ABSTRACT
Oral squamous cell carcinoma (OSCC) is one of the most common digestive tumors, which has high mortality rate. Long non-coding RNAs (lncRNA) and MicroRNAs (miRNAs) are associated with the cell cycle and differentiation during the occurrence and development of malignant tumors. This research aimed to investigate the effects of lncRNA SNHG20 on the progress of oral squamous cell carcinoma (OSCC) cells. Ninety pairs of tumor tissues and paracancerous tissues were collected from patients with OSCC and the CAL27 and SCC25 OSCC cells were selected for the following experiments. RT-qPCR was used for detecting the expression of SNHG20, miR-19b-3p, and RAB14. Western blotting was used to detect the protein levels of RAB14. MTT assay was employed to assess cell proliferation. Transwell assay was used to determine the cell migration and invasion abilities. Furthermore, luciferase reporter and RNA pull-down assays were used to verify the binding of SNHG20/RAB14 to miR-19b-3p. Then, the function of the SNHG20/miR-19b-3p/RAB14 axis in OSCC was explored. The results indicated that lncRNA SNHG20 was upregulated in the tissues. Furthermore, bioinformatic analysis showed that both SNHG20 and RAB14 could bind to miR-19b-3p. RAB14 was upregulated, and miR-19b-3p was downregulated in the tissues. The knockdown of SNHG20 inhibited cell proliferation, migration, and invasion. Contrarily, the knockdown of miR-19b-3p reversed the effects of si-SNHG20 on cell proliferation, migration, and invasion, and the overexpression of RAB14 reversed the effects of miR-19b-3p mimic on the cell biological functions. LncRNA SNHG20 affects cell proliferation, migration, and invasion via the miR-19b-3p/RAB14 axis in OSCC.
Introduction
Oral cancer is one of the most common digestive tumors and is ranked as the thirteenth most common cancer in the world [Citation1]. It was reported that the exposure to tobacco smoke, chemicals, radiation, and air pollution could induce the occurrence of oral cancer [Citation2,Citation3]. Oral squamous cell carcinoma (OSCC) makes up the majority of the oral cancer cases [Citation4,Citation5]. The generally used OSCC treatment approaches are chemotherapy and targeted therapy [Citation6]. However, due to late diagnosis and drug resistance, the 5-year survival rate of patients remains below 17% [Citation7]. Therefore, it is critical to develop novel anti-OSCC therapies to improve the life quality and survival rate of patients.
Long noncoding RNAs (lncRNAs) are significant regulators in different cellular processes [Citation8]. Furthermore, most lncRNAs are transcribed by RNA polymerase II; therefore, they are similar to messenger RNAs (mRNAs), involving a 5'-7-methylguanosine cap and a 3' poly(A) tail. However, lncRNAs lack a coding capacity [Citation9]. Meanwhile, lncRNAs are known as gene regulators that participate in different molecular events, such as chromatin modifications, chromatin structure, DNA methylation, genome organization, and interactions with transcription factors, at various levels, to influence the transcription of chromatin-related proteins and gene expression [Citation10]. LncRNA small nucleolar RNA host gene 20 (GenBank Accession ID NR_027058.1), which belongs to the SNHG lncRNA family, was first reported in hepatocellular carcinoma (HCC) [Citation11]. A previous study showed that the upregulation of lncRNA SNHG20 in OSCC could promote cell proliferation and migration [Citation12]. However, the specific mechanism of lncRNA SNHG20 in OSCC remains unclear.
Rab protein, as a small GTP binding protein, circulates between the active GTP binding state and the inactive GDP binding state [Citation13]. Rab protein is a molecular switch for intracellular and extracellular transport in all eukaryotes . RAB14 is a member of Rab GTPase family. RAB14 is involved in autophagy, lysosome activity, cell migration and membrane transport between Golgi and endosomes under normal physiological conditions [Citation14]. In various tumor cells, RAB14 affects the development of tumor by regulating the proliferation, migration and invasion of tumor cells. Previous study confirmed that the mRNA expressions levels of RAB14 in gastric cancer was significantly higher than that in healthy gastric mucosa and the abnormal expression of RAB14 in gastric cancer promotes the malignant development of tumor by activating Akt signaling pathway [Citation15]. In kidney cancer cells, Rab14 expression is significantly up-regulated, which promotes the proliferation, invasion, and metastasis of tumor cells and enhances the chemotherapy resistance of tumor cells [Citation16]. In addition, the increased expression of RAB14 in ovarian cancer promotes tumor cell proliferation and invasion through Wnt signaling pathway, which is associated with poor prognosis [Citation17]. However, the reports about RAB14 on the progress of OSSC is still limited.
Therefore, this research aimed to elucidate the specific mechanism of lncRNA SNHG20 during the progress of OSCC. We hypothesized that LncRNA SNHG20 affects cell proliferation, migration, and invasion via the miR-19b-3p/RAB14 axis.
Materials and methods
Patients and specimens
A total number of 90 patients with OSCC were recruited from the People Hospital of Lishui between October 2017 and October 2019. In brief, the para-carcinoma tissues that did not contain obvious cancer cells were collected 5–10 cm away from the border of the tumor. Subsequently, the tumor and the para-carcinoma tissues were fixed in 10% formalin. Additionally, an informed consent was signed by each patient. This study has been approved by the Ethics Committee of the People Hospital of Lishui.
Cell lines
The NHOK, TSCCA, SCC15, SCC25, and CAL27 cell lines were collected from the Cell Bank of Chinese Academy of Sciences. All cell lines were kept in RPMI-1640 medium (Thermo Fisher Scientific, MA USA) containing 10% FBS (Biosera, MO, USA) and 100 U/mL streptomycin/penicillin (Thermo Fisher Scientific, MA USA) in a humid atmosphere containing 5% CO2 at 37°C.
Cell culture and transfection
SiRNA targeting SNHG20 (si-SNHG20), miR-19b-3p inhibitor, miR-19b-3p mimic, overexpressed RAB14 (OE-RAB14), si-NC, inhibitor NC, mimic NC, and OE-NC were obtained from GenePharma (China, Soochow). Subsequently, the cells were inoculated on a 6-well plate and cultured in RPMI-1640 containing 5% CO2 at 37°C until the cells achieved 60% confluence. In brief, the plasmids were transfected into cells (1 × 105 cells/well) using LipofectamineTM 2000 Transfection Reagent (Thermo Fisher Scientific, MA USA) based on the manufacturer’s instructions.
MTT assay
Cells in the logarithmic growth phase were seeded on a 96-well plate. After incubation for 0 h, 24 h, 48 h, and 72 h, cells were treated with MTT in fresh medium for 4 h. Dimethyl sulfoxide was added to dissolve the formazan product. Furthermore, the optical absorbance was measured at 490 nm using a microplate reader.
Luciferase reporter assay
The luciferase reporter assay was performed according to previous study [Citation18]. The gene fragments containing the SNHG20/RAB14 binding site (HOTAIR-wild type (WT)) and the mutated SNHG20/RAB14 binding site (HOTAIR-MUT) were synthesized and cloned into the fluorescent vector psiCHECK2 to construct the luciferase reporter gene. Then, cells were co-transfected with mimic NC and miR-19b-3p mimic, respectively. According to the dual luciferase reporter gene test kit requirements, the luciferase activity of renilla and firefly was detected.
RNA pull-down assay
The RNA pull-down assay was performed as described by Tang et al. [Citation19]. CAL27 and SCC25 cells were lysed and incubated with biotinylated miR-19b-3p or biotinylated NC. The final concentration of each biotinylated miRNA was 20 nM. Two days later, the cell lysates were collected and then cultured with M-280 Streptavidin magnetic beads for 3 h. Then, they were washed three times. In the qRT-PCR analysis, TRIzol was employed to purify the bound RNA.
RNA isolation and real-time quantitative PCR
We used TRIzol to collect the total RNA from tissues and cells based on the manufacturer’s instructions. The RNA concentration was analyzed using NanoDrop 2000 (Thermo, America). The cDNAs were synthesized by using the PrimeScript RT reagent kit transcriptase, Random 6mers, RNase inhibitor, Oligo dT primer, a dNTP mixture, and reaction buffer. Quantitative real-time PCR (qRT-PCR) was performed using an SYBR Premix EX Taq™ kit (TAKARA, Tokyo, Japan). The cycle conditions used were: 95°C for 30 s, followed by 95°C for 5 s and 40 cycles at 60°C for 30 s each. We used the 2−ΔΔCt method to analyze the data [Citation20]. GAPDH or U6 was selected as the internal reference gene.
The primers used in qRT-PCR were the following:
LncRNA SNHG20: forward, 5'-ATGGCTATAAATAGATACACGC-3'; reverse, 5'-GGTACAAACAGGGAGGGA-3'; miR-19b-3p: forward, 5'-ACACTCCAGCTGGGTGTGCAAATCCATGCAA-3',
reverse: 5'-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGTCAGTTT-3';
RAB14: forward, 5'-TATGGCTGATTGTCCTCACACA-3';
reverse: 5'-CTGTCCTGCCGTATCCCAAAT-3';
GAPDH forward: 5'-TGTGGGCATCAATGGATTTGG-3';
reverse: 5'-ACACCATGTATTCCGGGTCAAT-3'; and
U6: forward, 5'-AAAGCAAATCATCGGACGACC-3';
reverse: 5'-GTACAACACATTGTTTCCTCGGA-3'.
Transwell assays
The Transwell method was employed to determine the growth and metastasis abilities of the OSCC cells described as a previous study [Citation21]. At first, cell culture Transwell inserts (8 mm pore size; Falcon; BD Biosciences) have been placed into 48-well plates to generate upper and lower chambers. Furthermore, cells were seeded on the upper chamber with a Matrigel-coated membrane, for the migration assays, or in chambers not coated with Matrigel, for the invasion assays, while the lower wells were filled with culture media containing serum. After 24 h of culture, we used a counting chamber to count the number of migrating and invading cells.
Western blot
The western blot was conducted according to a previous research [Citation22]. Protein was collected from SCC25 and CAL27 cells using RIPA buffer (Sigma-Aldrich, MO, USA) and the concentration of total protein was determined using a BCA kit (Sigma-Aldrich, MO, USA). Additionally, the proteins (20 µg/lane) were isolated using a 15% SDS-PAGE gel, and then transferred to PVDF membranes (Bio-rad, California, USA), which were blocked with 5% skimmed milk for 2 h before the blotting procedure. Cells were incubated with the primary anti-RAB14 antibody overnight at 4°C. On the next day, the membranes were treated with the secondary antibodies. Subsequently, the membrane was stained using an ECL western blotting kit. At the same time, we used GAPDH as a loading control. The results were analyzed using Image J.
Statistical methods
Data were examined using SPSS 21.0. All experiments were repeated three times and all data were presented as the mean ± standard deviation (X ± s). Furthermore, comparisons between two groups were analyzed using a t-test. Multiple-group comparisons were investigated using one-way ANOVA. A P < 0.05 was defined as a meaningful statistical difference.
Results
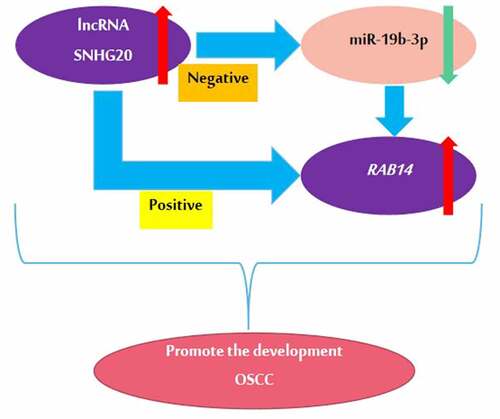
The aim of this study was to explore the specific mechanism of lncRNA SNHG20 during the progress of OSCC. Through bioinformatic analysis, we found that LncRNA SNHG20 regulated OSCC cell functions by sponging miR-19b-3p and RAB14 is the target gene of miR-19b-3p in OSCC via. We hypothesized that LncRNA SNHG20 affects cell proliferation, migration, and invasion via the miR-19b-3p/RAB14 axis
LncRNA SNHG20 was upregulated in OSCC tissues and SCC25 and CAL27 cells
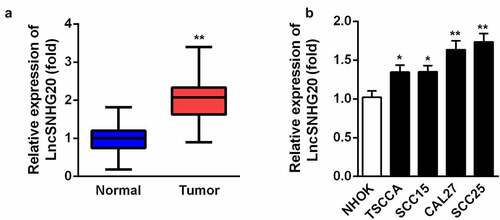
Firstly, we determined the expression of lncRNA SNHG20 in the OSCC samples by using RT-qPCR. Our data suggested that, compared with the adjacent non-tumor tissues, SNHG20 was dramatically upregulated in the OSCC tissues (). Then, we examined the level of lncRNA SNHG20 in OSCC cell lines (TSCCA, SCC15, SCC25, and CAL27) and used NHOK cells as control cells. We found that the expression of lncRNA SNHG20 was increased in TSCCA, SCC15, SCC25, and CAL27 cells; moreover, it was the highest in SCC25 and CAL27 cells (). Therefore, we used SCC25 and CAL27 cells in the following experiments.
The knockdown of lncRNA SNHG20 inhibited the proliferation, migration, and invasion of SCC25 and CAL27 cells
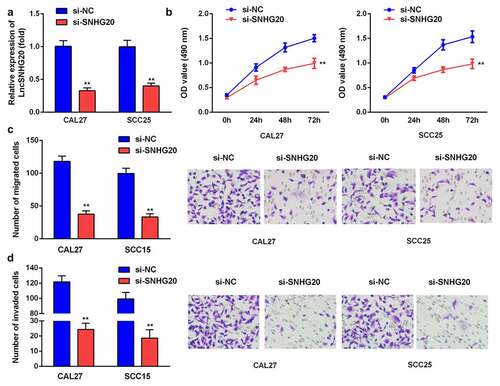
To explore the impact of lncRNA SNHG20 on SCC25 and CAL27 cells, we knocked down the lncRNA SNHG20 using siRNA. Compared with the si-NC group, the expression of SNHG20 was remarkably downregulated in the si-SNHG20 group (). In addition, compared with the si-NC group, the proliferation of SCC25 and CAL27 cells in the si-SNHG20 group was significantly decreased (). Similarly, the knockdown of lncRNA SNHG20 dramatically downregulated the growth and metastasis of SCC25 and CAL27 cells ().
Figure 2. The knockdown of lncRNA SNHG20 decreased the proliferation, migration, and invasion of SCC25 and CAL27 cells
LncRNA SNHG20 regulated OSCC cell functions by sponging miR-19b-3p
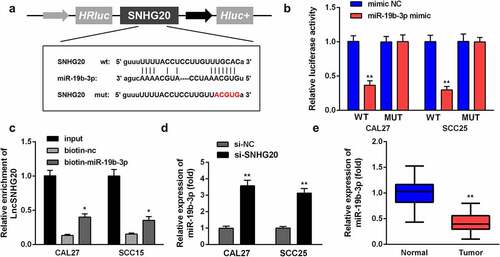
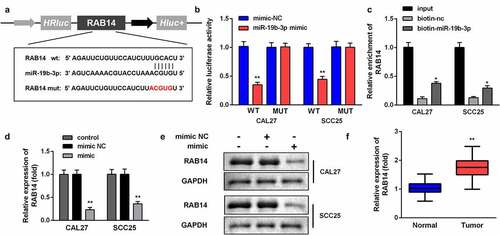
We predicted the targets of lncRNA CRNDE and searched miR-19b-3p for further analysis. The targeting sites of LncRNA SNHG20 and miR-19b-3p were estimated by using the online starBase software (). The luciferase reporter data showed that miR-19b-3p mimic sharply inhibited the luciferase activity in cells transfected with WT LncRNA SNHG20, rather than with Mut-LncRNA SNHG20, in SCC25 and CAL27 cells (). In addition, the luciferase reporter results were confirmed by using an lncRNA SNHG20 probe pull-down assay (). Moreover, we found that the knockdown of lncRNA SNHG20 can promote miR-19b-3p expression (). The RT-qPCR results showed that miR-19b-3p was dramatically downregulated in OSCC tissues, compared to the adjacent non-tumor tissues ().
Figure 3. LncRNA SNHG20 regulates the OSCC cell functions by sponging miR-19b-3p
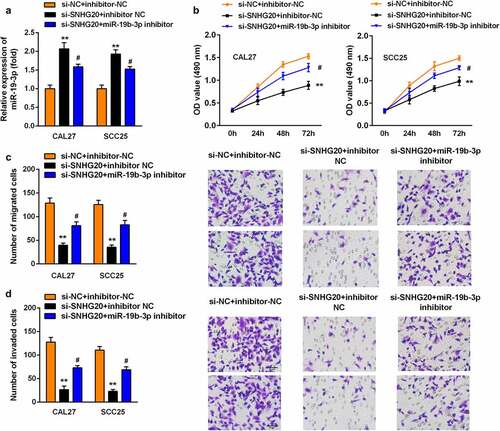
Knockdown of miR-19b-3p reversed the effects of si-SNHG20 on the proliferation, migration, and invasion of SCC25 and CAL27 cells
Subsequently, si-SNHG20 and the miR-19b-3p inhibitor have been co-transfected into SCC25 and CAL27 cells. The expression of miR-19b-3p was sharply downregulated after miR-19b-3p inhibitor transfection (). Furthermore, the knockdown of miR-19b-3p promoted the proliferation of SCC25 and CAL27 cells (). Similarly, the knockdown of miR-19b-3p upregulated the growth and metastasis of SCC25 and CAL27 cells ().
Figure 4. The knockdown of miR-19b-3p reversed the effect of si-SNHG20 on the proliferation, migration, and invasion of SCC25 and CAL27 cells
RAB14 is the target gene of miR-19b-3p in OSCC
Furthermore, the targeting sites of miR-19b-3p and RAB14 were estimated by using the online TargetScan software () and validated using a dual luciferase reporter gene system and a pull-down assay. The data showed that miR-19b-3p mimic sharply downregulated the luciferase activity in cells transfected with WT-RAB14, in place of Mut-RAB14, in SCC25 and CAL27 cells (). In addition, the luciferase reporter assay results were validated by the RAB14 probe pull-down assay (), indicating that RAB14 is a direct target of miR-19b-3p. Moreover, compared with the adjacent non-tumor tissues, RAB14 expression was dramatically increased in OSCC tissues and the overexpression of miR-19b-3p promoted the mRNA and protein expression of RAB14 ().
Figure 5. RAB14 is the target gene of miR-19b-3p in OSCC
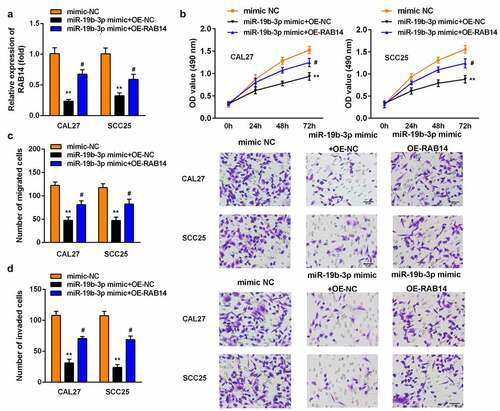
Overexpression of RAB14 reversed the effect of miR-19b-3p mimic on the proliferation, migration, and invasion of SCC25 and CAL27 cells
Last, the miR-19b-3p mimic and OE-RAB14 were transfected into SCC25 and CAL27 cells. The level of RAB14 was significantly lower after OE-RAB14 transfection (). The overexpression of RAB14 promoted the proliferation of SCC25 and CAL27 cells (). Similarly, the Transwell assay results suggested that the overexpression of RAB14 promoted the growth and metastasis of SCC25 and CAL27 cells ().
Figure 6. Overexpression of RAB14 reversed the effect of miR-19b-3p mimic on the proliferation, migration, and invasion of SCC25 and CAL27 cells
Discussion
In this study, we found that the expression of lncRNA SNHG20 was increased in OSCC tissues and that the knockdown of lncRNA SNHG20 could effectively suppress the biological activity of SCC25 and CAL27 cells. Our study indicated that lncRNA SNHG20 suppresses the biological activities of OSCC by regulating the miR-19b-3p/RAB14 axis.
Previous studies have shown that lncRNA can not only change the chromosome concept through the interaction of chromosome-modifying enzymes, but also silence the expression of a variety of miRNAs [Citation23]. Therefore, lncRNA can also be used as an intervention target in tumor therapy. LncRNA SNHG20 was first isolated from the cDNA subtractive library of tumor suppressor gene by Zhang et al. and it was located on chromosome [Citation24]. There is growing evidence indicating that lncRNA SNHG20 is closely involved in the development of many cancer processes. However, the research on lncRNA SNHG20 in OSCC is still limited. In this study, we found that, compared with the adjacent non-tumor tissues, the level of lncRNA SNHG20 was increased in OSCC tissues. In addition, we found that the knockdown of lncRNA SNHG20 can sharply inhibit the growth and metastasis of SCC25 and CAL27 cells, which indicates that lncRNA SNHG20 plays a role in promoting cancer during the progression of OSCC. Li et al. [Citation25] also found that lncRNA SNHG20 can promote the growth of laryngeal squamous cell carcinoma cells, similar to our results.
Additionally, lncRNA can affect tumor progression by regulating miRNA expression [Citation26,Citation27]. In order to further analyze the functional mechanism of lncRNA SNHG20 in OSCC, here, using bioinformatics, we predicted that lncRNA SNHG20 could target and negatively regulate miR-19b-3p. miRNAs belong to a group of small noncoding RNAs that regulate gene expression at the post-transcriptional level. Previous evidence has confirmed that lncRNA SNHG20 increases the proliferation of laryngeal squamous cell carcinoma cells by regulating miR-140 [Citation25]; Wu et al. [Citation28] also found that lncRNA SNHG20 increases the proliferation and invasion of prostate cancer cells by targeting miR-6516-5p. In this study, we predicted that lncRNA SNHG20 has a binding site where it binds to miR-19b-3p, and the luciferase reporter pull-down assay confirmed that lncRNA SNHG20 can target and regulate miR-19b-3p. Then, we detected the level of miR-19b-3p in OSCC tissues and found that miR-19b-3p was downregulated. miR-19b-3p has been explored in various cancers, such as breast, pancreatic, and colon cancer [Citation29–31]. MicroRNA profiling in patients with gastric cancer showed that the level of miR-19b-3p was remarkably decreased, similar to our results [Citation32]. Furthermore, the knockdown of lncRNA SNHG20 can downregulate the level of miR-19b-3p in CAL27 and SCC25 cells. In the rescue experiment, the knockdown of miR-19b-3p reversed the effect of si-SNHG20 on the growth and metastasis of SCC25 and CAL27 cells. These results demonstrated the possible mechanism by which lncRNA SNHG20 regulates the biological behavior of OSCC cells by affecting the level of miR-19b-3p.
As reported in gastric cancer, miR-19b-3p was downregulated and negatively regulated neuropilin-1 [Citation33]. In this study, through bioinformatics prediction and luciferase reporter and pull-down assay, RAB14 was found to be a potential target gene of miR-19b-3p. At the same time, the level of RAB14 was increased in OSCC tissues. Rab family proteins attach an essential part in carcinogenesis and promote the malignant development of tumors by influencing cell proliferation, differentiation, apoptosis, and various cell signaling pathways [Citation15,Citation16]. According to previous research, the mRNA expression of RAB14 in gastric cancer is dramatically higher than that in healthy gastric mucosa; the abnormal expression of RAB14 in gastric tumors promotes the malignant development of tumors by activating the Akt signaling pathway [Citation15]. In kidney cancer cells, RAB14, which is a target gene of miR-148a, promotes the growth and metastasis of tumor cells, and enhances the chemotherapy resistance of tumor cells [Citation16]. The increased expression of RAB14 in ovarian cancer promotes tumor cell proliferation and invasion through the Wnt signaling pathway [Citation17]. In this study, the overexpression of miR-19b-3p dramatically decreased the mRNA and protein expression of RAB14. However, the overexpression of RAB14 reversed the effect of miR-19b-3p mimic on the proliferation, migration, and invasion of SCC25 and CAL27 cells, further demonstrating that RAB14 is a vital target gene for lncRNA SNHG20 and miR-19b-3p.
Conclusion
In brief, our data showed that lncRNA SNHG20 is significantly upregulated in OSCC and promotes the growth and metastasis of SCC25 and CAL27 cells by absorbing miR-19b-3p to upregulate the RAB14 level. This study provided a new idea and theoretical basis for the diagnosis and treatment of OSCC.
Article highlights
SNHG20 was up-regulated in OSCC
SNHG20 regulated OSCC cell functions by sponging miR-19b-3p
RAB14 is the target gene of miR-19b-3p in OSCC
Ethical certification
This study has been approved by the Ethics Committee of the People Hospital of Lishui.
Data availability statement
The datasets used and/or analyzed during the current study are available from the corresponding author upon reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Morgensztern D, Campo MJ, Dahlberg SE, et al. Molecularly targeted therapies in non–small-cell lung cancer annual update 2014. J Thorac Oncol. 2015;10:S1–S63.
- Alberg AJ, Brock MV, Ford JG, et al. Epidemiology of lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143:e1S–e29S.
- Shtivelman E, Hensing T, Simon GR, et al. Molecular pathways and therapeutic targets in lung cancer. Oncotarget. 2014;5(6):1392–1433.
- Wu X, Wu G, Yao X, et al. The clinicopathological significance and ethnic difference of FHIT hypermethylation in non-small-cell lung carcinoma: a meta-analysis and literature review. Drug Des Devel Ther. 2016;10:699–709.
- Lemjabbar-Alaoui H, Hassan OU, Yang Y, et al. Lung cancer: biology and treatment options, Biochimica et biophysica acta. Rev Cancer. 2015;1856:189–210.
- Socinski MA, Evans T, Gettinger S, et al. Treatment of stage IV non-small cell lung cancer: diagnosis and management of lung cancer, 3rd ed: American College of chest physicians evidence-based clinical practice guidelines. Chest. 2013;143(5):e341S–e368S.
- Farhat FS, Houhou W. Targeted therapies in non-small cell lung carcinoma: what have we achieved so far? Ther Adv Med Oncol. 2013;5(4):249–270.
- Sun M, Kraus WL. From discovery to function: the expanding roles of long noncoding RNAs in physiology and disease. Endocr Rev. 2015;36:25–64.
- Marchese FP, Raimondi I, Huarte M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017;18(1):206.
- Schmitz SU, Grote P, Herrmann BG. Mechanisms of long noncoding RNA function in development and disease. Cell Mol Life Sci. 2016;73(13):2491–2509.
- Li C, Zhou L, He J, et al. Increased long noncoding RNA SNHG20 predicts poor prognosis in colorectal cancer. BMC Cancer. 2016;16(1):655.
- Wu J, Zhao W, Wang Z, et al. Long non‐coding RNA SNHG20 promotes the tumorigenesis of oral squamous cell carcinoma via targeting miR‐197/LIN28 axis. J Cell Mol Med. 2019;23(1):680–688.
- Seixas E, Ramalho JS, Mota LJ, et al. Bacteria and protozoa differentially modulate the expression of Rab proteins. PLoS One. 2012;7(7):e39858.
- Lu R, Johnson DL, Stewart L, et al. Rab14 regulation of claudin-2 trafficking modulates epithelial permeability and lumen morphogenesis. Mol Biol Cell. 2014;25(11):1744–1754.
- Guo B, Wang W, Zhao Z, et al. RAB14 act as oncogene and induce proliferation of gastric cancer cells via AKT signaling pathway. PLoS One. 2017;12:e170620.
- Kim EA, Kim TG, Sung EG, et al. miR-148a increases the sensitivity to cisplatin by targeting RAB14 in renal cancer cells. Int J Oncol. 2017;50(3):984–992.
- Hou R, Jiang L, Yang Z, et al. Rab14 is overexpressed in ovarian cancers and promotes ovarian cancer proliferation through Wnt pathway. Tumor Biol. 2016;37(12):16005–16013.
- Shang J, Sun S, Zhang L, et al. miR-211 alleviates ischaemia/reperfusion-induced kidney injury by targeting TGFβR2/TGF-β/SMAD3 pathway. Bioengineered. 2020;11(1):547–557.
- Tang C, Feng W, Bao Y, et al. Long non-coding RNA TINCR promotes hepatocellular carcinoma proliferation and invasion via STAT3 signaling by direct interacting with T-cell protein tyrosine phosphatase (TCPTP). Bioengineered. 2021;12(1):2119–2131.
- Liu K, Zhao D, Wang D. LINC00528 regulates myocardial infarction by targeting the miR-143-3p/COX-2 axis. Bioengineered. 2020;11(1):11–18.
- Shang D, Liu Y, Zhang J, et al. Peroxisome proliferator-activated receptor γ (PPARγ) suppresses the proliferation and metastasis of patients with urothelial carcinoma after renal transplantation by inhibiting LEF1/β-catenin signaling. Bioengineered. 2020;11(1):1350–1367.
- Zhang N, Liu JF. MicroRNA (MiR)-301a-3p regulates the proliferation of esophageal squamous cells via targeting PTEN. Bioengineered. 2020;11(1):972–983.
- Chen J, Zhang K, Song H, et al. Long noncoding RNA CCAT1 acts as an oncogene and promotes chemoresistance in docetaxel-resistant lung adenocarcinoma cells. Oncotarget. 2016;7(38):62474–62489.
- Zhang D, Cao C, Liu L, et al. Up-regulation of LncRNA SNHG20 predicts poor prognosis in hepatocellular carcinoma. J Cancer. 2016;7(5):608–617.
- Li Y, Xu J, Guo Y, et al. LncRNA SNHG20 promotes the development of laryngeal squamous cell carcinoma by regulating miR-140. Eur Rev Med Pharmacol Sci. 2019;23:3401–3409.
- Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
- Wang J, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacother. 2020;121:109627.
- Wu X, Xiao Y, Zhou Y, et al. lncRNA SNHG20 promotes prostate cancer migration and invasion via targeting the miR-6516-5p/SCGB2A1 axis. Am J Transl Res. 2019;11:5162–5169.
- Song M, Sun M, Xia L, et al. miR-19b-3p promotes human pancreatic cancer Capan-2 cells proliferation by targeting phosphatase and tension homolog. Ann Transl Med. 2019;7(11):236.
- Jin J, Sun Z, Yang F, et al. miR-19b-3p inhibits breast cancer cell proliferation and reverses saracatinib-resistance by regulating PI3K/Akt pathway. Arch Biochem Biophys. 2018;645:54–60.
- Jiang T, Ye L, Han Z, et al. miR-19b-3p promotes colon cancer proliferation and oxaliplatin-based chemoresistance by targeting SMAD4: validation by bioinformatics and experimental analyses. J Exp Clin Cancer Res. 2017;36(1):131.
- Zhang T, Liu C, Huang S, et al. A downmodulated microRNA profiling in patients with gastric cancer. Gastroenterol Res Pract. 2017;2017:1526981–1526988.
- Wei Y, Guo S, Tang J, et al. MicroRNA-19b-3p suppresses gastric cancer development by negatively regulating neuropilin-1. Cancer Cell Int. 2020;20(1):193.