ABSTRACT
Long noncoding RNA small nucleolar RNA host gene 10 (SNHG10) has been suggested to function as tumor promoter in various human cancer types. Herein, the role of SNHG10 in colorectal cancer (CRC) was explored. Expression levels of genes in colorectal cancer tissues and cell lines were detected by Starbase and reverse transcription quantitative PCR (RT-qPCR) Cell Counting Kit-8 (CCK-8), the BrdU incorporation assay and Transwell assays were explored to study the function of SNHG10 in HCT116 and DXH-1 cells. In addition, the interaction of SNHG10 and miR-3690 was analyzed by dual-luciferase reporter assays. SNHG10 had a high expression level in CRC tissues and cell lines. Meanwhile, knockdown of SNHG10 reduced cell viability, inhibited cell proliferation and decreased cell migration and invasion. Moreover, bioinformatics analysis revealed that one potential target gene of SNHG10 was miR-3690. Dual-luciferase reporter assay confirmed that miR-3690 directly targeted SNHG10. Importantly, SNHG10 could decrease the expression of miR-3690 in HCT116 and DXH-1 cells. More importantly, the silencing of miR-3690 reversed the effect of the SNHG10 knockdown on the cell viability, proliferation, migration and invasion of HCT116 and DXH-1 cells. The present results demonstrated that SNHG10 promotes colorectal cancer cells the malignant progression by targeting miR-3690.
Abbreviations: CRC: Colorectal cancer; Lnc RNA: Long noncoding RNA; microRNAs: miRNAs/miRs; RT-qPCR: reverse transcription quantitative polymerase chain reaction; CCK-8: Cell Counting Kit-8
KEYWORDS:
Introduction
Colorectal cancer (CRC) is one of the most common malignant tumors and the second most common cause of death from malignant cancers all around the world [Citation1]. Meanwhile, in China, there were about ~388,000 cases of colorectal cancer and 187,000 colorectal cancer death cases were reported in 2015 [Citation2]. The development and promotion of colorectal malignancy is multifactor, involve an unhealthy diet, smoking, physical inactivity, high alcohol consumption, excess body weight and so on [Citation3]. However, the essential reason of colorectal malignancy is the abnormal expression of certain genes including protein coding genes [Citation4], microRNAs [Citation5] and long noncoding RNAs [Citation6].
Long noncoding RNAs (lncRNAs) are non-coding RNAs with a length of over 200 nucleotides that modulate genes expression by act as trans regulators scaffolds [Citation7] or microRNAs sponges [Citation8]. During the past few decades, a great amount of reports has shown that lncRNAs play some vital roles in cell proliferation [Citation9], differentiation [Citation10] or apoptosis [Citation11]. Numerous studies [Citation12–14] have provided clear evidence that lncRNAs play tumor-suppressive or oncogenic roles in human colorectal cancer and the abnormally expressed lncRNAs could serve as potential prognostic and diagnostic biomarkers for colorectal cancer, as well as potential therapeutic targets. The RNA-seq data and miRNA-seq data from The Cancer Genome Atlas (TCGA, https://tcga-data.nci.nih.gov/tcga/) showed that Long noncoding RNA small nucleolar RNA host gene 10 (SNHG10) was differentially expressed in lung adenocarcinoma [Citation15]. Then, SNHG10 was observed in several other tumors, for instance, SNHG10 was significantly up-regulation in hepatocellular carcinoma [Citation16] and bladder cancer [Citation17]. The investigation of the function and molecular mechanism showed SNHG10 sponges miR-543 to upregulate tumor suppressive SIRT1 to suppress cell proliferation in non-small cell lung cancer [Citation18].
In this study, we checked the expression of SNHG10 in colorectal cancer using ENSEMBL, and revealed the function of SNHG10 in the pathogenesis of colorectal cancer. Our findings showed that knockdown SNHG10 reduced cell viability, inhibited cell proliferation and decreased cell migration and invasion in HCT116 and DXH-1 cells. Moreover, SNHG10 down-regulated miR-3690 expression by direct interaction. Meanwhile, the silencing of miR-3690 reversed the effect of the SNHG10 knockdown on the cell viability, proliferation, migration and invasion of HCT116 and DXH-1 cells. These results indicated the role of SNHG10 as a potential therapeutic target in CRC.
Materials and methods
Cell culture and transfections
The human colorectal cancer cell lines, HCT116 and DXH-1 cells along with the normal fetal human colon epithelial cell line FHC were bought from Procell Life Science&Technology Co.,Ltd. HCT116 and DXH-1 cells were maintained in McCoy’s 5A (PM150710A, ThermoFly) supplemented with 10% fetal bovine serum (FBS; S9020; Solarbio), 1% Penicillin-Streptomycin Liquid (P/S; P1400; Solarbio). FHC cells were maintained in Dulbecco’s modified Eagle’s medium (DMEM; 11,995; Solarbio) supplemented with 10% FBS and 1% P/S. All the cells were cultured in a humidified incubator with 5% CO2 at 37°C. Cells were harvested at 85%–95% confluence to perform the following experiments.
MiR-3690 mimic, miR-3690 inhibitor and negative control (NC) miRNA mimic were synthesized in Shanghai GenePharma Co., Ltd. (Shanghai, China). The SNHG10 expression vector was constructed by cloning the fragment of SNHG10 gene (NCBI Reference Sequence: NR_003138.3) into pcDNA3.1 plasmid. Cell transfection was performed on HCT116 and DXH-1 cells with the lipofectamine 3000 reagent (L3000008; Invitrogen; Thermo Fisher Scientific, Inc.) at the ratio of 10 nM vector and/or 40 nM miRNA per 106 cells. The following experiments were performed at 24 hours after transfections.
Rreal-time quantitative PCR (RT-qPCR)
Total RNA from HCT116, DXH-1 and FHC cells was isolated by Trizol® reagent (Thermo Fisher Scientific, Inc.). The total RNA was reverse-transcripted into cDNA with PrimeScript™ RT reagent Kit (RR037Q; Takara). For lncRNA and miRNA express measure, real-time quantitative PCR analysis was performed with Mir-X miRNA qRT-PCR TB Green® Kit (638,314; Clontech). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and U6 were used as normalization controls. 2−ΔΔCt method was used for gene expression quantify [Citation18]. The primers information provided as follows:
SNHG10-Forward: 5ʹ- CCAGCTTAGATTCATTGATTCC-3ʹ;
SNHG10-Reverse: 5ʹ- TTAAGTGCACCAGATGCTG-3ʹ;
miRNA-3690-Forward: 5ʹ- AGAGGATACCCTTTGTATG-3ʹ;
miRNA-3690- Reverse: 5ʹ- CAGTGCGTGTCGTGGAGT-3ʹ;
U6-Forward: 5ʹ- CTCGCTTCGGCAGCACA-3ʹ;
U6-Reverse: 5ʹ- AACGCTTCACGAATTTGCGT-3ʹ;
GAPDH-Forward: 5ʹ- GAACGGGAAGCTCACTGG-3ʹ;
GAPDH-Reverse: 5ʹ- GCCTGCTTCACCACCTTCT-3ʹ.
All of the primers were synthesized from GenePharma (Shanghai, China).
Cell viability assay
The Cell Counting Kit-8 (CCK-8; Beyotime Institute of Biotechnology) colorimetric assay was used to measure cell viability according to manufacturer’s instruction. Briefly, 12 hours before transfection, HCT116 and DXH-1 cells were seeded into 96-well plates at a density of 1 × 104 cells/well. 48 hours after transfection by the lipofectamine 3000 reagent, the cell supernatant was removed, and the cells were incubated with 100 µl/well CCK-8 solution for 3 hours at 37°C. The optical density value at 450 nm was measured by a fluorescence spectrophotometer (F-7000; Hitachi-Hightech). Cell viability was assessed by the values constructed as growth curve.
Colony proliferation assay
According to manufacturer’s instruction, the cell proliferation was measured using Cell Proliferation ELISA BrdU (colorimetric) kit (11,647,229,001; Roche). Briefly as describe, HCT116 or DXH-1 cells were seeded into 96-well plates at a density of 1 × 104 cells/well. 48 hours after transfection, the cells were incubated with BrdU-labeling solution (100 μM) for 4 hours at 37°C. The absorbance was measured at 450 nm by a fluorescence spectrophotometer (F-7000; Hitachi-Hightech).
Transwell assay
A transwell assay was performed to evaluate the ability of CRC cells to migration and invade according to the previously described [Citation19]. HCT116 and DXH-1 cells (1 × 105) were seeded in the upper chamber containing McCoy’s 5A. Meanwhile, the lower chamber was filled with complete culture medium. About 48 hours after transfection, cells in the lower chambers were incubated with 4% paraformaldehyde for 30 minutes at 4°C. After that, the cells were stained with 5% crystal violet for 30 minutes at 4°C. Finally, cell colonies were counted via photographing under a light microscope.
Dual-luciferase reporter assay
Dual-luciferase reporter assay (Promega Corporation) was performed to study the interaction between SNHG10 and miR-3690 according to the previously described [Citation20]. The Dual-Luciferase system was generated by inserting the cDNA fragments of the wild-type and mutant SNHG10 into pmirGLO Dual-Luciferase miRNA Target Expression Vectors (Promega, Madison, WI, USA). The wild-type and mutant SNHG10 reporter plasmids were co-transfected into HCT116 and DXH-1 cells along with miR-3690 mimic or miR-NC, respectively. Meanwhile, Renilla luciferase expression plasmid was transfected into the HCT116 and DXH-1 cells as reference control. About 48 hours after transfection, the cells were lysed and collected to measure Firefly and Renilla luciferase activities by dual luciferase reporter assays. Finally, the relative luciferase activity of reporter plasmids was calculated with the ratio of Firefly and Renilla luciferase activities.
Online analysis
The profiling expression data of SNHG10 and miR-3690 in colorectal cancer and tumor-adjacent normal tissues in ENSEMBL (http://asia.ensembl.org/). The prediction of SNHG10 and miR-3690 interaction was performed by the starBase of ENCORI (http://starbase.sysu.edu.cn/).
Statistical analysis
All experiments were performed at least in triplicate. The statistical analysis was performed by GraphPad Prism (version 6.01 for Windows; GraphPad Software, Inc.) statistical software. The differences between two groups were measured by student t-tests. P < 0.01 was considered as statistically significant difference.
Results
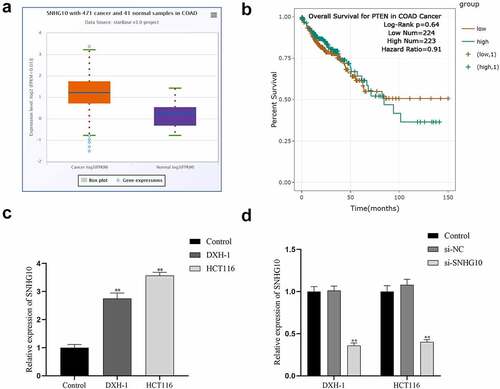
Upregulation of SNHG10 in colorectal cancer To confirm whether SNHG10 participated in CRC progression, we analyzed the profiling expression data of SNHG10 (ENSG00000247092.6) in colorectal cancer and tumor-adjacent normal tissues in ENSEMBL (http://asia.ensembl.org/). The results showed SNHG10 was dramatically up-regulated in colorectal cancer samples compared to normal control samples (). Besides that, Patients with high expression of SNHG10 showed a significantly poorer survival than those with low expression of SNHG10 (). Meanwhile, RT-qPCR was used to measure the expression level of SNHG10 in HCT116, DXH-1 and FHC cells. The results indicated that SNHG10 expressed significantly higher level in HCT116, DXH-1 cells than that in FHC cells (P < 0.01; ).
Figure 1. SNHG10 is up-regulated in colorectal cancer. (a) Starbase analysis of the expression of SNHG10 in human colorectal cancer and tumor-adjacent normal tissues in the database. (b) The survival rate of patients with high and low expression of SNHG10. (c) RT-qPCR was used to assess the expression of SNHG10 in different human colorectal cancer (HCT116 and DXH-1 cells) and the normal fetal human colon epithelial cell line FHC cells. (d) HCT116 and DXH-1 cells were transfected with si-SNHG10 or NC. The results are representative of three independent experiments. The data are presented as the mean ± SD. n = 3 for each group. **P < 0.01 vs. si-NC group
Knockdown of SNHG10 inhibits cell progression of HCT116 and DXH-1 cells
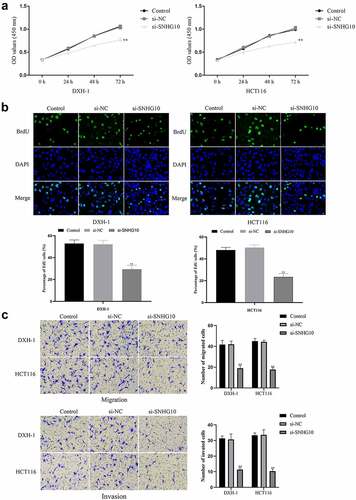
To evaluate the effect of SNHG10 in CRC progression, SNHG10 or NC siRNA were transfected into HCT116 and DXH-1 cells. The data from RT-qPCR showed that SNHG10 siRNA could significantly down-regulate SNHG10 in HCT116 and DXH-1 cells (P < 0.01; ). Firstly, the CCK-8 assay showed that si-SNHG10 could reduce the viability of HCT116 and DXH-1 cells (P < 0.05; ). Then, the BrdU incorporation assay found that si-SNHG10 reduced the proliferation of HCT116 and DXH-1 cells (P < 0.01; ). In addition, Transwell assays was performed to further investigate the effect of SNHG10 on the migration and invasion in CRC cells. The result of Transwell assay demonstrated that si-SNHG10 inhibited cell migration and invasion in HCT116 and DXH-1 cells (P < 0.001; ). In combination, these data proved that knockdown of SNHG10 expression could inhibit cell proliferation, migration and invasion in CRC.
Figure 2. Knockdown of SNHG10 inhibits cell progression of HCT116 and DXH-1 cells. (a) Cell Counting Kit-8 assay was explored to measure cell viability. (b) The BrdU incorporation assay was performed to assess cell proliferation. Magnification ×200. (c) Transwell assay was performed to assess cell migration. The results were representative of three independent experiments. The results were representative of three independent experiments. Magnification ×200. The data are presented as the mean ± SD. n = 3 for each group. **P < 0.01 vs. control group
SNHG10 reduces expression of miR-3690 by interaction
The mechanism studies of LncRNAs showed that lncRNAs could regulate genes expression as miRNA sponges [Citation21,Citation22]. Therefore, the potential target miRNAs of SNHG10 were predicted by bioinformatics analysis. Interestingly, the results of bioinformatics analysis showed that one of the target miRNAs of SNHG10 was miR-3690 (). Next, dual-luciferase reporter assay was performed to verify this prediction. The data of dual-luciferase reporter assay showed that the luciferase activity of SNHG10-WT reporter vector was significantly inhibited by miR-3690 mimic (). However, when the predicted binding site in SNHG10 was mutated, the inhibitory effect of miR-3690 mimic to the SNHG10 reporter vector’s luciferase activity was abrogated (). Meanwhile, using ENSEMBLanalysis the profiling the expression data of miR-3690 (ENSG00000265658) in colorectal cancer and tumor-adjacent normal tissues in the database, we found that miR-3690 was significantly down-regulated in colorectal cancer samples compared to normal control samples (). Besides that, the detection of RT-qPCR indicated that the expression of miR-3690 was significantly down-regulated in HCT116 and DXH-1 cells compared to FHC cells (). In addition, our results show that the expression levels of miR-3690 in HCT116 and DXH-1 cells transfected with si-SNHG10 were upregulated, and the downregulation of miR-3690 in HCT116 and DXH-1 cells transfected with miR-3690 inhibitor was reversed by co-transfected si-SNHG10 in (). Taken together, all of the above results indicate that SNHG10 interacts with miR-3690 and regulates its expression in human CRC cells.
Figure 3. miR-3690 serves as a direct SNHG10 target in HCT116 and DXH-1 cells. (a) The putative binding sites between miR-3690 and SNHG10 were predicted by StarBase. (b) Dual-luciferase reporter assay was performed to reveal the relationship between miR-3690 and SNHG10. (c) Starbase analysis of the expression of miR-3690 in human colorectal cancer and tumor-adjacent normal tissues in the database. (d) RT-qPCR was performed to assess the mRNA expression of miR-3690 in CRC cell lines. The results were representative of three independent experiments. The data are presented as the mean ± SD. **P < 0.01 vs. control group
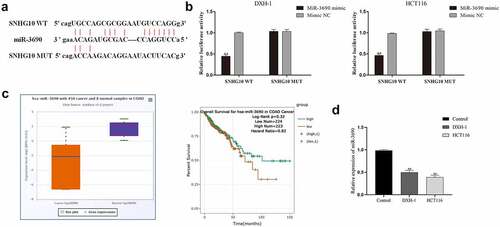
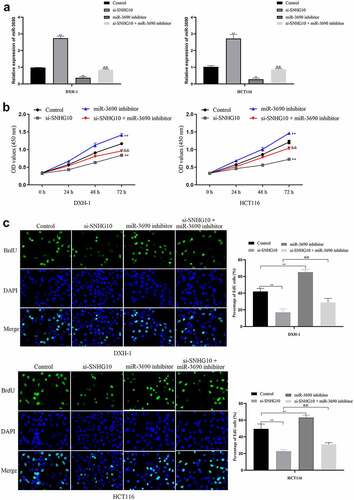
Figure 4. miR-3690 downregulation reverses the effect of SNHG10 downregulation in HCT116 and DXH-1 cells. (a) HCT116 and DXH-1 cells were co-transfected miR-3690 inhibitor along with si-SNHG10, respectively. (b) Cell Counting Kit-8 assay was explored to measure cell viability. (c) the BrdU incorporation assay was performed to assess cell proliferation. Magnification ×200. The data are presented as the mean ± SD. n = 3 for each group. **P < 0.01 vs. control group, &P < 0.05, &&P < 0.05 vs. si- SNHG10 group
miR-3690 overexpression reverses the effect of SNHG10 in HCT116 and DXH-1 cells
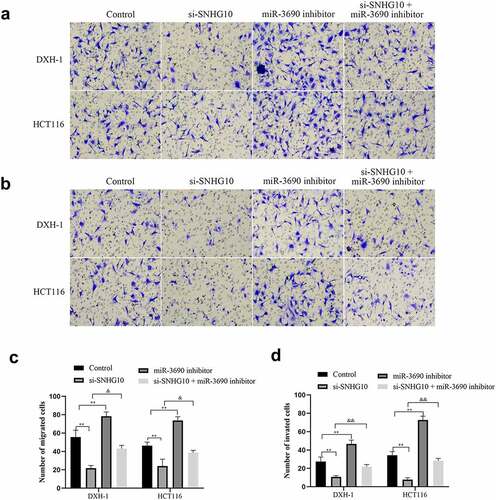
To confirm the interaction of SNHG10 and miR-3690, miR-3690 inhibitor or NC were co-transfected into HCT116 and DXH-1 cells with si-SNHG10, respectively. The data from CCK-8 assay showed that the viability of HCT116 and DXH-1 cells transfection of miR-3690 inhibitor was enhanced compared with the NC cells group, while the viability of cells co-transfection of miR-3690 inhibitor and si-SNHG10 was enhanced compared with the cells co-transfection of si-SNHG10 (P < 0.05; ). Then, the BrdU incorporation assay found that miR-3690 inhibitor increased the proliferation of HCT116 and DXH-1 cells. As the proliferation of HCT116 and DXH-1 cells transfected miR-3690 inhibitor was higher than the control cells, meanwhile, the proliferation of cells co-transfection of miR-3690 inhibitor and si-SNHG10 was higher compared with the cells transfection of si-SNHG10 (P < 0.01; ). Besides that, the results of Transwell assays demonstrated that miR-3690 inhibitor reversed the effect of si-SNHG10 in HCT116 and DXH-1 cells migration and invasion ability. As the number of HCT116 and DXH-1 cells transfected si-SNHG10 migrated and invaded the chamber was lower than the control cells, while the migration and invasion of the cells co-transfected si-SNHG10 and miR-3690 inhibitor was higher than the cells co-transfected si-SNHG10 (P < 0.001; ). All of these results suggested that silenced of miR-3690 partially reversed the effect of SNHG10 down-regulation on CRC.
Figure 5. miR-3690 downregulation reverses the effect of SNHG10 in HCT116 and DXH-1 cells. (a, b) Transwell assay was performed to assess cell migration and invasion. The results were representative of three independent experiments. Magnification ×200. The data are presented as the mean ± SD. n = 3 for each group. **P < 0.01 vs. control group, &P < 0.05, &&P < 0.05 vs. si- SNHG10 group
Discussion
Colorectal cancer is the third most common cause of cancer in men and women in the world [Citation23]. To reduce the harm of colorectal cancer, prevention strategies such as eating habits improvement, high-quality screening and diagnosis have been suggested [Citation24,Citation25]. Since colorectal cancer is a multifactorial disease, studies aiming to reveal the molecular changes associated with this type of cancer can provide potential preventative and therapeutic strategies for this disease [Citation26–28]. In colorectal cancer, a few lncRNAs have been identified as potential diagnostic and therapeutic targets [Citation29,Citation30]. In present study, we found that SNHG10 expression was up-regulated in colorectal cancer, and downregulation of SNHG10 inhibited the ability of HCT116 and DXH-1 cells proliferation, migration and invasion. This preliminary study indicated that SNHG10 might be a promising candidate in colorectal cancer treatment.
Increasing evidence indicates that lncRNAs are abnormal expression and plays important roles in human cancers progression and metastasis [Citation9,Citation31,Citation32]. In colorectal cancer, several lncRNAs have been identified with high prognostic value. For examples, serum lncRNA NEAT1 was reported to have strong diagnostic values for predicting the existence of human colorectal cancer, as the CRC, especially in metastatic CRC were detected higher expression levels of serum NEAT1 than colorectal adenoma and healthy controls. Meanwhile, the CRC with poor clinical outcome was significantly associated with high expression level of serum NEAT1 [Citation33]. Meanwhile, in non-small cell lung cancer, lncRNA MAGI2-AS3 was detected with lower expression level which may be a sponge of miR-25 [Citation34]. The study of lncRNA SNHG10 in cancers progression and metastasis has been reported in several kinds of human tumors [Citation18,Citation20,Citation35,Citation36]. These reports showed that the expression level of SNHG10 in different tumors was diverse. In gastric cancer, SNHG10 was up-regulated in gastric cancer cells, and SNHG10 promoted the gastric cancer cell proliferation and migration [Citation37]. However, SNHG10 was down-regulated in non-small cell lung cancer (NSCLC) compared with non-tumor tissues, and overexpression of SNHG10 promoted cell proliferation [Citation38]. In present study, we demonstrated that expression of lncRNA SNHG10 was up-regulated in colorectal cancer samples and HCT116 and DXH-1 cells compared to normal control samples and FHC cells. Besides that, knockdown SNHG10 could inhibit the ability of HCT116 and DXH-1 cells proliferation, migration and invasion.
Increasing evidence suggests that lncRNAs could play as trans regulators scaffolds [Citation7] or microRNAs sponges [Citation8]. LncRNA SNHG10 has been reported dual-faced effects on cancer progress via different microRNAs in different human tumor [Citation18,Citation37,Citation39]. SNHG10 was reported as oncogenic functions in glioma via targeting miR-532-3p/FBXL19 axis [Citation39]. Meanwhile, in non-small cell lung cancer, SNHG10 was reported suppress cancer cell proliferation by upregulating tumor suppressive SIRT1 as miR-543 sponge [Citation18]. Here, bioinformatics analysis identified miR-3690 as the target miRNAs of SNHG10, as they have complementary binding sites, and the direct binding relationship was validated with the luciferase assay. Besides that, si-SNHG10 could up-regulate the expression of miR-3690 by interaction in HCT116 and DXH-1 cells.
MiR-3690 has been studied as potential biomarker and therapeutic target for human thyroid cancer, as MiR-3690 promoted thyroid cancer cell proliferation and cell cycle progression by altering DKK3 expression [Citation40]. In this study, miR-3690 was found be down-regulated in colorectal cancer samples and HCT116 and DXH-1 cells compared to normal control samples and FHC cells. Knockdown miR-3690 could promote HCT116 and DXH-1 cells proliferation, migration and invasion. Besides that, the knockdown the expression of miR-3690 could reverse the effect of SNHG10 down-expression.
Regrettably, our experiment did not analyzed the expression of SNHG10 and miR-3690 in a cohort of patients affected by colorectal cancer. Our results demonstrated the effect of SNHG10 on promotes colorectal cancer cells malignant progression by targeting miR-3690 in vitro. In the future research, we will focus on the underlying mechanisms of miR-3690 regulate mRNAs involved in cancer cells malignant progression. As there are little reports about the function of miR-3690 in cancer.
Conclusion
The present study provides evidence that knockdown of lncRNA SNHG10 could upregulate the expression of miR-3690 to inhibit the malignant progression of colorectal cancer cells. Our data suggests that SNHG10/miR-3690 signal axis may provide a new therapeutic target for colorectal cancer prevention.
Highlights
SNHG10 had a high expression level in CRC tissues and cell lines.
Knockdown of SNHG10 reduced cell viability, inhibited cell proliferation and decreased cell migration and invasion in HCT116 and DXH-1 cells.
SNHG10 promotes colorectal cancer cells the malignant progression by targeting miR-3690.
Disclosure statement
No potential conflict of interest was reported by the author(s).
References
- Zheng Y, Wang ZZ. Interpretation of global colorectal cancer statistics. Zhonghua Liu Xing Bing Xue Za Zhi. 2021;42(1):149–152.
- Zheng RS, Sun KX, Zhang SW, et al. Report of cancer epidemiology in China, 2015. Zhonghua Zhong Liu Za Zhi. 2019;41(1):19–28.
- Mizuno S, Seishima R, Okabayashi K, et al. Sarcopenic obesity is a postoperative prognostic factor for stage II and III colorectal cancer. J Gastrointest Surg. 2021. DOI:10.1007/s11605-021-04965-8
- Wu M, Wang J, Tang W, et al. FOXK1 interaction with FHL2 promotes proliferation, invasion and metastasis in colorectal cancer. Oncogenesis. 2016;5(11):e271.
- Liu DR, Guan QL, Gao MT, et al. miR-1260b is a potential prognostic biomarker in colorectal cancer. Med Sci Monit. 2016;22:2417–2423.
- Zhang Z, Zhou C, Chang Y, et al. Long non-coding RNA CASC11 interacts with hnRNP-K and activates the WNT/beta-catenin pathway to promote growth and metastasis in colorectal cancer. Cancer Lett. 2016;376(1):62–73.
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6(7):784–801.
- Zhang CY, Chen CY, Wen HX, et al. miR-182-5p enhances cisplatin resistance in epithelial ovarian cancer by downregulating GRB2. EUROPEAN JOURNAL OF GYNAECOLOGICALONCOLOGY.
- Xu C, Tian LH. LncRNA XIST promotes proliferation and epithelial-mesenchymal transition of retinoblastoma cells through sponge action of miR-142-5p. Eur Rev Med Pharmacol Sci. 2020;24(18):9256–9264.
- Zhang ZK, Li J, Guan D, et al. A newly identified lncRNA MAR1 acts as a miR-487b sponge to promote skeletal muscle differentiation and regeneration. J Cachexia Sarcopenia Muscle. 2018;9(3):613–626.
- Lu C, Li Z, Hu S, et al. LncRNA PART-1 targets TGFBR2/Smad3 to regulate cell viability and apoptosis of chondrocytes via acting as miR-590-3p sponge in osteoarthritis. J Cell Mol Med. 2019;23(12):8196–8205.
- Hu H, Shu M, He L, et al. Epigenomic landscape of 5-hydroxymethylcytosine reveals its transcriptional regulation of lncRNAs in colorectal cancer. Br J Cancer. 2017;116(5):658–668.
- Wang J, Du S, Wang J, et al. The prognostic value of abnormally expressed lncRNAs in colorectal cancer: a meta-analysis. PLoS One. 2017;12(6):e0179670.
- Tian Y, Xu Y, Wang H, et al. Comprehensive analysis of microarray expression profiles of circRNAs and lncRNAs with associated co-expression networks in human colorectal cancer. Funct Integr Genomics. 2019;19(2):311–327.
- Li DS, Ainiwaer JL, Sheyhiding I, et al. Identification of key long non-coding RNAs as competing endogenous RNAs for miRNA-mRNA in lung adenocarcinoma. Eur Rev Med Pharmacol Sci. 2016;20(11):2285–2295.
- Zhu Q, Yang H, Cheng P, et al. Bioinformatic analysis of the prognostic value of the lncRNAs encoding snoRNAs in hepatocellular carcinoma. Biofactors. 2019;45(2):244–252.
- Jiang B, Hailong S, Yuan J, et al. Identification of oncogenic long noncoding RNA SNHG12 and DUXAP8 in human bladder cancer through a comprehensive profiling analysis. Biomed Pharmacother. 2018;108:500–507.
- Zhang Z, Nong L, Chen ML, et al. Long noncoding RNA SNHG10 Sponges miR-543 to upregulate tumor suppressive SIRT1 in nonsmall cell lung cancer. Cancer Biother Radiopharm. 2020;35(10):771–775.
- Liao Z, Zhang H, Su C, et al. Long noncoding RNA SNHG14 promotes hepatocellular carcinoma progression by regulating miR-876-5p/SSR2 axis. J Exp Clin Cancer Res. 2021;40(1):36.
- Zhu S, Liu Y, Wang X, et al. lncRNA SNHG10 promotes the proliferation and invasion of osteosarcoma via Wnt/beta-catenin signaling. Mol Ther Nucleic Acids. 2020;22:957–970.
- Zhang H, Wei N, Zhang W, et al. lncRNA SNHG3 promotes breast cancer progression by acting as a miR326 sponge. Oncol Rep. 2020;44(4):1502–1510.
- Zhu B, Liu J, Zhao Y, et al. lncRNA-SNHG14 promotes atherosclerosis by regulating RORalpha expression through sponge miR-19a-3p. Comput Math Methods Med. 2020;2020:3128053.
- Sterpetti AV, Costi U, D’Ermo G. National statistics about resection of the primary tumor in asymptomatic patients with Stage IV colorectal cancer and unresectable metastases. Need for improvement in data collection. A systematic review with meta-analysis. Surg Oncol. 2020;33:11–18.
- Zhou E, Rifkin S. Colorectal cancer and diet: risk versus prevention, is diet an intervention? Gastroenterol Clin North Am. 2021;50(1):101–111.
- Van Blarigan EL, Meyerhardt JA. Role of physical activity and diet after colorectal cancer diagnosis. J Clin Oncol. 2015;33(16):1825–1834.
- Vekic B, Dragojevic-Simic V, Jakovljevic M, et al. A correlation study of the colorectal cancer statistics and economic indicators in selected Balkan countries. Front Public Health. 2020;8:29.
- Zhou W, Yang W, Duan L, et al. MicroRNA-483 functions as an oncogene in colorectal cancer. Ann Clin Lab Sci. 2021;51(1):30–37.
- Sasaki T, Shigeta K, Okabayashi K, et al. Horizontal spread of pericolic lymph node metastasis as a prognostic factor for recurrence in stage III colorectal cancer. Colorectal Dis. 2021. DOI:10.1111/codi.15586.
- Zhang J, Li K, Zheng H, et al. Research progress review on long non-coding RNA in colorectal cancer. Neoplasma. 2021;68(2):240–252.
- Liao Z, Nie H, Wang Y, et al. The emerging landscape of long non-coding RNAs in colorectal cancer metastasis. Front Oncol. 2021;11:641343.
- Zheng Z, Li X, You H, et al. LncRNA SOCS2-AS1 inhibits progression and metastasis of colorectal cancer through stabilizing SOCS2 and sponging miR-1264. Aging (Albany NY). 2020;12(11):10517–10526.
- Zhang Y, Huang C, Zhu Z, et al. lncRNA NEAT1 regulates the proliferation and migration of hepatocellular carcinoma cells by acting as a miR320a molecular sponge and targeting L antigen family member 3. Int J Oncol. 2020;57(4):1001–1012.
- Wang Y, Zhang D, Zhang C, et al. The diagnostic and prognostic value of serum lncRNA NEAT1 in colorectal cancer. Cancer Manag Res. 2020;12:10985–10992.
- Sui Y, Chi W, Feng L, et al. LncRNA MAGI2-AS3 is downregulated in non-small cell lung cancer and may be a sponge of miR-25. BMC Pulm Med. 2020;20(1):59.
- Xiao S, Zha Y, Zhu H. miR-621 may suppress cell proliferation via targeting lncRNA SNHG10 in acute myeloid leukemia. Cancer Manag Res. 2021;13:2117–2123.
- Zhang Y, Guo H, Zhang H. SNHG10/DDX54/PBX3 feedback loop contributes to gastric cancer cell growth. Dig Dis Sci. 2020; 66(6):1875-1884.
- Yuan X, Yang T, Xu Y, et al. SNHG10 promotes cell proliferation and migration in gastric cancer by targeting miR-495-3p/CTNNB1 axis. Dig Dis Sci. 2020; 66(8):2627-2636.
- Liang M, Wang L, Cao C, et al. LncRNA SNHG10 is downregulated in non-small cell lung cancer and predicts poor survival. BMC Pulm Med. 2020;20(1):273.
- Jin L, Huang S, Guan C, et al. ETS1-activated SNHG10 exerts oncogenic functions in glioma via targeting miR-532-3p/FBXL19 axis. Cancer Cell Int. 2020;20(1):589.
- Shen F, Gan XX, Deng XY, et al. MicroRNA-3690 promotes cell proliferation and cell cycle progression by altering DKK3 expression in human thyroid cancer. Oncol Lett. 2020;20(5):223.
