X, Topal B, et al. Gastric cancer. Lancet. 2016;388(10060):2654–2664.
- Karimi P, Islami F, Anandasabapathy S, et al. Gastric cancer: descriptive epidemiology, risk factors, screening, and prevention. Cancer Epidemiol Biomarkers Prev. 2014;23(5):700–713.
- Kamran SC, Hong TS, Wo JY. Advances in the Management of Gastric and Gastroesophageal Cancers. Curr Oncol Rep. 2016;18(2):13.
- Chivu-Economescu M, Matei L, Necula LG, et al. New therapeutic options opened by the molecular classification of gastric cancer. World J Gastroenterol. 2018;24(18):1942–1961.
- Lv C, Shi C, Li L, et al. Chinese herbal medicines in the prevention and treatment of chemotherapy-induced nausea and vomiting. Curr Opin Support Palliat Care. 2018;12(2):174–180.
- Tao WW, Jiang H, Tao XM, et al. Effects of Acupuncture, Tuina, Tai Chi, Qigong, and Traditional Chinese Medicine Five-Element Music Therapy on Symptom Management and Quality of Life for Cancer Patients: a Meta-Analysis. J Pain Symptom Manage. 2016;51(4):728–747.
- Liao YH, Li CI, Lin CC, et al. Traditional Chinese medicine as adjunctive therapy improves the long-term survival of lung cancer patients. J Cancer Res Clin Oncol. 2017;143(12):2425–2435.
- Yuan K, Li X, Lu Q, et al. Application and Mechanisms of Triptolide in the Treatment of Inflammatory Diseases-A Review. Front Pharmacol. 2019;10:1469.
- Qiu D, Kao PN. Immunosuppressive and anti-inflammatory mechanisms of triptolide, the principal active diterpenoid from the Chinese medicinal herb Tripterygium wilfordii Hook. f. Drugs R D. 2003;4(1):1–18.
- Hu H, Zhu S, Tong Y, et al. Antitumor activity of triptolide in SKOV3 cells and SKOV3/DDP in vivo and in vitro. Anticancer Drugs. 2020;31(5):483–491.
- Hinshaw DC, Shevde LA. The Tumor Microenvironment Innately Modulates Cancer Progression. Cancer Res. 2019;79(18):4557–4566.
- Noel P, Von Hoff DD, Saluja AK, et al. Its Derivatives as Cancer Therapies. Trends Pharmacol Sci. 2019;40(5):327–341.
- Hou ZY, Tong XP, Peng YB, et al. Broad targeting of triptolide to resistance and sensitization for cancer therapy. Biomed Pharmacother. 2018;104:771–780.
- Yoshihara K, Shahmoradgoli M, Martinez E, et al. Inferring tumour purity and stromal and immune cell admixture from expression data. Nat Commun. 2013;4(1):2612.
- Wang T, Chen B, Meng T, et al. Identification and immunoprofiling of key prognostic genes in the tumor microenvironment of hepatocellular carcinoma. Bioengineered. 2021;12(1):1555–1575.
- Islam T, Rahman MR, Aydin B, et al. Integrative transcriptomics analysis of lung epithelial cells and identification of repurposable drug candidates for COVID-19. Eur J Pharmacol. 2020;887:173594.
- Yan H, Zheng G, Qu J, et al. Identification of key candidate genes and pathways in multiple myeloma by integrated bioinformatics analysis. J Cell Physiol. 2019;234(12):23785–23797.
- Zhang YF, Huang Y, Ni YH, et al. Systematic elucidation of the mechanism of geraniol via network pharmacology. Drug Des Devel Ther. 2019;13:1069–1075.
- von Mering C, Huynen M, Jaeggi D, et al. STRING: a database of predicted functional associations between proteins. Nucleic Acids Res. 2003;31(1):258–261.
- Shannon P, Markiel A, Ozier O, et al. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13(11):2498–2504.
- Ru J, Li P, Wang J, et al. TCMSP: a database of systems pharmacology for drug discovery from herbal medicines. J Cheminform. 2014;6(1):13.
- Bi KW, Wei XG, Qin XX, et al. BTK Has Potential to Be a Prognostic Factor for Lung Adenocarcinoma and an Indicator for Tumor Microenvironment Remodeling: a Study Based on TCGA Data Mining. Front Oncol. 2020;10:424.
- Guo R, Li X, Chen X, et al. An ultrasonic-extracted arabinoglucan from Tamarindus indica L. pulp: a study on molecular and structural characterizations. Int J Biol Macromol. 2020;164:3687–3697.
- Wu B, Chien EY, Mol CD, et al. Structures of the CXCR4 chemokine GPCR with small-molecule and cyclic peptide antagonists. Science. 2010;330(6007):1066–1071.
- Zhao Q, Cao L, Guan L, et al. Immunotherapy for gastric cancer: dilemmas and prospect. Brief Funct Genomics. 2019;18(2):107–112.
- Schlosser HA, Drebber U, Kloth M, et al. Immune checkpoints programmed death 1 ligand 1 and cytotoxic T lymphocyte associated molecule 4 in gastric adenocarcinoma. Oncoimmunology. 2016;5(5):e1100789.
- Li Z, Lai Y, Sun L, et al. PD-L1 expression is associated with massive lymphocyte infiltration and histology in gastric cancer. Hum Pathol. 2016;55:182–189.
- Wang H, Wu X, Chen Y. Stromal-Immune Score-Based Gene Signature: a Prognosis Stratification Tool in Gastric Cancer. Front Oncol. 2019;9:1212.
- Jia D, Li S, Li D, et al. TCGA database for genes of prognostic value in glioblastoma microenvironment. Aging (Albany NY). 2018;10(4):592–605.
- Mao M, Yu Q, Huang R, et al. Stromal score as a prognostic factor in primary gastric cancer and close association with tumor immune microenvironment. Cancer Med. 2020;9(14):4980–4990.
- Lippitz BE. Cytokine patterns in patients with cancer: a systematic review. Lancet Oncol. 2013;14(6):e218–228.
- Saxena R, Wang Y, Mier JW. CXCR4 inhibition modulates the tumor microenvironment and retards the growth of B16-OVA melanoma and Renca tumors. Melanoma Res. 2020;30(1):14–25.
- Izumi D, Ishimoto T, Miyake K, et al. CXCL12/CXCR4 activation by cancer-associated fibroblasts promotes integrin beta1 clustering and invasiveness in gastric cancer. Int J Cancer. 2016;138(5):1207–1219.
- Perrot-Applanat M, Vacher S, Pimpie C, et al. Differential gene expression in growth factors, epithelial mesenchymal transition and chemotaxis in the diffuse type compared with the intestinal type of gastric cancer. Oncol Lett. 2019;18(1):674–686.
- Yang J, Kumar A, Vilgelm AE, et al. Loss of CXCR4 in Myeloid Cells Enhances Antitumor Immunity and Reduces Melanoma Growth through NK Cell and FASL Mechanisms. Cancer Immunol Res. 2018;6(10):1186–1198.
- Liu Y, Zhang H, Wang H, et al. Long non-coding RNA DUXAP8 promotes the cell proliferation, migration, and invasion of papillary thyroid carcinoma via miR-223-3p mediated regulation of CXCR4. Bioengineered. 2021;12(1):496–506.
- Zou D, Wang Y, Wang M, et al. Bioinformatics analysis reveals the competing endogenous RNA (ceRNA) coexpression network in the tumor microenvironment and prognostic biomarkers in soft tissue sarcomas. Bioengineered. 2021;12(1):496–506.
- Zhang J, Pang Y, Xie T, et al. CXCR4 antagonism in combination with IDO1 inhibition weakens immune suppression and inhibits tumor growth in mouse breast cancer bone metastases. Onco Targets Ther. 2019;12:4985–4992.
- Zeng Y, Li B, Liang Y, et al. Dual blockade of CXCL12-CXCR4 and PD-1-PD-L1 pathways prolongs survival of ovarian tumor-bearing mice by prevention of immunosuppression in the tumor microenvironment. FASEB J. 2019;33(5):6596–6608.
- Masuda T, Nakashima Y, Ando K, et al. Nuclear expression of chemokine receptor CXCR4 indicates poorer prognosis in gastric cancer. Anticancer Res. 2014;34(11):6397–6403.
- Xiang Z, Zhou ZJ, Xia GK, et al. A positive crosstalk between CXCR4 and CXCR2 promotes gastric cancer metastasis. Oncogene. 2017;36(36):5122–5133.
- Mortezaee K. CXCL12/CXCR4 axis in the microenvironment of solid tumors: a critical mediator of metastasis. Life Sci. 2020;249:117534.
- Liu B, Zhang H, Li J, et al. Triptolide downregulates Treg cells and the level of IL-10, TGF-beta, and VEGF in melanoma-bearing mice. Planta Med. 2013;79(15):1401–1407.
- Wei J, Yan Y, Chen X, et al. The Roles of Plant-Derived Triptolide on Non-Small Cell Lung Cancer. Oncol Res. 2019;27(7):849–858.
- Isharwal S, Modi S, Arora N, et al. Minnelide Inhibits Androgen Dependent, Castration Resistant Prostate Cancer Growth by Decreasing Expression of Androgen Receptor Full Length and Splice Variants. Prostate. 2017;77(6):584–596.
- Oliveira A, Beyer G, Chugh R, et al. Triptolide abrogates growth of colon cancer and induces cell cycle arrest by inhibiting transcriptional activation of E2F. Lab Invest. 2015;95(6):648–659.
- Wang H, Ma D, Wang C, et al. Triptolide Inhibits Invasion and Tumorigenesis of Hepatocellular Carcinoma MHCC-97H Cells Through NF-kappaB Signaling. Med Sci Monit. 2016;1:1827–1836.
- Nagarsheth N, Wicha MS, Zou W. Chemokines in the cancer microenvironment and their relevance in cancer immunotherapy. Nat Rev Immunol. 2017;17(9):559–572.
- Katoh M, Katoh M. Integrative genomic analyses of CXCR4: transcriptional regulation of CXCR4 based on TGFbeta, Nodal, Activin signaling and POU5F1, FOXA2, FOXC2, FOXH1, SOX17, and GFI1 transcription factors. Int J Oncol. 2010;36(2):415–420.
- Wang J, Wang H, Cai J, et al. Artemin regulates CXCR4 expression to induce migration and invasion in pancreatic cancer cells through activation of NF-kappaB signaling. Exp Cell Res. 2018;365(1):12–23.
- Ma YK, Chen YB, Li P. Quercetin inhibits NTHi-triggered CXCR4 activation through suppressing IKKalpha/NF-kappaB and MAPK signaling pathways in otitis media. Int J Mol Med. 2018;42(1):248–258.
- Guo Z, Chen W, Dai G, et al. Cordycepin suppresses the migration and invasion of human liver cancer cells by downregulating the expression of CXCR4. Int J Mol Med. 2020;45(1):141–150.
- Oh YS, Kim HY, Song IC, et al. Hypoxia induces CXCR4 expression and biological activity in gastric cancer cells through activation of hypoxia-inducible factor-1alpha. Oncol Rep. 2012;28(6):2239–2246.
- Liaw K, Sharma R, Sharma A, et al. Appiani La Rosa S, Kannan RM. Systemic dendrimer delivery of triptolide to tumor-associated macrophages improves anti-tumor efficacy and reduces systemic toxicity in glioblastoma. J Control Release. 2021;329:434–444.
- Chugh R, Sangwan V, Patil SP, et al. A preclinical evaluation of Minnelide as a therapeutic agent against pancreatic cancer. Sci Transl Med. 2012;4(156):156ra139–156ra139. 156ra139.