ABSTRACT
Dexmedetomidine (Dex) has been reported to exhibit neuroprotective effects through various regulatory mechanisms. This study aims to investigate the role and molecular mechanism of SNHG11 in Dex-mediated neuroprotection. The ischemic stroke (IS) model was established in vivo by middle cerebral artery occlusion (MCAO) and in vitro by oxygen-glucose deprivation and reperfusion (OGD/R)-treated SH-SY5Y. SNHG11 was highly expressed after OGD/R, and Dex improved OGD/R-induced neurological injury. Additionally, Dex reversed the effects of SNHG11 on OGD/R-induced neurological injury. Furthermore, we found that SNHG11 upregulated vascular endothelial growth factor A (VEGFA) expression by targeting miR-324-3p. Through rescue assays, it was confirmed that SNHG11 regulated OGD/R-induced neurological injury through increasing VEGFA expression. At last, Dex was also discovered to improve neurological injury through regulating SNHG11 in the rat model. In conclusion, our work demonstrated that Dex improved OGD/R-induced neurological injury via SNHG11/miR-324-3p/VEGFA axis. These findings may offer a novel therapeutic strategy for IS treatment.
Introduction
Ischemic stroke (IS) is a common cerebrovascular disease with a high incidence rate among the elderly [Citation1,Citation2]. IS can decrease blood flow to the brain and trigger a chain reaction that may lead to severe neuronal damage [Citation3,Citation4]. Despite the great progress in IS treatment, the patients’ prognosis is still far from satisfactory due to the narrow treatment window [Citation5]. Therefore, it is important to develop effective therapeutic strategies for IS.
Dexmedetomidine (Dex) is a type of alpha2 adrenergic receptor agonist, which has pharmacological properties of analgesia and retarding sympathetic activity [Citation6–8]. A large number of researches have illustrated that Dex exhibits neuroprotective effects through various regulatory mechanisms [Citation9,Citation10]. For example, Dex activates the PI3K/Akt/mTOR pathway to promote the neuroprotective effect in traumatic brain injury [Citation11]. MiR-128 targets WNT1 to strengthen neuroprotective effects of Dex in hypoxic-ischemic brain damage [Citation12]. Additionally, Dex regulates SHNG16/miR-10b-5p/BDNF axis to exert neuroprotective effects in hippocampal neuronal cells [Citation13]. However, the molecular mechanisms of Dex in IS need further investigation.
Long non-coding RNAs (lncRNAs) are widely reported to participate in the pathogenesis of various diseases, including IS [Citation14]. For instance, lncRNA H19 drives M1 microglial polarization to facilitate neuroinflammation in IS [Citation15]. LncRNA MALAT1 regulates the apoptosis in IS via the miR-205-3p/PTEN axis [Citation16]. LncRNA GAS5 facilitates the progression of IS by targeting miR-137 [Citation17]. Furthermore, lncRNA-N1LR suppresses p53 phosphorylation to promote neuroprotection against IS [Citation18]. Small nucleolar RNA host gene (SNHG) family members, such as SNHG15 [Citation19], SNHG12 [Citation20], and SNHG1 [Citation21], have been reported to exhibit neuroprotective effects in cerebral ischemic injury. Nevertheless, the role of SNHG11 keeps unknown in IS progression.
This study aimed to investigate the molecular mechanisms of SHNG11 in Dex-mediated neuroprotection. We hypothesized that Dex improved neurological injury by regulating SNHG11/miR-324-3p/VEGFA axis. Our study revealed the vital roles of SNHG11 in neurological injury, which might offer a novel therapeutic strategy for IS.
Materials and methods
Animals
The Sprague-Dawley rats (8-week-old, n = 24), obtained from Vital River Co. Ltd. (Beijing, China), were divided into 4 groups (n = 6): the Sham group, the MCAO group, the MCAO +oe-SNHG11 group, and the MCAO +oe-SNHG11+ Dex group. Then, a 6/0 surgical nylon monofilament was utilized to block the middle cerebral artery. 2 h after middle cerebral artery occlusion (MCAO), the surgical nylon monofilament was retracted to allow reperfusion. The Sham-operated rats were manipulated in the same way but without the insertion. In the MCAO +oe-SNHG11+ Dex group, Dex (100 μmol/kg) was administered intravenously outside the jugular at the initiation of reperfusion [Citation22]. The animal experiments were approved by the Animal Care and Use Committee of Shanghai Ninth people’s Hospital
Cell culture
SH-SY5Y cells are widely used as an in vitro model to study neuronal function [Citation23–25]. Hence, SH-SY5Y cells were bought from American Type Culture Collection (ATCC) and cultivated in DMEM supplemented with 10% FBS (Thermo Fisher Scientific), 100 mg/mL streptomycin, and 100 U/mL penicillin at 37°C with 5% CO2.
OGD/R treatment
SH-SY5Y cells were maintained in glucose-free DMEM medium for 4 h with the hypoxic atmosphere (95% N2, 5% CO2). Next, the medium was substituted with fresh DMEM medium with 4.5 g/L glucose and 10% FBS, and incubated for 12, 24, and 48 h under normoxic conditions (95% air and 5% CO2) at 37°C [Citation26].
Transfection
Short hairpin RNA targeting SNHG11 (sh-SNHG11), shNC, NC mimics, miR-324-3p mimics, pcDNA3.1 vectors, SNHG11 overexpression plasmid (oe-SNHG11), and VEGFA overexpression plasmid (oe-SNHG11) were synthesized by GenePharma (Shanghai, China). The transfection was performed using Lipofectamine 2000 (Invitrogen).
RT-qPCR
Total RNA was extracted from SH-SY5Y cells using TRIzol reagent (Invitrogen), and reverse-transcribed to cDNA using a PrimeScript RT Reagent Kit (Takara). RT-qPCR was then performed using the SYBR Green kit (Applied Biosystems) on the ABI 7500 Real-time PCR system (Applied Biosystems). GAPDH or U6 was used as the internal control. The 2−ΔΔCt method [Citation27] was used to calculate the relative gene expression.
CCK-8 assay
SH-SY5Y cells (5 × 103 cells/well) were seeded onto the 96-well plate. Then, CCK-8 solution (10 μL) was added to each well and incubated for another 2 h. The optical density (OD) value at the wave of 450 nm was assessed under a microplate reader (Bio-Rad) [Citation28].
TUNEL assay
Cell apoptosis was inspected with the In Situ Cell Death Detection kit (Roche) [Citation29]. Samples were immobilized with 4% paraformaldehyde and permeabilized with 0.2% Triton X-100. Next, samples were dyed with the TUNEL reaction mixture (50 μl) and counterstained with DAPI. The fluorescence microscope (Leica, Wetzlar, Germany) was utilized to observe TUNEL-positive cells.
Detection of oxidative stress markers
Oxidative stress markers, such as catalase (CAT), glutathione peroxidase (GSH-PX), superoxide dismutase (SOD), and malondialdehyde (MDA), were detected using ELISA kits (Jiancheng Bioengineering Institute, China) according to the manufacturer’s instruction [Citation30].
RNA pull-down
Biotinylated SNHG11 sense (Bio-SNHG11 sense) and SNHG11 antisense (Bio-SNHG11 antisense) probes were acquired from Sangon (Shanghai, China). After mixing cell lysate, these above probes, and Dynabeads M-280 Streptavidin (Invitrogen), the eluted RNAs were measured by RT-qPCR [Citation31].
Luciferase reporter assay
The wild-type (Wt) or mutant-type (Mut) sequences of SNHG11 and VEGFA were sub-cloned into pmirGLO vectors (Promega). Then, NC mimics or miR-324-3p mimics were co-transfected with the above vectors into SH-SY5Y cells. The luciferase activity was measured by Dual-Luciferase Reporter System (Promega) [Citation32].
RIP assay
The EZ-Magna RIP Kit (Millipore) was employed to conduct RIP assay [Citation33]. The cell lysates and magnetic beads coated with anti-Ago2 or anti-IgG were mixed. After the incubation, the extracted RNAs were assessed with RT-qPCR.
Statistical analysis
Data were exhibited as mean± standard deviation (SD) and analyzed through SPSS 17.0 (SPSS). The comparisons in groups were assessed using student’s t-test or ANOVA followed by Tukey’s post hoc test. P < 0.05 represented statistical significance.
Results
In our study, the IS model was established in vivo by MCAO and in vitro by OGD/R-treated SH-SY5Y. It was found that SNHG11 expression was up-regulated in IS. Further exploration uncovered that Dex improved neurological injury by regulating the SNHG11/miR-324-3p/VEGFA axis.
Dexmedetomidine improved OGD/R-induced neurological injury
An in vitro cell model of IS was firstly established through treating SH-SY5Y cells with OGD/R. As displayed in , the cell viability was reduced after OGD/R treatment in a time-dependent manner. Additionally, SNHG11 expression was up-regulated with the increased treatment time of OGD/R (). OGD 4 h/reperfusion 24 h had the most significant changes. Therefore, OGD 4 h/reperfusion 24 h was used for subsequent experiments. Under treating with Dex with a dose-dependent effect, the cell viability was increased (). Besides, SNHG11 expression was decreased after Dex treatment (). These findings suggested that Dex (200 ng/ml) treatment had the most obvious effect. Next, we discovered that cell apoptosis was enhanced after OGD/R induction, but this effect could be restored by Dex treatment (). Moreover, OGD/R treatment considerably reduced the levels of SOD, CAT, and GSH-PX, and increased the levels of MDA. However, these effects were reversed by Dex treatment (). Taken together, SNHG11 was highly expressed in OGD/R-treated SH-SY5Y cells, and Dex improved OGD/R-treated neurological injury.
Figure 1. Dexmedetomidine improved OGD/R-induced neurological injury. (a) The cell viability of SH-SY5Y cells was tested through CCK-8 assay in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (b) SNHG11 expression was examined through RT-qPCR in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (c) The cell viability was measured through CCK-8 assay after treating with Dex (0, 50, 100, 200, and 400 ng/ml). (d) SNHG11 expression was assessed through RT-qPCR after in the Control, OGD/R-6 h, OGD/R-12 h, OGD/R-24 h, OGD/R-48 h group. (e) The cell apoptosis was measured through TUNEL assay in the Control, OGD/R, OGD/R+ saline, and OGD/R+ Dex groups. (f) The levels of SOD, CAT, GSH-Px, and MDA were verified through the appropriate commercial kits in the Control, OGD/R, OGD/R+ saline, and OGD/R+ Dex groups. *P < 0.05
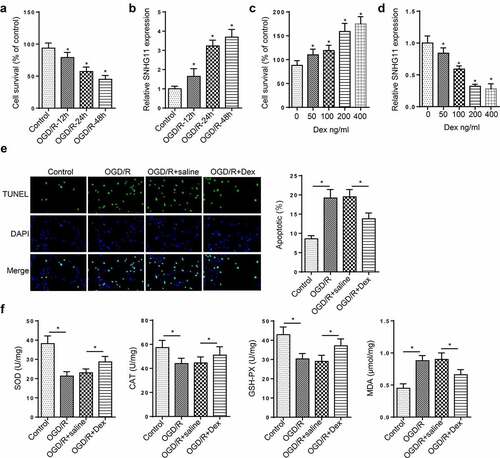
Dexmedetomidine reversed the effects of SNHG11 on neurological injury
To probe the role of SNHG11 in neurological injury, more experiments were carried out. CCK-8 indicated that the up-regulation of SNHG11 weakened the cell viability, but co-treating with Dex could relieve this effect (). Dex treatment reversed the enhanced cell apoptosis induced by SNHG11 overexpression (). Moreover, Dex treatment partially abolished the effect of SNHG11 up-regulation on the levels of OD, CAT, GSH-PX, and MDA (). These data demonstrated that Dex treatment reversed the effects of SNHG11 on the neurological injury.
Figure 2. Dexmedetomidine reversed the effects of SNHG11 on OGD/R-induced neurological injury. Groups were divided into the OGD/R+ oe-NC, OGD/R+ oe-SNHG11, and OGD/R+ oe-SNHG11+ Dex groups. (a) The cell viability was measured through CCK-8 assay. (b) The cell apoptosis was detected through TUNEL assay. (c) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits. *P < 0.05
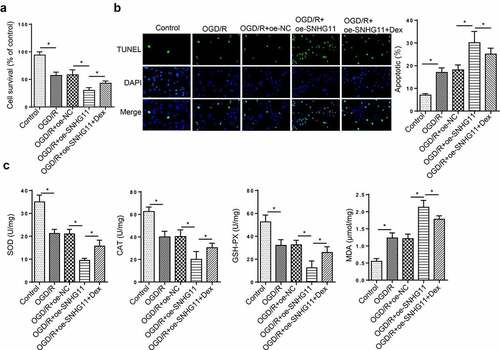
miR-324-3p was a target of SNHG11
Subsequently, starBase website was used to predict the downstream miRNAs of SNHG11. RNA pull-down assay indicated that miR-324-3p had the strongest binding ability to SNHG11 (). Moreover, miR-324-3p was lowly expressed in OGD/R-induced SH-SY5Y cells (). RT-qPCR indicated that miR-324-3p expression was up-regulated in OGD/R-treated SH-SY5Y cells transfected with miR-324-3p mimics (). In addition, miR-324-3p overexpression decreased the luciferase activity of SNHG11-Wt reporters, but no change was observed in the luciferase activity of SNHG11-Mut reporters (). The above results demonstrated that SNHG11 could bind with miR-324-3p.
Figure 3. miR-324-3p is a target of SNHG11. (a) The potential miRNAs that could bind with SNHG11 were predicted through starBase website with the condition (CLIP Data: high stringency (≥3)). The binding ability of miRNAs to SNHG11 was confirmed through RNA pull-down assay. (b) MiR-324-3p expression was tested through RT-qPCR. (c) The overexpression efficiency of miR-324-3p was verified through RT-qPCR. (d) The binding ability between SNHG11 and miR-324-3p was detected through the luciferase reporter assay. *P < 0.05
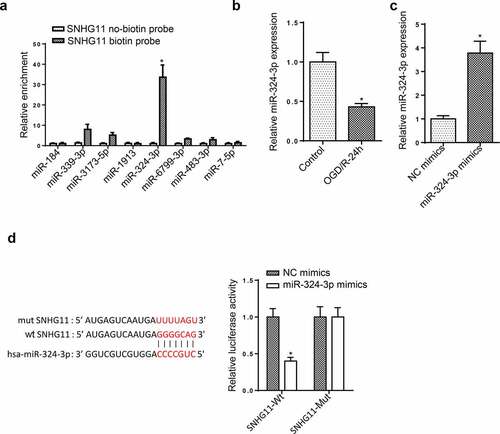
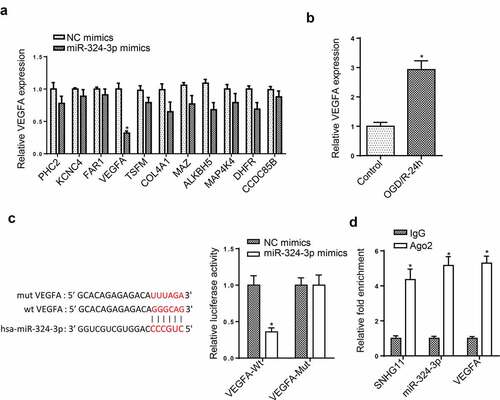
SNHG11 regulated VEGFA level by targeting miR-324-3p
Through using starBase, the downstream targets of miR-324-3p were screened. VEGFA expression was obviously decreased after overexpressing miR-324-3p among these target genes (). Besides, the up-regulated VEGFA level was found in OGD/R-mediated SH-SY5Y (). Furthermore, the luciferase activity of VEGFA-Wt reporters was reduced by miR-324-3p overexpression, but that of VEGFA-Mut reporters had no change (). At last, RIP assay was performed, and it was uncovered that the enrichment of SNHG11, miR-324-3p, and VEGFA was discovered in the Ago2 group (). These data revealed that SNHG11 upregulated VEGFA expression by targeting miR-324-3p.
Figure 4. SNHG11 regulated VEGFA level by targeting miR-324-3p. (a) The downstream target genes (n = 11) were selected through starBase website with the condition (Degradome Data: high stringency (≥3)). The levels of target genes were confirmed after miR-324-3p mimics through RT-qPCR. (b) VEGFA expression was measured through RT-qPCR. (c) The binding ability between miR-324-3p and VEGFA was verified through the luciferase reporter assay. (d) The enrichment of SNHG11, miR-324-3p, and VEGFA was evaluated in RISC complex through RIP assay. *P < 0.05
SNHG11 affected OGD/R-induced neurological injury by regulating VEGFA
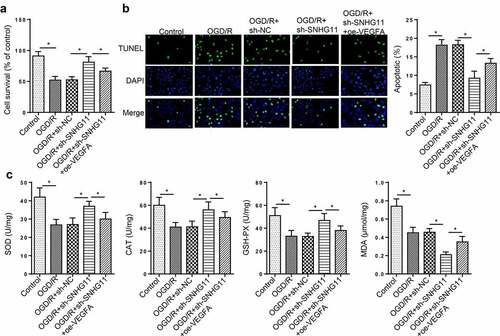
To confirm whether SNHG11 regulated OGD/R-induced neurological injury via VEGFA, rescue assays were performed. It was found that overexpression of VEGFA offset the increased cell viability mediated by silencing SNHG11 (). The cell apoptosis was reduced by SNHG11 suppression, but this effect was rescued by overexpressing VEGFA (). Additionally, VEGFA up-regulation attenuated the increased SOD, CAT, and GSH-PX levels as well as the decreased MDA levels induced by SNHG11 knockdown (). These results testified that SNHG11 regulated OGD/R-induced neurological injury through regulating VEGFA.
Figure 5. SNHG11 affected OGD/R-induced neurological injury by regulating VEGFA. Groups were divided into the OGD/R+ sh-NC, OGD/R+ sh-SNHG11, and OGD/R+ sh-SNHG11+ oe-VEGFA groups. (a) The cell viability was measured through CCK-8 assay. (b) The cell apoptosis was confirmed through TUNEL assay. (c) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits. *P < 0.05
Dexmedetomidine improved neurological injury by regulating SNHG11 in vivo
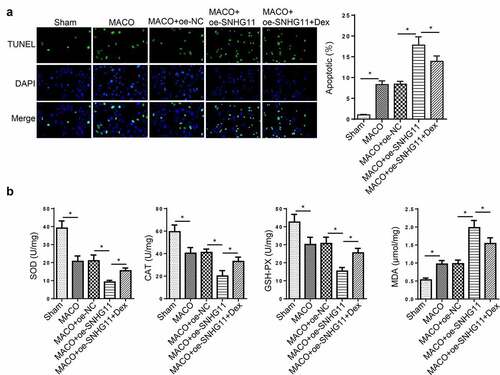
To further explore the function of SNHG11 in Dex-mediated neuroprotection in IS, the IS model was established in vivo by MCAO. The cell apoptosis was enhanced in the MCAO group, and further strengthened in the MCAO +oe-SNHG11 group. Dex treatment could reverse the increased cell apoptosis induced by SNHG11 overexpression (). Moreover, the MCAO group showed a distinct reduction in SOD, CAT, and GSH-PX levels and a significant increase in MDA level, which was further strengthened by overexpressing SNHG11; however, Dex treatment partly attenuated the effects of SNHG11 up-regulation on SOD, CAT, GSH-PX, and MDA levels (). To sum up, Dex improved neurological injury through regulating SNHG11 in the rat model.
Figure 6. Dexmedetomidine improved neurological injury by regulating SNHG11 in vivo. Groups were divided into the Sham, MCAO, MCAO +oe-SNHG11 and MCAO +oe-SNHG11+ Dex group. (a) The cell apoptosis was examined through TUNEL assay in the hippocampus of rat model. (b) The levels of SOD, CAT, GSH-PX, and MDA were confirmed through the appropriate commercial kits in the hippocampus of rat model. *P < 0.05
Discussion
Increasing evidence illustrates that Dex possesses neuroprotective effects in nervous system diseases [Citation11–13]. Herein, we further explored the effects of Dex on IS-induced neuronal damage in vitro and in vivo. Our findings confirmed that Dex significantly improved neuronal injury. Previous studies indicated SNHG11 participated in the pathogenesis of various diseases [Citation34–36], but its role in IS progression is undefined. In our work, we found that SNHG11 was highly expressed in OGD/R-treated SH-SY5Y cells. Furthermore, Dex reversed the effects of SNHG11 on OGD/R-induced neurological injury.
MicroRNAs (miRNAs), a group of highly conserved ncRNAs with 20–25 nucleotides in length [Citation37,Citation38], also play a crucial role in IS progression. For instance, repression of miR-497 promotes neuronal autophagy to improve functional outcome after IS [Citation39]. Suppression of miR-19a modulates neuronal apoptosis and glucose metabolism to protect neurons against IS [Citation40]. MiR-145 regulates the MAPK pathway to protect neuronal stem cells in cerebral IS rat [Citation41]. MiR-324-3p has been disclosed to be involved in many diseases progression. For example, miR-324-3p targets TGF-β1 to modulate fibroblast proliferation in atrial fibrillation [Citation42]. In addition, miR-324-3p targets WNT2B to retard nasopharyngeal carcinoma progression [Citation43]. Besides, SNHG22 contributes to the malignant phenotypes of breast cancer by regulating miR-324-3p [Citation44]. However, the association between SNHG11 and miR-324-3p in IS is unknown. In our study, miR-434-3p was verified to bind with SNHG11.
Accumulating studies have proved that lncRNAs act as competing endogenous RNAs (ceRNAs) by specifically adsorbing miRNAs, and then regulate the expression of target genes [Citation45–47]. For instance, LncRNA LOC100912373 acts as a ceRNA to contribute to fibroblast-like synoviocyte proliferation in rheumatoid arthritis via miR-17-5p/PDK1 axis [Citation48]. LncRNA NEAT1 regulates miR-124/BACE1 axis in Alzheimer’s disease progression [Citation49]. LncRNA GAS5 sponges miR-221 to modulate SIRT1 and suppresses diabetic nephropathy progression [Citation50]. Herein, VEGFA was confirmed as a downstream target of miR-324-3p. Additionally, previous researches reported that VEGFA was implicated in the pathogenesis of various diseases, including IS [Citation51–53]. In terms of ceRNA regulatory mechanisms, we discovered that SNHG11 up-regulated VEGFA expression by targeting miR-324-3p. Through rescue assays, it was verified that SNHG11 regulated OGD/R-induced neurological injury via VEGFA. At last, Dex was also revealed to improve neurological injury through regulating SNHG11 in the rat model.
Conclusion
This study demonstrated that Dex improved OGD/R-induced neurological injury by regulating SNHG11/miR-324-3p/VEGFA axis. However, there were several limitations in this study. Firstly, the downstream effectors or signaling pathways related to the SNHG11/miR-324-3p/VEGFA axis must be further investigated. Secondly, the interaction between Dex and SNHG11 will be further explored in future study.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Rodrigo R, Fernandez-Gajardo R, Gutierrez R, et al. Oxidative stress and pathophysiology of ischemic stroke: novel therapeutic opportunities. CNS Neurol Disord Drug Targets. 2013;12(5):698–714.
- Barthels D, Das H. Current advances in ischemic stroke research and therapies. Biochim Biophys Acta Mol Basis Dis. 2020;1866(4):165260.
- Radak D, Katsiki N, Resanovic I, et al. Apoptosis and acute brain ischemia in ischemic stroke. Curr Vasc Pharmacol. 2017;15(2):115–122.
- Uzdensky AB. Apoptosis regulation in the penumbra after ischemic stroke: expression of pro- and antiapoptotic proteins. Apoptosis. 2019;24(9–10):687–702.
- Hacke W, Brott T, Caplan L, et al. Thrombolysis in acute ischemic stroke: controlled trials and clinical experience. Neurology. 1999;53(7 Suppl 4):S3–14.
- Mahmoud M, Mason KP. Dexmedetomidine: review, update, and future considerations of paediatric perioperative and periprocedural applications and limitations. Br J Anaesth. 2015;115(2):171–182.
- Lee S. Dexmedetomidine: present and future directions. Korean J Anesthesiol. 2019;72(4):323–330.
- Liaquat Z, Xu X, Zilundu PLM, et al. The current role of dexmedetomidine as neuroprotective agent: an updated review.Brain Sci. 2021; 11(7)
- Li G, LeiQian, Gu P, et al. Dexmedetomidine post-conditioning attenuates cerebral ischemia following asphyxia cardiac arrest through down-regulation of apoptosis and neuroinflammation in rats.BMC Anesthesiol. 2021;21(1):180.
- Ding XD, Cao -Y-Y, Li L, et al. Dexmedetomidine Reduces the Lidocaine-Induced Neurotoxicity by Inhibiting Inflammasome Activation and Reducing Pyroptosis in Rats. Biol Pharm Bull. 2021;44(7):902–909.
- Shen M, Wang S, Wen X, et al. Dexmedetomidine exerts neuroprotective effect via the activation of the PI3K/Akt/mTOR signaling pathway in rats with traumatic brain injury. Biomed Pharmacother. 2017;95:885–893.
- Fang H, Li H-F, Yang M, et al. microRNA-128 enhances neuroprotective effects of dexmedetomidine on neonatal mice with hypoxic-ischemic brain damage by targeting WNT1. Biomed Pharmacother. 2019;113:108671.
- Wang L, Liu W, Zhang Y, et al. Dexmedetomidine had neuroprotective effects on hippocampal neuronal cells via targeting lncRNA SHNG16 mediated microRNA-10b-5p/BDNF axis. Mol Cell Biochem. 2020;469(1–2):41–51.
- Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. 2019;21(5):542–551.
- Wang J, Zhao H, Fan Z, et al. Long noncoding RNA H19 promotes neuroinflammation in ischemic stroke by driving histone deacetylase 1-dependent M1 microglial polarization. Stroke. 2017;48(8):2211–2221.
- Gao Q, Wang Y. Long noncoding RNA MALAT1 regulates apoptosis in ischemic stroke by sponging miR-205-3p and modulating PTEN expression. Am J Transl Res. 2020;12(6):2738–2748.
- Chen F, Zhang L, Wang E, et al. LncRNA GAS5 regulates ischemic stroke as a competing endogenous RNA for miR-137 to regulate the Notch1 signaling pathway. Biochem Biophys Res Commun. 2018;496(1):184–190.
- Wu Z, Wu P, Zuo X, et al. LncRNA-N1LR enhances neuroprotection against ischemic stroke probably by inhibiting p53 phosphorylation. Mol Neurobiol. 2017;54(10):7670–7685.
- Guo T, Liu Y, Ren X, et al. Promoting role of long non-coding RNA small nucleolar RNA host gene 15 (SNHG15) in neuronal injury following ischemic stroke via the MicroRNA-18a/CXC chemokine ligand 13 (CXCL13)/ERK/MEK Axis. Med Sci Monit. 2020;26:e923610.
- Cheng Y, Jiang Y, Sun Y, et al. The role of long non-coding RNA SNHG12 in neuroprotection following cerebral ischemic injury. Neuroreport. 2019;30(14):945–952.
- Zhang L, Luo X, Chen F, Yuan W, et al. LncRNA SNHG1 regulates cerebrovascular pathologies as a competing endogenous RNA through HIF-1α/VEGF signaling in ischemic stroke.J Cell Biochem. 2018;119(7):5460–5472.
- Liu T, et al. IL-17A-mediated excessive autophagy aggravated neuronal ischemic injuries via Src-PP2B-mTOR pathway. Front Immunol. 2019;10:2952.
- Cai HA, Tao X, Zheng LJ, et al. Ozone alleviates ischemia/reperfusion injury by inhibiting mitochondrion-mediated apoptosis pathway in SH-SY5Y cells. Cell Biol Int. 2020;44(4):975–984.
- Zhu Q, Zhang Y, Liu Y, et al. MLIF alleviates SH-SY5Y neuroblastoma injury induced by oxygen-glucose deprivation by targeting eukaryotic translation elongation factor 1A2. PLoS One. 2016;11(2):e0149965.
- Wang T, Zhu L, Liu H, Yu G, et al. Picroside II protects SH-SY5Y cells from autophagy and apoptosis following oxygen glucose deprivation/reoxygen injury by inhibiting JNK signal pathway. Anat Rec (Hoboken). 2019;302(12):2245–2254.
- Zhi W, Li K, Wang H, Lei M, et al. Melatonin elicits protective effects on OGD/R‑insulted H9c2 cells by activating PGC‑1α/Nrf2 signaling. Int J Mol Med. 2020;45(5):1294–1304.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Gao F, Wu H, Wang R, et al. MicroRNA-485-5p suppresses the proliferation, migration and invasion of small cell lung cancer cells by targeting flotillin-2. Bioengineered. 2019;10(1):1–12.
- Chen W, Su J, Cai S, et al. Cullin3 aggravates the inflammatory response of periodontal ligament stem cells via regulation of SHH signaling and Nrf2. Bioengineered. 2021;12(1):3089–3100.
- Wen Y, Zhang X, Liu X, Huo Y, et al. Suppression of lncRNA SNHG15 protects against cerebral ischemia-reperfusion injury by targeting miR-183-5p/FOXO1 axis. Am J Transl Res. 2020;12(10):6250–6263.
- Tang G, Liu L, Xiao Z, Wen S, et al. CircRAB3IP upregulates twist family BHLH transcription factor (TWIST1) to promote osteosarcoma progression by sponging miR-580-3p. Bioengineered. 2021;12(1):3385–3397.
- Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
- Long H, Li Q, Xiao Z, Yang B. LncRNA MIR22HG promotes osteoarthritis progression via regulating miR-9-3p/ADAMTS5 pathway. Bioengineered. 2021;12(1):3148–3158.
- Cheng R, Lu X, Xu C, et al. SNHG11 contributes to NSCLC cell growth and migration by targeting miR-485-5p/BSG axis. Biomed Pharmacother. 2020;128:110324.
- Wu Q, Ma J, Wei J, Meng W, et al. lncRNA SNHG11 promotes gastric cancer progression by activating the Wnt/β-Catenin pathway and oncogenic autophagy. Mol Ther. 2020;29:1258–1278.
- Xu L, Huan L, Guo T, et al. LncRNA SNHG11 facilitates tumor metastasis by interacting with and stabilizing HIF-1α. Oncogene. 2020;39(46):7005–7018.
- Ge Y, Zhang L, Nikolova M, et al. Strand-specific in vivo screen of cancer-associated miRNAs unveils a role for miR-21(*) in SCC progression. Nat Cell Biol. 2016;18(1):111–121.
- Yoon JH, Abdelmohsen K, Gorospe M. Functional interactions among microRNAs and long noncoding RNAs. Semin Cell Dev Biol. 2014;34:9–14.
- Chen X, Lin S, Gu L, Zhu X, et al. Inhibition of miR-497 improves functional outcome after ischemic stroke by enhancing neuronal autophagy in young and aged rats. Neurochem Int. 2019;127:64–72.
- Ge XL, Wang J-L, Liu X, et al. Inhibition of miR-19a protects neurons against ischemic stroke through modulating glucose metabolism and neuronal apoptosis. Cell Mol Biol Lett. 2019;24(1):37.
- Xue WS, Wang N, Wang NY, et al. miR-145 protects the function of neuronal stem cells through targeting MAPK pathway in the treatment of cerebral ischemic stroke rat. Brain Res Bull. 2019;144:28–38.
- Xu J, Lei S, Sun S, et al. MiR-324-3p regulates fibroblast proliferation via targeting TGF-β1 in atrial fibrillation. Int Heart J. 2020;61(6):1270–1278.
- Liu C, Li G, Yang N, et al:, et al. miR-324-3p suppresses migration and invasion by targeting WNT2B in nasopharyngeal carcinoma. Cancer Cell Int. 2017;17(1):2.
- Fang X, Zhang J, Li C, et al. Long non-coding RNA SNHG22 facilitates the malignant phenotypes in triple-negative breast cancer via sponging miR-324-3p and upregulating SUDS3. Cancer Cell Int. 2020;20(1):252.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Smillie CL, Sirey T, Ponting CP. Complexities of post-transcriptional regulation and the modeling of ceRNA crosstalk. Crit Rev Biochem Mol Biol. 2018;53(3):231–245.
- Dey BK, Mueller AC, Dutta A. Long non-coding RNAs as emerging regulators of differentiation, development, and disease. Transcription. 2014;5(4):e944014.
- Fan C, Cui X, Chen S, Huang S, Jiang H. LncRNA LOC100912373 modulates PDK1 expression by sponging miR-17-5p to promote the proliferation of fibroblast-like synoviocytes in rheumatoid arthritis. Am J Transl Res. 2020;12(12):7709–7723.
- Zhao MY, Wang G-Q, Wang -N-N, et al. The long-non-coding RNA NEAT1 is a novel target for Alzheimer’s disease progression via miR-124/BACE1 axis. Neurol Res. 2019;41(6):489–497.
- Ge X, Xu B, Xu W, et al. Long noncoding RNA GAS5 inhibits cell proliferation and fibrosis in diabetic nephropathy by sponging miR-221 and modulating SIRT1 expression. Aging (Albany NY). 2019;11(20):8745–8759.
- Claesson-Welsh L, Welsh M. VEGFA and tumour angiogenesis. J Intern Med. 2013;273(2):114–127.
- Ma L, Zheng Y, Tang X, et al. miR-21-3p inhibits autophagy of bovine granulosa cells by targeting VEGFA via PI3K/AKT signaling. Reproduction. 2019;158(5):441–452.
- Gao J, Ailifeire M, Wang C, et al. miR-320/VEGFA axis affects high glucose-induced metabolic memory during human umbilical vein endothelial cell dysfunction in diabetes pathology. Microvasc Res. 2020;127:103913.
