?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.
?Mathematical formulae have been encoded as MathML and are displayed in this HTML version using MathJax in order to improve their display. Uncheck the box to turn MathJax off. This feature requires Javascript. Click on a formula to zoom.ABSTRACT
Lung adenocarcinoma (LUAD) has been the major cause of tumor-associated mortality in recent years and has a poor prognosis. Pyroptosis is regulated via the activation of inflammasomes and participates in tumorigenesis. However, the effects of pyroptosis-related lncRNAs (PRlncRNAs) on LUAD have not yet been completely elucidated. Therefore, we attempted to systematically explore patterns of cell pyroptosis to establish a novel signature for predicting LUAD survival. Based on TCGA database, we set up a prognostic model by incorporating PRlncRNAs with differential expression using Cox regression and LASSO regression. Kaplan–Meier analysis was conducted to compare the survival of LUAD patients. We further simplified the risk model and created a nomogram to enhance the prediction of LUAD prognosis. Altogether, 84 PRlncRNAs with differential expression were discovered. Subsequently, a new risk model was constructed based on five PRlncRNAs, GSEC, FAM83A-AS1, AL606489.1, AL034397.3 and AC010980.2. The proposed signature exhibited good performance in prognostic prediction and was related to immunocyte infiltration. The nomogram exactly forecasted the overall survival of patients and had excellent clinical utility. In the present study, the five-lncRNA prognostic risk signature and nomogram are trustworthy and effective indicators for predicting the prognosis of LUAD.
Introduction
Lung cancer represents a fatal malignancy and a primary cause of cancer-associated mortality, with 2,206,771 new lung cancer cases and 1,796,144 deaths occurring in 2020 worldwide [Citation1]. The most frequently observed histological subtype of lung cancer is non-small cell lung cancer (NSCLC), which is primarily classified into squamous cell carcinoma (LUSC) and adenocarcinoma (LUAD) [Citation2]. Despite new developments in cancer therapeutic treatments for LUAD in recent years, including surgical resection, immunotherapy, chemotherapy and radiotherapy, the prognosis of LUAD continues to be frustrating, and its 5-year survival is less than 20% [Citation3]. Therefore, there is an urgent need to identify robust biomarkers for predicting the prognosis of LUAD patients.
Long noncoding RNAs (lncRNAs) comprise at least 200 nucleotides [Citation4]. The close interaction between lncRNAs and other cellular molecules, including DNA, mRNA, and microRNA, has been reported to be involved in multiple diseases, including cardiovascular diseases, cancers, and metabolic diseases [Citation5–8], and has attracted increasing attention. For instance, aging-regulated lncRNAs were found to act as antiapoptotic agents in cardiomyocytes [Citation9], and more recently, Lin et al. discovered that several lncRNAs are involved in papillary thyroid carcinoma by sponging microRNAs and modulating the PI3K/Akt and Wnt signaling pathways [Citation10]. Moreover, accumulating evidence indicates the prognostic value of aberrantly expressed lncRNAs in multiple biological and pathological processes. However, the pyroptosis-related lncRNAs associated with cancer diagnosis and prognosis remain poorly understood, making it very urgent to identify novel biomarkers based on pyroptosis-related lncRNA expression profiles.
Pyroptosis, a newly observed proinflammatory form of programmed cell death (PCD), is distinct from apoptosis, autophagy and ferroptotic cell death [Citation11]. The pyroptotic process is hallmarked by the rapid rupture of the plasma membrane and the release of proinflammatory cytokines. Pyroptotic cells first generate numerous vesicles under an electron microscope. After the development of these vesicles, pores are formed in the cell membrane through which the intracellular contents flow out [Citation12]. The typical pyroptotic pathway is sparked by the activation of nucleotide-binding domain leucine-rich repeats family protein 3 (NLRP3) inflammasomes as well as the associated inflammatory response [Citation13,Citation14]. Several reports have verified that pyroptosis plays a critical role in tumorigenesis and cancer treatment [Citation15–19]. Notably, various cancers, including colon cancer [Citation20,Citation21], gastric cancer [Citation22], hepatocellular carcinoma [Citation23,Citation24], breast cancer [Citation25,Citation26] and lung cancer [Citation27,Citation28], are sensitive to pyroptosis. Pyroptosis-related genes (PRGs), such as NLRP3 [Citation29,Citation30], Caspase 1 (CASP1) [Citation31], Gasdermin D (GSDMD) [Citation11,Citation27] and Gasdermin E (GSDME) [Citation32], are strongly implicated in oncogenesis and tumor progression. For example, GSDMD represses the proliferation of lung cancer cells by inhibiting EGFR/Akt signaling and inducing the intrinsic mitochondrial apoptotic pathway [Citation27]. In colon cancer, Dupaul-Chicoine et al. revealed that knocking out NLRP3 and CASP1 favored the development of cancer in a transgenic mouse model compared to wild type mice [Citation33]. Numerous cellular molecules, such as lncRNAs, are also involved in the regulation of pyroptosis. Liu et al. suggested that lncRNA-XIST knockdown abolished the development of NSCLC by promoting pyroptotic cell death [Citation34]. LncRNA ADAMTS9-AS2 restrained gastric cancer advancement through the pyroptosis pathway [Citation35]. However, the effect of pyroptosis-associated lncRNAs in LUAD has not been completely clarified. Consequently, we attempted to identify a pyroptosis-related lncRNA (PRlncRNA) signature and to investigate its clinical correlation with LUAD.
While several previous reports have investigated the relationship between pyroptosis-associated lncRNAs and prognosis in patients with lung cancer, there has yet to be much systematic analysis with respect to the context of pyroptosis in LUAD, and the underlying mechanism of LUAD remains poorly understood. Here, we identified pyroptosis-associated lncRNAs and generated a novel risk signature for the prognostic prediction of LUAD, opening up new perspectives for promoting individualized treatment for LUAD patients.
Materials and methods
Data collection
We obtained both clinical and FPKM RNA-seq data from LUAD cases using TCGA database (https://tcga-data.nci.nih.gov/tcga/), including 535 cancer samples and 59 noncancerous samples. Next, based on patient ID, we compared the clinical data of patients to their transcriptome data. Patients who had unmatched IDs were excluded from this work. Our inclusion criteria for patients were as follows [Citation1]: histologically diagnosed with LUAD [Citation2]; available expression profiles; and [Citation3] OS time greater than 30 days. Consequently, we extracted 504 patients who had sufficient gene expression profiling along with overall survival (OS) data from the TCGA dataset for subsequent analysis. Next, a total of 14 pyroptosis-related genes (CASP1, CASP3, CASP4, CASP5, PYCARD, IL18, IL1B, NLRP3, NLRC4, GSDMA, GSDMB, GSDMC, GSDMD and GSDME) were retrieved from previous research and the literature [Citation36–38].
Identification of differentially expressed PRlncRNAs
To identify PRlncRNAs, we used Pearson correlation to assess the associations between lncRNAs and PRGs. Typically, we selected PRlncRNAs using the thresholds of p < 0.001 and correlation coefficient |R2| > 0.3. Based on the Bioconductor limma package in R software [Citation39], we compared LUAD samples and non-carcinoma samples and selected the differentially expressed lncRNAs (DElncRNAs). DElncRNAs were determined based on |log2 (fold change, FC) | >1 and false discovery rate (FDR) <0.05 thresholds. Next, differentially expressed PRlncRNAs were extracted from the DElncRNAs.
Construction of the PRlncRNAs prognostic model
To develop an optimal PRlncRNAs prognostic model, we randomly and evenly separated patients from the entire set (n = 504) into training or internal test sets at a 1:1 ratio. First, potential prognostic lncRNAs were identified by univariate Cox regression from the training set using the threshold of p < 0.05. Subsequently, overfitting genes were reduced by least absolute shrinkage and selection operator (LASSO) regression. Finally, we established a prognostic model by multivariate Cox regression prognostic outcomes of LUAD. The risk score for LUAD cases was calculated as follows: risk score = (PRlncRNA 1 expression × coefficient) + (PRlncRNA 2 expression× coefficient) + … + (PRlncRNA n expression× coefficient). Meanwhile, the cases were classified into low- or high-risk groups based on the median value. In addition, the entire set and test set were used to validate our signature. Moreover, we utilized the R package ‘rms’ to construct a nomogram that integrated the risk score of the signature and clinical factors (age, clinical stage, T stage and N stage) [Citation40]. Calibration curves were plotted to determine the discrimination power of the nomogram.
Gene set enrichment analysis (GSEA)
GSEA was employed to detect the high-risk group correlated pathways and biological processes. The expressed gene sets of the low- or high-risk group together with hallmark gene sets collected based on Molecular Signatures Database v7.1 were analyzed using GSEA software. Gene sets conforming to NOM p < 0.05 and | NES |> 1 were deemed to be significant based on the User Guide of GSEA [Citation41].
Infiltrating immune cell analysis of the prognostic signature
We evaluated the association between the risk model and immunocyte infiltration according to the Tumor Immune Estimation Resource (TIMER, https://cistrome.shinyapps.io/timer/), which can be utilized to detect the infiltration fraction of six immune cells, including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, and dendritic cells, in the tumor microenvironment [Citation42]. Spearman’s test was used to analyze the correlations of the risk score with the infiltrating immunocytes. The significance level was set at P < 0.05.
Tumor mutation burden analysis
LUAD mutation data (TCGA.LUAD.varscan.acb6852e-dd48-4ca5-80f2-3d1a2c7d7ceb.DR-10.0.somatic) were also acquired from TCGA database. Somatic mutations in LUAD were evaluated based on the Mutation Annotation Format (MAF) and assessed using MAF tools [Citation43]. The tumor mutation burden (TMB) score for each LUAD case was generated using the following formula [Citation44].
Analysis of the risk model performance in clinical chemotherapy
To assess the signature in the clinical utility of LUAD treatment, we analyzed the half inhibitory centration (IC50) of typical chemotherapy drugs in the TCGA dataset using pRRophetic R software [Citation45]. AJCC guidelines recommend antineoplastic agents, such as cisplatin, docetaxel, doxorubicin, gemcitabine and paclitaxel, for the treatment of hepatocellular carcinoma. Thereafter, we utilized the Wilcoxon signed-rank test to detect the heterogeneous IC50 in the low-risk group relative to the high-risk group.
Cell culture and transfection
We obtained a human lung epithelial cell line (BEAS-2B) together with human LUAD cell lines (A549 and NCI-H460) from Shanghai Institute of Biochemistry and Cell Biology, the Chinese Academy of Sciences. Then, we cultivated cells in RPMI-1640 medium that contained 10% fetal bovine serum (FBS, Gibco Company) and 10% penicillin-streptomycin (Sigma‐Aldrich), followed by incubation at 37°C and 5% CO2. siRNA negative control (si‐NC) and si‐GSEC were chemically synthesized by RiboBio (Guangzhou, China). We transfected si‐GSEC and its negative control (si-NC) into lung adenocarcinoma cell lines. The sense sequence of si-GSEC was 5ʹ-GGUCACAACAGUACAAAGA-3ʹ. Subsequently, Lipofectamine 3000 (Invitrogen) was utilized to transfect cells with siRNAs in line with specific protocols. Forty-eight hours post-transfection, we collected cells for subsequent experiments.
RNA extraction and quantitative real-time PCR (qRT-PCR)
Total cellular RNA was isolated using TRIzol (Vazyme Biotech, Nanjing, China). Then, the extracted RNA was measured for concentration and purity using a BioSpec-nano spectrophotometer (Shimadzu, Japan) and was reverse transcribed to synthesize complementary DNA (cDNA) utilizing the Prime Script RT Master Mix reagent (Takara Bio, Dalian, China). Subsequently, we performed qRT-PCR using the StepOnePlus real-time PCR system (Thermo Fisher Science) and polymerase chain reaction system TB Green®PreMix Ex Taq™ (Takara Bio, Dalian, China), and the 2-ΔΔCT method was used to calculate the relevant gene expression. The expression of GSEC was detected using 5ʹ-GAGTTCATTTGCTCTCTCTGGCAC-3ʹ (forward) and 5ʹ-AAGAGGAGGCCTGATGGGGATA-3ʹ (reverse) primers. GAPDH was used as a reference gene.
Western blot analysis
Western blot analysis was conducted to determine NLRP3, cleaved caspase‑1 and GAPDH levels. Antibodies against NLRP3 (#15,101, 1:1,000), cleaved caspase‑1 (#4199, 1:1,000) and GAPDH (#5174, 1:1,000) were provided by Cell Signaling Technology (CST, Danvers, MA, US). Each protein’s expression was detected using Super ECL Plus Detection Reagent (Millipore) on a Bio-Imaging System (Bio-Rad, USA).
Cell counting kit-8 (CCK-8) assay
We measured cell proliferation using a CCK-8 assay (Beyotime, Shanghai, China) following specific protocols. Thereafter, cells (2000/well) were inoculated into 96-well plates and cultured in RPMI-1640 medium containing 10% FBS. At a fixed time of day, we added CCK-8 solution to each well and incubated the cells at 37°C for an additional 2 h. The absorbance values were measured at 450 nm using a microplate spectrophotometer (Thermo, USA) and were used to determine the capability of LUAD cell proliferation.
Colony formation assay
To perform the colony formation assay, transfected cells (300/well) were plated into 6-well plates and cultured for 14 days in RPMI-1640 medium containing 10% FBS. Next, 1% formaldehyde was used to fix proliferating cell colonies, whereas 1% crystal violet was applied for staining. The number of colonies that contained at least 50 cells was calculated, and photos were taken.
Statistical analysis
R software (3.6.3) and GraphPad (8.0) were employed for all statistical data analyses. Differences in OS between high-risk patients and low-risk patients were assessed using Kaplan–Meier analysis as well as log-rank tests. Additionally, independent predictors were identified by univariate and multivariate Cox regression. Moreover, we plotted time-dependent receiver operating characteristic (t-ROC) curves to evaluate the predictive performance of our established prognostic signature. P < 0.05 was set as the significance level.
Results
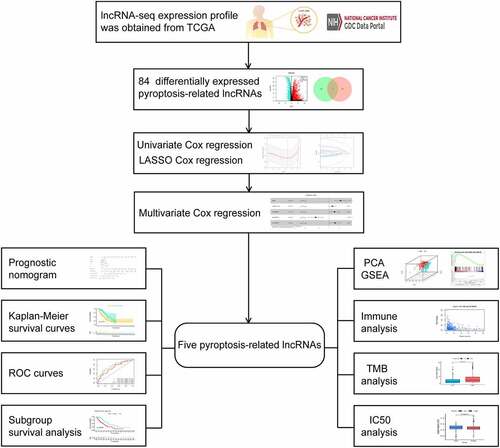
In the present study, our proposed novel pyroptosis-related lncRNAs signature was composed of five lncRNAs: GSEC, FAM83A-AS1, AL606489.1, AL034397.3 and AC010980.2. The pyroptosis-related risk model could ameliorate the prediction of LUAD prognosis. Cox relative regression methods indicated the independence of risk score generated from the risk model. Moreover, we simplified the constructed signature to set up a nomogram by combining risk score and other clinical traits. Immune microenvironment analysis, GSEA, and chemotherapy drugs analysis were utilized to exploit the clinical potency of the risk signature. Finally, we selected the lncRNA GSEC to valid our signature by in vitro experiments, including qRT-PCR, CCK-8 assay, colony formation assay and western blot analysis.
Identification of differentially expressed PRlncRNAs
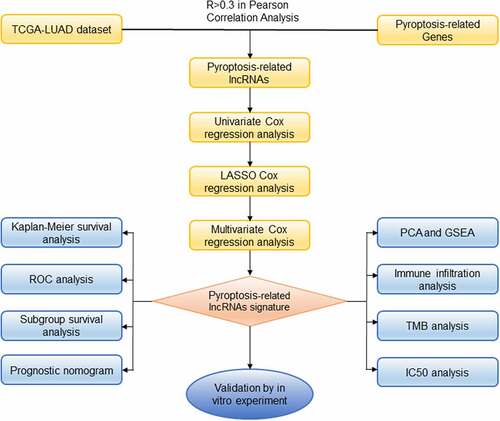
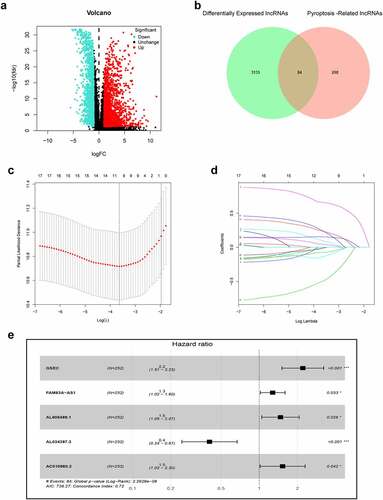
The infographic flowchart of the whole study is shown in . Based on the 14 pyroptosis-related genes (PRGs), we identified 382 PRlncRNAs by Pearson correlation analysis of lncRNA levels and PRG levels in the LUAD samples (Pearson correlation coefficient > 0.3, p < 0.001). Using cutoff values of |log2FC| > 1 and P < 0.05, 3219 DElncRNAs were identified between 535 LUAD and 59 noncancerous samples (). By overlapping the PRlncRNAs and DElncRNAs of LUAD, we identified 84 significantly differentially expressed pyroptosis-related lncRNAs (DEPRlncRNAs) for subsequent analysis ().
Figure 2. Construction of prognostic pyroptosis-related risk model composed of five lncRNAs. (a) Volcano plot presenting differentially expressed lncRNAs (DElncRNAs) discovered from LUAD tissues compared with non-carcinoma samples from TCGA dataset; (b) The Venn diagram of genes among DElncRNAs list and pyroptosis-related lncRNAs; (c-d) Lasso Cox regression analysis showing that 11 out of the 84 pyroptosis-related lncRNAs were good candidates for constructing the prognostic signature; (e) Forest plot presenting the HRs for the pyroptosis-associated prognosis model containing five lncRNAs
Establishment and validation of the PRlncRNA prognostic model
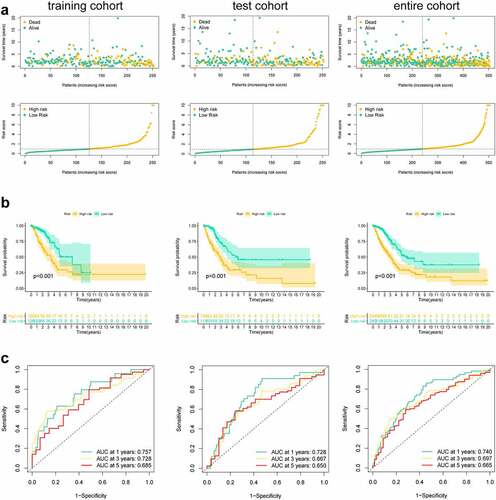
First, this study classified the included cases (n = 504) into training (n = 252) and validation (n = 252) cohorts at a 1:1 ratio (). Univariate Cox regression was employed to identify 18 PRlncRNAs correlated with OS in the training cohort. Then, LASSO Cox regression was adopted to reduce the risk of overfitting using the R software ‘glmnet’ package (). Finally, multivariate Cox regression was performed to establish a prognostic pyroptosis-related risk model composed of five lncRNAs (GSEC, FAM83A-AS1, AL606489.1, AL034397.3 and AC010980.2) (). As shown in Supplementary Figure 1, the expression levels of five signature lncRNAs were further confirmed by paired differentiation analysis. The risk score was determined according to the following formula: Risk Score = [GSEC expression × (0.7919)] + [FAM83A-AS1 expression × (0.2422)] + [AL606489.1 expression× (0.3857)] + [AL034397.3 expression × (−0.9045)] + [AC010980.2 expression × (0.4247)]. All patients were divided into low- or high-risk groups based on the median value of the risk score. shows the predictive performance of our constructed five-lncRNA pyroptosis-associated prognostic model for predicting the OS of patients. The Kaplan–Meier survival curves illustrated that the OS time of high-risk LUAD patients was significantly shorter than that of low-risk patients (). ROC analysis was used to evaluate the predictive reliability of our prognostic signature (). In addition, we evaluated prognostic power in the validation cohort and the entire cohort to verify its accuracy ().
Table 1. Clinicopathologic characteristics of LUAD patients
Figure 3. Risk score of the pyroptosis-related signature for overall survival (OS). (a) Distribution of survival and (b) risk scores of high- and low-risk patients; (c) Kaplan-Meier survival curve analysis suggests that the notable difference in OS between low-risk and high-risk score group; (d) ROC analysis for verifying model performance in the prediction of LUAD survival rates at 1, 3 and 5 years in the training cohort, validation cohort and entire cohort
Subgroup analysis of the PRlncRNA prognostic model
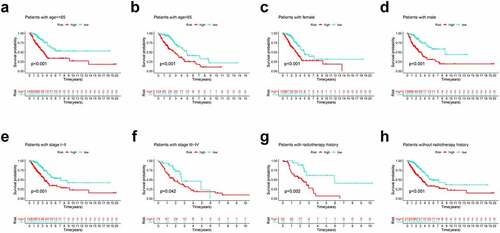
We further performed subgroup survival analysis to determine whether the prognostic model could predict OS for patients based different clinical features. These subgroups were separated by age (≤ 65 or > 65), gender (male or female), radiotherapy history and clinical stage (stage I–II or stage III–IV). As shown in , high-risk patients exhibited inferior 5-year OS rates compared to low-risk patients according to age, gender, radiotherapy history and clinical stage.
Construction and validation of a prognostic nomogram
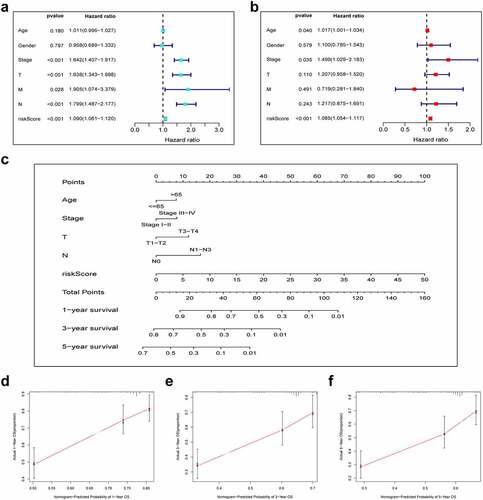
To verify that our constructed prognostic signature could independently predict the prognosis of LUAD cases, univariate and multivariate Cox regression analyses were performed on the entire cohort. As revealed by univariate analysis, clinical stage (P < 0.001), T stage (P < 0.001), risk score (P < 0.001), N stage (P < 0.001) and M stage (P = 0.028) predicted dismal OS (). Moreover, our multivariate Cox regression results validated the independence of our constructed prognostic model for predicting LUAD prognosis (). Next, we combined the risk score and other clinicopathologic parameters to develop a novel nomogram to predict OS rates for LUAD cases at 1, 3 and 5 years, aiming to optimize the predictive accuracy of the risk model (). The 1-, 3- and 5-year calibration curves of our constructed nomogram fit the nomogram well for the entire cohort ().
Figure 5. Combination of pyroptosis-related lncRNAs (PRlncRNAs) and clinical characteristics in predicting LUAD prognosis. (a) Univariate and (b) multivariate Cox regression methods for independent prognostic analysis of risk model; (c) Nomogram constructed to predict OS rates at 1, 3 and 5 years; (d-f) The nomogram calibration curves on consistency between predicted and observed 1‐, 3‐, and 5-year survival
Functional analysis of the PRlncRNA prognostic model
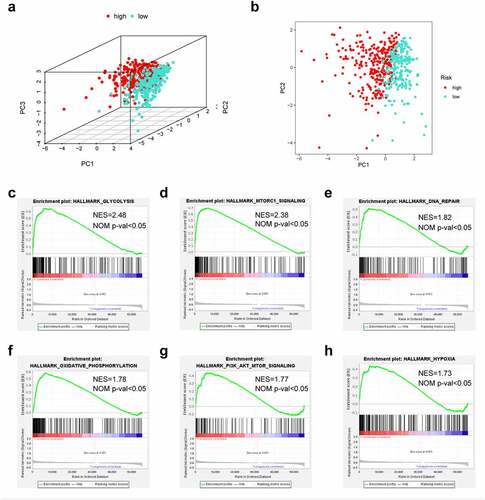
PCA suggested that LUAD cases of diverse groups were classified into 2 clusters (). To detect the possible biological signaling pathways associated with high-risk patients, we further applied GSEA to compare the two groups. As a result, ‘glycolysis’, ‘mTORC1 signaling’, ‘DNA repair’, ‘oxidative phosphorylation’, ‘PI3K/AKT/mTOR signaling’ and ‘hypoxia’ were significantly activated in high-risk patients (). Taken together, these findings indicate that high risk was closely correlated with processes that facilitate tumor growth and development.
Figure 6. The low-risk and high-risk groups exhibited different distribution statuses and gene-set enrichment analysis (GSEA). (a) Principal components analysis (PCA) of risk groups based on the pyroptosis-associated gene sets; (b) GSEA on glycolysis; (c) GSEA on mTORC1 pathway; (d) GSEA on DNA repair; (e) GSEA on oxidative phosphorylation; (f) GSEA on PI3K/AKT/mTOR pathway; (g) GSEA on hypoxia
Immune infiltration analysis and mutation profile of the prognostic signature
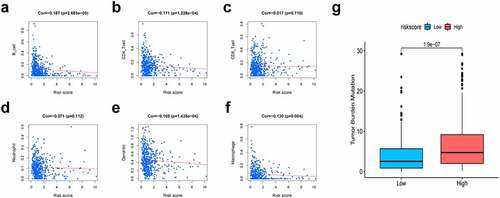
To explore the association of the constructed nomogram with the tumor immune microenvironment, this study evaluated the association of the immunocyte infiltration level with the risk score based on the TIMER database. As a result, the greater risk scores were inversely proportional to CD4 + T cells (cor = −0.171), B cells (cor = −0.187), dendritic cells (cor = −0.169, P < 0.05), and macrophages (cor = −0.130), suggesting that immune cell infiltration levels were decreased overall (). TMB has been demonstrated to be a crucial indicator for predicting the clinical benefits of immunotherapy [Citation46]. To investigate our present model’s clinical utility for LUAD immunotherapy, the TMB of high- and low-risk patients was analyzed. We observed that high-risk cases exhibited increased TMB relative to those with low risk ().
Figure 7. Association of risk scores with diverse immune cells and tumor mutation burden (TMB). (a) B cells (cor = −0.187); (b) CD4 + T cells (cor = −0.171); (c) CD8 + T cells (cor = −0.017); (d) Dendritic cells (cor = −0.169); (e) Macrophages (cor = −0.130); (f) Neutrophil (cor = −0.071); (g) The TMB of LUAD cases of in the high-risk and low-risk groups. Cor means the correlation value of risk scores with each immune cells
Correlation analysis between the risk group and chemotherapeutics
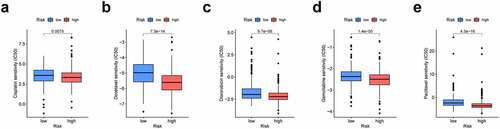
Chemotherapy still plays a vital role in treating LUAD patients. Based on the LUAD dataset of TCGA, we investigated the associations between risk group and the efficacy of common chemotherapeutics in treating patients. The findings indicated that the high-risk group exhibited a lower IC50 for cisplatin, docetaxel, doxorubicin, gemcitabine and paclitaxel (P < 0.05), suggesting that our proposed risk signature can be used as a potential indicator of drug sensitivity ().
Knockdown of GSEC attenuates LUAD cell proliferation and promotes pyroptosis
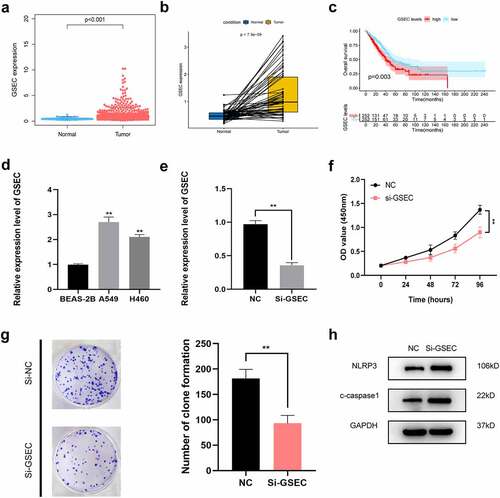
We chose GSEC to confirm our signature. First, the expression profile and prognostic value of GSEC were evaluated using the TCGA dataset. As a result, GSEC expression was remarkably increased in LUAD samples compared to non-carcinoma samples (). According to Kaplan–Meier analysis results, increased GSEC levels predicted poor OS in LUAD (). Next, PCR assays demonstrated that GSECs were highly expressed in LUAD cell lines (A549 and H460) compared to BEAS-2B cells (). Next, we used A549 cells to explore the functional role of GSEC. As shown in , GSEC expression was markedly decreased in A549 cells in response to si-GSEC transfection. CCK-8 assays demonstrated that si-GSEC-transfected A549 cells exhibited a markedly decreased growth rate relative to the negative control (). Moreover, colony formation assays also demonstrated that silencing GSEC significantly suppressed the proliferation of A549 cells (). Western blot analysis illustrated that downregulation of GSEC increased NLRP3 and cleaved caspase-1, indicating the potential role of GSEC on LUAD cell pyroptosis ().
Figure 9. Effects of inhibiting the expression of GSEC on LUAD cell proliferation and pyroptosis. (a) Differential expression of GSEC between cancer and non-carcinoma samples; (b) Paired differentiation analysis on GSEC expression between cancer and non-carcinoma samples collected in one patient; (c) Kaplan-Meier survival analysis for LUAD patients with different GSEC expression; (d) GSEC expression increased within H460 and A549 cells relative to BEAS-2B; (e) GSEC was downregulated in A549 using siRNAs; (f-g) The proliferation of A549 cells transfected with siRNA against GSEC were measured using CCK8 assays and colony formation assays; (h) Western Blot was carried out to examine NLRP3 and cleaved caspase-1 (*p < 0.05; **p < 0.01; ***p < 0.001)
Discussion
Lung cancer ranks second among health problems and is the major cause of cancer-associated mortality worldwide. LUAD is considered to be the most common subtype in nonsmokers. The prevalence of LUAD is rapidly increasing with the development of anti-smoking movements. Although headway has been made in cancer treatment, the overall survival of LUAD remains disappointing due to a lack of reliable early prognostic indicators. Pyroptosis, a novel accepted form of non-apoptotic cell death, was proved to be a double-edged sword for tumor progression and cancer therapy. Under the stimulation of a great deal of inflammatory cytokines released by pyroptotic cell, normal cells could be transformed into cancer cells [Citation36]. Alternatively, the development of tumor pyroptotic death could make pyroptosis a treatment target for cancer [Citation19]. Numerous studies suggest that lncRNAs play central part in the sustainment of various biological activities in tumor, such as cell pyroptosis. As discovered by Tan et al., HOTTIP, a crucial oncogenic driver in various cancers, could block pyroptosis by binding with miR-148a-3p and further positively enhancing AKT2 [Citation47]. In liver cancer, NLRP3-related pyroptosis pathway suppressed by SNHG7 through regulation of miR-34a/SIRT1 ceRNA axis [Citation48]. In addition, MEG3 was found to activate cisplatin-induced cellular pyroptosis by promoting NLRP3/caspase-1/GSDMD axis, implying that MEG3 could be effective therapeutic target of breast cancer [Citation49]. Therefore, it is essential to identify robust pyroptosis-related signatures to enhance the prognostic prediction of LUAD patients. This work successfully established a prognostic risk signature based on PRlncRNAs for predicting the overall survival in patients with LUAD. Furthermore, we initially investigated the oncogenic role of GSEC in LUAD and found that suppression of GSEC may inhibit proliferation and facilitate pyroptosis in LUAD cells.
In this study, the Pearson correlation method and differential expression analysis were used to identify 84 DEPRlncRNAs. Next, these lncRNAs were selected to develop a five-PRlncRNA signature based on the training set. Then, we adopted ROC analysis to evaluate the predictive performance of our constructed risk model. The AUCs of the ROC curves for 1-, 3-, and 5-year OS in our proposed model were 0.757, 0.728 and 0.848, respectively. Furthermore, we utilized ROC curves to compare the predictive ability of our present risk signature to other signatures. We observed that our risk signature attained consistently outstanding predictive power compared to other published pyroptosis‑based prognostic models in LUAD [Citation50]. Moreover, we confirmed that our signature exhibited strong independent prognostic ability for OS. Finally, we constructed a nomogram that integrated the risk score and clinical characteristics to enhance the prediction of LUAD prognosis.
Our present pyroptosis-related signature consists of five PRlncRNAs, which were remarkably correlated with OS in LUAD patients. Among these five lncRNAs, GSEC, FAM83A-AS1, AL606489.1 and AC010980.2 are potentially dangerous lncRNAs, but AL034397.3 is a potentially protective lncRNA. In this study, a number of lncRNAs in the risk model, such GSEC, FAM83A-AS1, AC010980.2 and AL034397.3, were suggested to exert vital roles in regulating different cancers, while AL606489.1 was identified for the first time. GSEC was shown to participate in cancer growth and development. In osteosarcoma, GSEC boosts proliferation and metastasis through the miR-588/EIF5A2 axis with sponge activity [Citation51]. As suggested by Matsumura et al., GSEC was highly expressed within colorectal cancer (CRC) samples and regulated tumor migration by targeting DHX36 [Citation52]. FAM83A-AS1 exerts a carcinogenic effect on LUAD, esophageal cancer and hepatocellular carcinoma. As discovered by Xiao et al., FAM83A-AS1 promotes the development of LUAD by regulating MMP14 expression and binding to miR-150-5p [Citation53]. In addition, FAM83A-AS1 may also strengthen the pre-mRNA stability of FAM83A to improve the metastatic ability of lung adenocarcinoma [Citation54]. In esophageal cell squamous carcinoma, FAM83A-AS1 downregulation regulates the miR-214/CDC25B axis to suppress tumor cell growth, invasion and migration [Citation55]. Furthermore, He and colleagues reported that FAM83A-AS1 was expressed in HCC cells and tissues, suggesting that increased FAM83A-AS1 expression enhances tumor proliferation and migration and represses apoptosis by interacting with NOP58 [Citation56]. Our results are in line with these studies, suggesting that GSEC and FAM83A-AS1 are risk factors (HR > 1) in LUAD. Interestingly, AC010980.2 was identified to be closely associated with immunity, autophagy and ferroptosis in LUAD and may represent an oncogene in a risk model for predicting the prognosis of patients with LUAD [Citation57–59]. In addition, Jin et al. used AL034397.3 to establish an immune-related risk model that improved the prediction of LUAD prognosis [Citation60].
To further detect the underlying functional mechanisms of the signature, we performed GSEA. The high-risk group exhibited significantly activated ‘glycolysis’, ‘mTORC1 signaling’, ‘DNA repair’, ‘oxidative phosphorylation’, ‘PI3K/Akt/mTOR signaling’, and ‘hypoxia’ compared to the low-risk group. Aerobic glycolysis is recognized as the characteristic metabolic pathway of cancer. Efficient glucose utilization by malignant tumors is associated with high proliferation, aggressiveness, and self-renewal capacity [Citation61]. Numerous studies have demonstrated a link between DNA damage repair and glycolysis, which are interdependent and promote the uncontrolled proliferation and survival of tumor cells [Citation62]. Thomas M Ashton et al. revealed that oxidative phosphorylation is commonly upregulated in tumor cells with high metastatic and tumorigenic potential, including LUAD [Citation63]. In a hypoxic microenvironment, activated hypoxic-inducible factor-1α may promote epithelial to mesenchymal transformation (EMT), thereby increasing invasion, tumor stem cell-like phenotypes, and chemoradiotherapy resistance [Citation64]. MTORC1 is the downstream effector in oncogenic pathways with frequent mutations, such as the MAPK pathway (excessively activated in diverse human cancers) and the PI3K/Akt pathway [Citation65]. Upregulation of the mTOR pathway is observed in up to 90% of lung adenocarcinoma patients [Citation66]. Immune cell infiltration has an important effect on LUAD survival. The results of immunocyte infiltration analysis revealed that the risk score exhibited a negative correlation with the infiltration of B cells, CD4 + T cells, DCs and macrophages. It has been previously shown that increased CD4 + T cells are a favorable independent prognostic factor for NSCLC [Citation67]. B cells may restrain tumor cells and reduce the incidence of occult micrometastases, resulting in prolonged survival by limiting further tumor spread [Citation67]. Romain Remark et al. summarized several works regarding the association of immune cells with NSCLC survival, suggesting that B-cell density is an indicator of better prognosis [Citation68]. With the increase in risk score, infiltration levels of B cells and CD4 + T cells decreased, consistent with the reduced survival times of high-risk patients.
To better assess the clinical feasibility of the risk model, we analyzed the efficacy of the presented model in immunotherapy according to the tumor mutation burden (TMB). Based on these findings, high-risk LUAD patients exhibit higher TMBs than low-risk patients, suggesting that our signature could be a potential index for evaluating the efficacy of immunotherapy in patients with LUAD. In addition to immunotherapy, we also identified the relationship between the signature and chemotherapy sensitivity of patients. Collectively, these discoveries may offer prospective treatment alternatives for LUAD patients.
Moreover, the functional phenotypic role of GSEC was investigated by experimental studies. We verified expression levels of GSEC between human lung epithelial cells and human lung adenocarcinoma cell lines. In vitro analysis showed that inhibition of GSEC blocked proliferation and activated pyroptosis in A549 cells by triggering the NLRP3 inflammasome and cleaved caspase-1, indicating that GSEC might be a potential pyroptosis-related lncRNA in LUAD.
In our analysis, we found that the integrated pyroptosis-related signature and clinical factors heightened the predictive reliability of prognosis relative to the TNM staging system, which may undergo routine application in the future. In addition, the proposed signature offers improved clinical utility for immunotherapy strategies and chemotherapy drug selection in LUAD patients.
Limitations
There are some shortcomings in our present study. First, the original dataset for setting up the lncRNA-related model was merely retrieved from the TCGA database. Our risk model needs to be validated for reliability and accuracy in other external datasets and large-scale clinical cohorts. Second, the mechanism by which pyroptosis regulates the precise process of LUAD is still not known and needs to be elucidated through additional studies.
Conclusions
The present study first developed a five-PRlncRNA signature that offers valuable clinical application for accurate prognostic forecasting. Our signature provides insights into personalized treatment for LUAD patients.
Highlights
A novel pyroptosis-related lncRNA signature was established.
Our signature improves the prediction of lung adenocarcinoma prognosis.
Pyroptosis might be associated with the immune response in lung adenocarcinoma.
Author’ s contributions
Ye Shi and Jiahang Song conceived and designed the original study. Jiahang Song, Zhengcheng Liu and Lei Xi collected the data. Jiahang Song, Hui Cao, Changqing Dong and Rusong Yang contributed to the interpretation of the data. Jiahang Song, and Yuanyuan Sun drafted the manuscript. Ye Shi revised the manuscript. All authors saw and approved the final version of the manuscript. These authors contributed equally: Jiahang Song, Yuanyuan Sun, Hui Cao.
Ethics approval and consent to participate
All data collection in studies involving human participants were in accordance with the ethical standards of the Cancer Genome Atlas Human Subjects Protection and Data Access Policies.
Supplemental Material
Download ()Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability
Publicly available datasets were analyzed in this study. These data can be found here: TCGA (https://portal.gdc.cancer.gov/).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Sung H, Ferlay J, Siegel RL, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2021;71(3):209–249. PubMed PMID: 33538338. Epub 2021/ 02/05.
- Travis WD. Pathology of lung cancer. Clin Chest Med. 2011;32(4):669–692. Epub 2011/ 11/08. PubMed PMID: 22054879.
- Hao CC, Xu CY, Zhao XY, et al. Up-regulation of VANGL1 by IGF2BPs and miR-29b-3p attenuates the detrimental effect of irradiation on lung adenocarcinoma. J Exp Clin Cancer Res. 2020;39(1):256. PubMed PMID: 33228740; PubMed Central PMCID: PMCPMC7687693. Epub 2020/ 11/25.
- Zehendner CM, Valasarajan C, Werner A, et al. Long Noncoding RNA TYKRIL plays a role in pulmonary hypertension via the p53-mediated Regulation of PDGFRβ. Am J Respir Crit Care Med. 2020;202(10):1445–1457. PubMed PMID: 32634060; PubMed Central PMCID: PMCPMC7786813. Epub 2020/ 07/08.
- Zhu G, Luo H, Feng Y, et al. HOXBLINC long non-coding RNA activation promotes leukemogenesis in NPM1-mutant acute myeloid leukemia. Nat Commun. 2021;12(1):1956. PubMed PMID: 33782403. Epub 2021/ 03/31.
- Monteiro JP, Rodor J, Caudrillier A, et al. MIR503HG loss promotes endothelial-to-mesenchymal transition in vascular disease. Circ Res. 2021;128(8):1173–1190. PubMed PMID: 33703914; PubMed Central PMCID: PMCPMC7610629. Epub 2021/ 03/12.
- Zhang Y, Zhang X, Cai B, et al. The long noncoding RNA lncCIRBIL disrupts the nuclear translocation of Bclaf1 alleviating cardiac ischemia–reperfusion injury. Nat Commun. 2021;12(1):522. PubMed PMID: 33483496; PubMed Central PMCID: PMCPMC7822959. Epub 2021/ 01/24.
- Ruan X, Li P, Ma Y, et al. Identification of human long noncoding RNAs associated with nonalcoholic fatty liver disease and metabolic homeostasis. J Clin Invest. 2021;131(1). Epub 2020/ 10/14. DOI:10.1172/JCI136336. PubMed PMID: 33048844; PubMed Central PMCID: PMCPMC7773374.
- Trembinski DJ, Bink DI, Theodorou K, et al. Aging-regulated anti-apoptotic long non-coding RNA Sarrah augments recovery from acute myocardial infarction. Nat Commun. 2020;11(1):2039. PubMed PMID: 32341350; PubMed Central PMCID: PMCPMC7184724. Epub 2020/ 04/29.
- Lin RX, Yang SL, Jia Y, et al. Epigenetic regulation of papillary thyroid carcinoma by long non-coding RNAs. Semin Cancer Biol. 2021 PubMed PMID: 33785446. DOI:10.1016/j.semcancer.2021.03.027. Epub 2021/ 04/01.
- Shi J, Gao W, Shao F. Pyroptosis: gasdermin-mediated programmed necrotic cell death. Trends Biochem Sci. 2017;42(4):245–254. Epub 2016/ 12/10. PubMed PMID: 27932073.
- Miao EA, Rajan JV, Aderem A. Caspase-1-induced pyroptotic cell death. Immunol Rev. 2011;243(1):206–214. Epub 2011/ 09/03. PubMed PMID: 21884178; PubMed Central PMCID: PMCPMC3609431.
- Yang Y, Liu PY, Bao W, et al. Hydrogen inhibits endometrial cancer growth via a ROS/NLRP3/caspase-1/GSDMD-mediated pyroptotic pathway. BMC Cancer. 2020;20(1):28. Epub 2020/ 01/12. PubMed PMID: 31924176; PubMed Central PMCID: PMCPMC6954594.
- Zhang J, Jiang N, Zhang L, et al. NLRP6 expressed in astrocytes aggravates neurons injury after OGD/R through activating the inflammasome and inducing pyroptosis. Int Immunopharmacol. 2020;80:106183. Epub 2020/ 01/14. PubMed PMID: 31927506.
- Xu R, Yang J, Qian Y, et al. Ferroptosis/pyroptosis dual-inductive combinational anti-cancer therapy achieved by transferrin decorated nanoMOF. Nanoscale Horiz. 2021;6(4):348–356. PubMed PMID: 33687417. Epub 2021/ 03/10.
- Wang Y, Gao W, Shi X, et al. Chemotherapy drugs induce pyroptosis through caspase-3 cleavage of a gasdermin. Nature. 2017;547(7661):99–103. PubMed PMID: 28459430. Epub 2017/ 05/02.
- Woo Y, Lee HJ, Jung YM, et al. Regulated necrotic cell death in alternative tumor therapeutic strategies. Cells. 2020;9(12):2709. Epub 2020/ 12/23. PubMed PMID: 33348858; PubMed Central PMCID: PMCPMC7767016.
- Fang Y, Tian S, Pan Y, et al. Pyroptosis: a new frontier in cancer. Biomed Pharmacother. 2020;121:109595. Epub 2019/ 11/12. PubMed PMID: 31710896.
- Ruan J, Wang S, Wang J. Mechanism and regulation of pyroptosis-mediated in cancer cell death. Chem Biol Interact. 2020;323:109052. Epub 2020/ 03/15. PubMed PMID: 32169591.
- Derangere V, Chevriaux A, Courtaut F, et al. Liver X receptor β activation induces pyroptosis of human and murine colon cancer cells. Cell Death Differ. 2014;21(12):1914–1924. PubMed PMID: 25124554; PubMed Central PMCID: PMCPMC4227150. Epub 2014/ 08/16.
- Dupaul-Chicoine J, Arabzadeh A, Dagenais M, et al. The Nlrp3 inflammasome suppresses colorectal cancer metastatic growth in the liver by promoting natural killer cell tumoricidal activity. Immunity. 2015;43(4):751–763. PubMed PMID: 26384545. Epub 2015/ 09/20.
- Wang WJ, Chen D, Jiang MZ, et al. Downregulation of gasdermin D promotes gastric cancer proliferation by regulating cell cycle-related proteins. J Dig Dis. 2018;19(2):74–83. PubMed PMID: 29314754. Epub 2018/ 01/10.
- Wei Q, Mu K, Li T, et al. Deregulation of the NLRP3 inflammasome in hepatic parenchymal cells during liver cancer progression. Lab Invest. 2014;94(1):52–62. PubMed PMID: 24166187. Epub 2013/ 10/30.
- Chen YF, Qi HY, Wu FL. Euxanthone exhibits anti-proliferative and anti-invasive activities in hepatocellular carcinoma by inducing pyroptosis: preliminary results. Eur Rev Med Pharmacol Sci. 2018;22(23):8186–8196. Epub 2018/ 12/18. PubMed PMID: 30556857.
- Pizato N, Luzete BC, Kiffer L, et al. Omega-3 docosahexaenoic acid induces pyroptosis cell death in triple-negative breast cancer cells. Sci Rep. 2018;8(1):1952. PubMed PMID: 29386662; PubMed Central PMCID: PMCPMC5792438. Epub 2018/ 02/02.
- Kolb R, Kluz P, Tan ZW, et al. Obesity-associated inflammation promotes angiogenesis and breast cancer via angiopoietin-like 4. Oncogene. 2019;38(13):2351–2363. PubMed PMID: 30518876; PubMed Central PMCID: PMCPMC6440811. Epub 2018/ 12/07.
- Gao J, Qiu X, Xi G, et al. Downregulation of GSDMD attenuates tumor proliferation via the intrinsic mitochondrial apoptotic pathway and inhibition of EGFR/Akt signaling and predicts a good prognosis in nonsmall cell lung cancer. Oncol Rep. 2018;40(4):1971–1984. PubMed PMID: 30106450; PubMed Central PMCID: PMCPMC6111570. Epub 2018/ 08/15.
- Wang Y, Kong H, Zeng X, et al. Activation of NLRP3 inflammasome enhances the proliferation and migration of A549 lung cancer cells. Oncol Rep. 2016;35(4):2053–2064. PubMed PMID: 26782741. Epub 2016/ 01/20.
- Lin TY, Tsai MC, Tu W, et al. Role of the NLRP3 Inflammasome: insights Into Cancer Hallmarks. Front Immunol. 2020;11:610492. Epub 2021/ 02/23. PubMed PMID: 33613533; PubMed Central PMCID: PMCPMC7886802.
- Moossavi M, Parsamanesh N, Bahrami A, et al. Role of the NLRP3 inflammasome in cancer. Mol Cancer. 2018;17(1):158. Epub 2018/ 11/19. PubMed PMID: 30447690; PubMed Central PMCID: PMCPMC6240225.
- Teng JF, Mei QB, Zhou XG, et al. Polyphyllin VI induces caspase-1-Mediated Pyroptosis via the Induction of ROS/NF-κB/NLRP3/GSDMD signal axis in non-small cell lung cancer. Cancers (Basel). 2020;12(1):193. PubMed PMID: 31941010; PubMed Central PMCID: PMCPMC7017302. Epub 2020/ 01/17.
- Zhang Z, Zhang Y, Xia S, et al. Gasdermin E suppresses tumour growth by activating anti-tumour immunity. Nature. 2020;579(7799):415–420. PubMed PMID: 32188940; PubMed Central PMCID: PMCPMC7123794. Epub 2020/ 03/20.
- Dupaul-Chicoine J, Yeretssian G, Doiron K, et al. Control of intestinal homeostasis, colitis, and colitis-associated colorectal cancer by the inflammatory caspases. Immunity. 2010;32(3):367–378. PubMed PMID: 20226691. Epub 2010/ 03/17.
- Liu J, Yao L, Zhang M, et al. Downregulation of LncRNA-XIST inhibited development of non-small cell lung cancer by activating miR-335/SOD2/ROS signal pathway mediated pyroptotic cell death. Aging (Albany NY). 2019;11(18):7830–7846. Epub 2019/ 09/26. PubMed PMID: 31553952; PubMed Central PMCID: PMCPMC6781979.
- Ren N, Jiang T, Wang C, et al. LncRNA ADAMTS9-AS2 inhibits gastric cancer (GC) development and sensitizes chemoresistant GC cells to cisplatin by regulating miR-223-3p/NLRP3 axis. Aging (Albany NY). 2020;12(11):11025–11041. PubMed PMID: 32516127; PubMed Central PMCID: PMCPMC7346038. Epub 2020/ 06/10.
- Karki R, Kanneganti TD. Diverging inflammasome signals in tumorigenesis and potential targeting. Nat Rev Cancer. 2019;19(4):197–214. Epub 2019/ 03/08. PubMed PMID: 30842595; PubMed Central PMCID: PMCPMC6953422.
- Man SM, Kanneganti TD. Regulation of inflammasome activation. Immunol Rev. 2015;265(1):6–21. Epub 2015/ 04/17. PubMed PMID: 25879280; PubMed Central PMCID: PMCPMC4400844.
- Wallach D, Kang TB, Dillon CP, et al. Programmed necrosis in inflammation: toward identification of the effector molecules. Science. 2016;352(6281):aaf2154. Epub 2016/ 04/02. PubMed PMID: 27034377.
- Ritchie ME, Phipson B, Wu D, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43(7):e47. PubMed PMID: 25605792; PubMed Central PMCID: PMCPMC4402510. Epub 2015/ 01/22.
- Iasonos A, Schrag D, Raj GV, et al. How to build and interpret a nomogram for cancer prognosis. J Clin Oncol. 2008;26(8):1364–1370. Epub 2008/ 03/08. PubMed PMID: 18323559.
- Subramanian A, Tamayo P, Mootha VK, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005;102(43):15545–15550. PubMed PMID: 16199517; PubMed Central PMCID: PMCPMC1239896. Epub 2005/ 10/04.
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells. Cancer Res. 2017;77(21):e108–e10. PubMed PMID: 29092952; PubMed Central PMCID: PMCPMC6042652. Epub 2017/ 11/03.
- Mayakonda A, Lin DC, Assenov Y, et al. Maftools: efficient and comprehensive analysis of somatic variants in cancer. Genome Res. 2018;28(11):1747–1756. Epub 2018/ 10/21. PubMed PMID: 30341162; PubMed Central PMCID: PMCPMC6211645.
- Robinson DR, Wu YM, Lonigro RJ, et al. Integrative clinical genomics of metastatic cancer. Nature. 2017;548(7667):297–303. PubMed PMID: 28783718; PubMed Central PMCID: PMCPMC5995337. Epub 2017/ 08/08.
- Geeleher P, Cox N, Huang RS. pRRophetic: an R package for prediction of clinical chemotherapeutic response from tumor gene expression levels. PLoS One. 2014;9(9):e107468. Epub 2014/ 09/18. PubMed PMID: 25229481; PubMed Central PMCID: PMCPMC4167990.
- Samstein RM, Lee CH, Shoushtari AN, et al. Tumor mutational load predicts survival after immunotherapy across multiple cancer types. Nat Genet. 2019;51(2):202–206. PubMed PMID: 30643254; PubMed Central PMCID: PMCPMC6365097. Epub 2019/ 01/16.
- Tan C, Liu W, Zheng ZH, et al. LncRNA HOTTIP inhibits cell pyroptosis by targeting miR-148a-3p/AKT2 axis in ovarian cancer. Cell Biol Int. 2021;45(7):1487–1497. Epub 2021/ 03/13. PubMed PMID: 33710684.
- Chen Z, He M, Chen J, et al. Long non‑coding RNA SNHG7 inhibits NLRP3‑dependent pyroptosis by targeting the miR‑34a/SIRT1 axis in liver cancer. Oncol Lett. 2020;20(1):893–901. Epub 2020/ 06/23. PubMed PMID: 32566017; PubMed Central PMCID: PMCPMC7285900.
- Yan H, Luo B, Wu X, et al. Cisplatin induces pyroptosis via activation of MEG3/NLRP3/caspase-1/GSDMD pathway in triple-negative breast cancer. Int J Biol Sci. 2021;17(10):2606–2621. PubMed PMID: 34326697; PubMed Central PMCID: PMCPMC8315016. Epub 2021/ 07/31.
- Lin W, Chen Y, Wu B, et al. Identification of the pyroptosis‑related prognostic gene signature and the associated regulation axis in lung adenocarcinoma. Cell Death Discov. 2021;7(1):161. Epub 2021/ 07/07. PubMed PMID: 34226539; PubMed Central PMCID: PMCPMC8257680.
- Liu R, Ju C, Zhang F, et al. LncRNA GSEC promotes the proliferation, migration and invasion by sponging miR-588/ EIF5A2 axis in osteosarcoma. Biochem Biophys Res Commun. 2020;532(2):300–307. PubMed PMID: 32868080. Epub 2020/ 09/02.
- Matsumura K, Kawasaki Y, Miyamoto M, et al. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. 2017;36(9):1191–1199. PubMed PMID: 27797375. Epub 2016/ 11/01.
- Xiao G, Wang P, Zheng X, et al. FAM83A-AS1 promotes lung adenocarcinoma cell migration and invasion by targeting miR-150-5p and modifying MMP14. Cell Cycle. 2019;18(21):2972–2985. Epub 2019/ 09/17. PubMed PMID: 31522616; PubMed Central PMCID: PMCPMC6791711.
- Wang W, Zhao Z, Xu C, et al. LncRNA FAM83A-AS1 promotes lung adenocarcinoma progression by enhancing the pre-mRNA stability of FAM83A. Thorac Cancer. 2021. Epub 2021/ 03/10. PubMed PMID: 33687144. DOI:10.1111/1759-7714.13928
- Jia J, Li H, Chu J, et al. LncRNA FAM83A-AS1 promotes ESCC progression by regulating miR-214/CDC25B axis. J Cancer. 2021;12(4):1200–1211. PubMed PMID: 33442418; PubMed Central PMCID: PMCPMC7797654. Epub 2021/ 01/15.
- He J, Yu J. Long noncoding RNA FAM83A-AS1 facilitates hepatocellular carcinoma progression by binding with NOP58 to enhance the mRNA stability of FAM83A. Biosci Rep. 2019;39(11). Epub 2019/ 11/07. DOI:10.1042/BSR20192550 PubMed PMID: 31696213; PubMed Central PMCID: PMCPMC6851519.
- You J, Fang W, Zhao Q, et al. Identification of a RNA-Seq based prognostic signature with seven immune-related lncRNAs for lung adenocarcinoma. Clin Lab. 2021;67(3). Epub 2021/ 03/20. DOI:10.7754/Clin.Lab.2020.200663 PubMed PMID: 33739047.
- Wu L, Wen Z, Song Y, et al. A novel autophagy-related lncRNA survival model for lung adenocarcinoma. J Cell Mol Med. 2021;25(12):5681–5690. Epub 2021/ 05/15. PubMed PMID: 33987935; PubMed Central PMCID: PMCPMC8184679.
- Guo Y, Qu Z, Li D, et al. Identification of a prognostic ferroptosis-related lncRNA signature in the tumor microenvironment of lung adenocarcinoma. Cell Death Discov. 2021;7(1):190. PubMed PMID: 34312372; PubMed Central PMCID: PMCPMC8313561. Epub 2021/ 07/28.
- Jin D, Song Y, Chen Y, et al. Identification of a Seven-lncRNA immune risk signature and construction of a predictive nomogram for lung adenocarcinoma. Biomed Res Int. 2020;2020:7929132. Epub 2020/ 07/01. PubMed PMID: 32596372; PubMed Central PMCID: PMCPMC7273488.
- Porporato PE, Filigheddu N, Pedro JMB, et al. Mitochondrial metabolism and cancer. Cell Res. 2018;28(3):265–280. Epub 2017/ 12/09. PubMed PMID: 29219147; PubMed Central PMCID: PMCPMC5835768.
- Sobanski T, Rose M, Suraweera A, et al. Cell metabolism and DNA repair pathways: implications for cancer therapy. Front Cell Dev Biol. 2021;9:633305. Epub 2021/ 04/10. PubMed PMID: 33834022; PubMed Central PMCID: PMCPMC8021863.
- Ashton TM, McKenna WG, Kunz-Schughart LA, et al. Oxidative Phosphorylation as an Emerging Target in Cancer Therapy. Clin Cancer Res. 2018;24(11):2482–2490. Epub 2018/ 02/09. PubMed PMID: 29420223.
- Madan E, Parker TM, Pelham CJ, et al. HIF-transcribed p53 chaperones HIF-1α. Nucleic Acids Res. 2019;47(19):10212–10234. PubMed PMID: 31538203; PubMed Central PMCID: PMCPMC6821315. Epub 2019/ 09/21.
- Saxton RA, Sabatini DM. mTOR signaling in growth, metabolism, and disease. Cell. 2017;169(2):361–371. Epub 2017/ 04/08. PubMed PMID: 28388417.
- Tan AC. Targeting the PI3K/Akt/mTOR pathway in non-small cell lung cancer (NSCLC). Thorac Cancer. 2020;11(3):511–518. Epub 2020/ 01/29. PubMed PMID: 31989769; PubMed Central PMCID: PMCPMC7049515.
- Bremnes RM, Al-Shibli K, Donnem T, et al. The role of tumor-infiltrating immune cells and chronic inflammation at the tumor site on cancer development, progression, and prognosis: emphasis on non-small cell lung cancer. J Thorac Oncol. 2011;6(4):824–833. PubMed PMID: 21173711. Epub 2010/ 12/22.
- Remark R, Becker C, Gomez JE, et al. The Non–small cell lung cancer immune contexture. A major determinant of tumor characteristics and patient outcome. Am J Respir Crit Care Med. 2015;191(4):377–390. PubMed PMID: 25369536; PubMed Central PMCID: PMCPMC5447326. Epub 2014/ 11/05.