ABSTRACT
MicroRNA-451a (miR-451a) has been implicated in the initiation and progression of multiple cancers. However, the regulatory mechanisms underlying its function in nasopharyngeal carcinoma (NPC) are poorly understood. Thus, we investigated in detail the role of the microRNA-451a/chromosome segregation 1-like (miR-45a/CSE1L) axis and its regulatory mechanism in NPC. We examined the levels of miR-451a and CSE1L in NPC, and assessed the effects of miR-451a and CSE1L on NPC by cell functional experiments. Furthermore, we elucidated the direct regulatory effect of miR-451a on CSE1L by the luciferase reporter assay, RNA pull-down assay, and RNA immunoprecipitation and validated our observations by calculating the Pearson’s correlation coefficient. We found that miR-451a was down-regulated in NPC cells, and its over-expression attenuated cell proliferation, migration, and invasion, and tumor growth in 5–8 F and SUNE-1 cells and promoted apoptosis. Moreover, CSE1L was the direct gene target of miR-451a, and its over-expression abrogated miR-451a-dependent inhibition of malignancy in 5–8 F and SUNE-1 cells. The Pearson’s correlation coefficient indicated a negative correlation between CSE1L and miR-451a. miR-451a serves as a tumor suppressor and targets CSE1L. miR-451a suppresses CSE1L expression, thereby reducing proliferation, invasion, and migration and increasing apoptosis of NPC cells.
Background
Nasopharyngeal carcinoma (NPC) originates from nasopharyngeal epithelial cells and is characterized by racial and geographical disparities [Citation1]. This unbalanced distribution mainly results from genetic susceptibility, viral infection, and lifestyle risks [Citation2–4]. An estimated 129,000 new NPC cases were reported worldwide in 2019 [Citation5]. Despite great improvements in treatment approaches, patients are frequently diagnosed with NPC at an advanced stage, when the cancer is highly aggressive, thereby reducing their chances of survival. Thus, better insights into the genetic heterogeneity and tumor biology of NPC might facilitate timely diagnosis and therapy in the future.
MicroRNAs (miRNAs) are approximately 20-nucleotide long single-stranded RNA molecules [Citation6]. Although they do not encode proteins, they can regulate gene expression by degrading or suppressing the translation of target mRNAs. Thus, miRNAs can alter several cellular functions, leading to the development of various diseases. Moreover, compelling evidences have indicated that miRNAs have potential functions in pan-cancers, including NPC [Citation7,Citation8]. For example, over-expression of miR-330-3p in ovarian cancer cells inhibits cell proliferation, migration, and invasion and halts ovarian cancer progression [Citation9]. On the other hand, miR-1269b in gastric cancer is linked to tumor enlargement and lymph node metastasis, and inhibits malignancy in gastric cancer cells [Citation10]. The tumor suppressor miR-3188 suppresses cell proliferation and sensitizes NPC cells to chemotherapeutics [Citation11]. Reportedly, miR-451a has tumor suppressor activities in various cancers; for instance, it sensitizes lung tumors to chemotherapy and inhibits epithelial-to-mesenchymal cell transition in lung cancer [Citation12]. Moreover, miR-451a inhibits cell proliferation and migration and in vitro cell invasion in papillary thyroid carcinoma [Citation13]. Remarkably, miR-451a suppresses tumorigenesis by regulating cell apoptosis and angiogenesis in hepatocellular carcinoma [Citation14]. However, the biological significance of miR-451a in NPC progression has not been investigated yet.
Chromosome segregation 1-like (CSE1L) encodes a cellular apoptosis susceptibility protein and orchestrates multiple biological processes, such as apoptosis, cell proliferation, and chromosome alignment and congression [Citation15]. As CSE1L regulates mitosis and cell survival, it is considered to be a putative oncogene and is highly amplified in pan-cancers [Citation16–18]. Furthermore, previous studies have revealed that high expression of CSE1L positively correlates with tumor invasion and distant metastasis in different cancers [Citation18–20]. Additionally, amplification of the CSE1L oncogene has been detected in NPC tissues and is associated with poor patient prognosis [Citation21,Citation22]. However, its function in NPC remains unclear.
In the current study, we aim to understand the role of miR-451a in propagating malignancy in NPC. Therefore, we have used bioinformatics analysis to reveal key roles of miR-451a and CSE1L in NPC. In addition, we have examined the potential regulatory functions of miRNA-451a via CSE1L in malignant NPC cells. Our findings highlight the potential of miR-451a/CSE1L as a novel target for treating patients with NPC.
Methods
Bioinformatics analysis
We retrieved the GSE118613 dataset from Gene Expression Omnibus (GEO), containing miRNA profiles obtained from NPC tumor and non-tumor samples. Subsequently, we screened these profiles to identify the miRNAs that were down-regulated in NPC with parameters kept as P < 0.05 and logFC≤-1.5. Additionally, we retrieved the GSE12452 dataset from GEO, consisting of mRNA profiles obtained from NPC tumor and non-tumor samples. We also screened these profiles to identify mRNAs that were up-regulated in NPC and kept the parameters as P < 0.01 and logFC ≥ 1.5. Target genes of miR-451a were predicted using the starBase algorithm (http://starbase.sysu.edu.cn/).
Human tissue samples and cell culture conditions
We acquired 32 pairs of tumor tissues and normal adjacent tissues from 32 NPC patients at our hospital. All patients were diagnosed with NPC by magnetic resonance imaging and endoscopy. Signed informed consent was obtained from all patients, and our study was approved by the ethics committee of our hospital. Clinical characteristics of these patients are listed in .
Table 1. Association of miR-451a and clinicopathological features in patients with NPC
We preserved the tumor tissues at −80°C before performing RNA extraction. Human NPC cell lines, 6–10B, 5–8 F, and SUNE-1, and the human immortalized nasopharyngeal epithelial cell line NP69 were obtained from Shanghai Kenqiang Company (China). All cells were maintained in Dulbecco’s modified Eagle’s medium containing 10% fetal bovine serum and 1% penicillin/streptomycin at 37°C in a 5% CO2 incubator.
Cell transfection
Synthetic miR-451a mimics and scrambled RNA (negative control, NC) were obtained from Suzhou GenePharma. Both synthetic miR-451a mimics and NC were transfected into 1 × 103 5–8 F and SUNE-1 cells using Lipofectamine 2000 (Thermo Fisher Scientific, China) as per the manufacturer’s instructions. To produce cells that over-expressed CSE1L, we transfected 5–8 F and SUNE-1 cells with the overexpression vector pcDNA 3.1-CSE1L (OE) and the empty vector (OE-NC), purchased from Sangon Biotech, using Lipofectamine 3000 (Thermo Fisher Scientific, USA). We calculated the transfection efficiency by quantitative reverse transcription polymerase chain reaction (qRT-PCR) after 48 h of transfection.
qRT-PCR
We extracted RNA from tissues and cells using the TRIzol reagent (Invitrogen). Subsequently we performed cDNA synthesis using the ImProm-II reverse transcription kit (Promega, USA) by mixing 1 μg of RNA (from tumor and transfected cells each) to 0.5 µL oligodT primers and 0.5 µL miRNA-specific stem-loop primers, respectively. SYBR Green Supermix (Bio-Rad, USA) was used for quantifying the mRNA expression of miR-451a and CSE1L. We used U6 and GAPDH as internal references and determined the mRNA expression levels of miR-451a and CSE1L using the 2−ΔΔCT method [Citation23]. The sequences for PCR primers used in this study are shown in .
Table 2. The sequence of PCR primers used in this study
Western blotting
Protein levels were measured as previously described [Citation24]. At 48 h post-transfection, 5–8 F and SUNE-1 cells were lysed using the radioimmunoprecipitation assay buffer (lysis buffer; Abcam, China). We quantified the supernatants using a bicinchoninic acid protein concentration kit (Beyotime, China). Subsequently, proteins (20 µg) were separated with 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis and were electro-transferred onto a polyvinylidene fluoride membrane. The membrane was incubated with 5% skimmed milk for 1 h at room temperature before being incubated with rat anti-CSE1L (1:1000, Cat. no. ABIN560494, Beijing 4A Biotech Co, Ltd., China), and rat anti-GAPDH (1:1000, Cat. no. AF1186, Beyotime, China) at 4°C for 24 h. The following day, the blot was probed with a mouse anti-rabbit horseradish peroxidase–conjugated secondary antibody (1:1000, Cat. no. 4060–01, Southern Biotech, USA), and protein bands were visualized by chemiluminescence using ECL (Beyotime, China).
Cell counting kit-8 (CCK-8) assay
We assessed the viability of NPC cells at 48 h post-transfection using the CCK-8 kit (4A Biotech Co. Ltd. China), as previously described [Citation10]. Briefly, 2 × 103 cells/well were seeded in a 96-well plate. Cells were cultured for 24 h, 48 h, 72 h and 96 h, following which 20 μL of CCK-8 was added to each well. Cells were incubated for additional 2 h, and the optical density (OD) of each well was measured at 450 nm using a spectrophotometer (BioTek, USA).
Bromodeoxyuridine (BrdU) cell proliferation assay
This assay was performed as described previously [Citation25]. We measured the cell proliferation rate at 48 h post-transfection in the 5–8 F and SUNE-1 cell lines using the BrdU cell proliferation assay kit (Frdbio, China) as per the manufacturer’s instructions. In brief, 1 × 103 cells/well were seeded in 24-well plates to achieve 60–70% confluency. Thereafter, cells were cultured in 10 μl BrdU solution for 4 h and were fixed with 5% paraformaldehyde for 20 min. Subsequently, cells were permeabilized in 0.5% Triton X-100 and incubated with a horseradish peroxidase-conjugated mouse anti-BrdU monoclonal antibody (1:1000) overnight at room temperature. Following this, 100 μL of a horseradish peroxidase substrate was added to the cells, and the cell plates were kept in the dark for 30 min. The OD was measured at 450 nm using a microplate reader (Biobase, China).
Flow cytometry analysis
At 48 h post-transfection, 5–8 F and SUNE-1 cells were collected, and cellular apoptosis was detected by the Annexin V-FITC/PI Apoptosis Detection kit (BD Biosciences, USA), performed as per the manufacturer’s protocol. Briefly, cells were diluted with 400 μL of Annexin V binding buffer. Subsequently, they were incubated with 5 μL of FITC and 5 μL of propidium iodide and kept in the dark for 15 min at room temperature. Cells were analyzed using a FACSAria flow cytometer (BD Biosciences) [Citation26].
Wound healing assay
A wound healing assay was performed as described previously [Citation27]. At 48 h post-transfection, 5 × 103 cells/well were seeded in 6-cm dishes to achieve 80–90% confluency. A pipette tip (10 μL) was used to produce a 400 μm gap in the confluent monolayer. After removing the detached cells, the adherent cells were cultured for an additional 24 h, following which they were fixed with 3.7% paraformaldehyde and stained with 1% crystal violet. Cell migration was observed under a phase-contrast microscope and the cell migration rate was calculated by Image J; it was calculated as (0 h width of gap-24 h gap width)/0 h gap width×100%.
Transwell invasion assay
Cell invasiveness was assessed using a Transwell insert. A Matrigel-precoated transwell insert was plated with 5 × 103 cells at 48 h post-transfection. This insert was added on a lower insert, containing 600 mL fresh medium. Cells were cultured for 24 h, following which the upper chamber was removed and the noninvasive cells were wiped out with a cotton swab. Next, the cells were fixed with formaldehyde for 20 min and subsequently stained with 0.1% crystal violet for 10 min. Invasive cells were visualized by phase-contrast microscopy, and the rate of invasiveness was calculated as described previously [Citation28].
Tumor xenograft model
All our experiments on animals were approved by the Institutional Animal Care and Use Ethics Committee of our hospital. Ten male BALB/c nude mice aged 4–6 weeks were purchased from the Medical Experimental Animal Center of Guangdong Province (Guangzhou, China). We suspended 1 × 106 of 5–8 F cells that stably over-expressed miR-451a or the NC in 200 μL of PBS. Subsequently, these cells were injected subcutaneously into the dorsal side of the nude mice. We measured the tumor size and calculated the tumor volume every 6 days. After 30 days, the mice were euthanized, and the tumors were dissected and weighed [Citation29].
Dual-luciferase reporter assay
To further verify whether the 3′ untranslated region (3΄ UTR) of CSE1L mRNA contained the miR-451a binding site, we inserted fragments of the wild-type 3′ UTR and mutant 3′ UTR regions into the pmirGLO luciferase reporter plasmid (Promega, USA). Subsequently, we cloned these fragments and obtained pmirGLO-CSE1L3′-UTR-WT (WT) and pmirGLO-CSE1L3′-UTR-MUT (MUT). Next, 1 × 105 cells were co-transfected with the WT or MUT vectors and with the synthetic miR-451a mimic or NC. At 48 h post-transfection, cells were lysed, and luminescent signals were detected using a Dual Luciferase Reporter Assay kit (Promega, USA). Relative luciferase activity was presented as firefly luciferase/Renilla luciferase [Citation30].
RNA pull-down assay
The assay was performed as described previously [Citation31]. Biotin-labeled miR-451a (Bio-miR-451a) and negative control (Bio-NC) were synthesized by GenePharma (Shanghai, China). Cells were transfected with Bio-NC and Bio-miR-451a probes for 48 h, following which they were lysed in lysis buffer and incubated with Dynabeads M-280 streptavidin (Invitrogen). Each streptavidin-bound biotin-labeled RNA was eluted using magnetic beads; the eluted RNA was analyzed by qRT-PCR.
RNA immunoprecipitation (RIP)
We performed RIP for the 5–8 F and SUNE-1 cell lines using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, USA). Briefly, cells were lysed with the RIP buffer, and after centrifugation the lysate was collected and re-suspended. Subsequently, it was incubated with Ago2 or IgG antibody-conjugated magnetic beads for 30 min. After isolating the precipitated RNA, the enriched CSE1L mRNA and miR-451a were detected by qRT-PCR [Citation32].
Statistical analysis
We statistically analyzed our data using GraphPad Prism version 9.0; these results were presented as mean±standard deviation (SD). All experiments were performed in triplicates. Either One-way ANOVA or Two-way ANOVA was used to analyze data between multiple groups, whereas the Paired student’s t-test was performed to analyze data between two groups. The Pearson’s correlation coefficient was calculated to correlate the expression of miR-451a with that of CSE1L. Statistical relevance was defined as p < 0.05.
Results
In this study, we investigated the effects of miR-451a and CSE1L on cell viability, proliferation, migration, invasion, and tumor growth in NPC. We observed that miR-451a expression was down-regulated, whereas that of CSE1L was up-regulated in NPC. Moreover, we observed that over-expression of miR-451a suppressed malignancy in NPC cells. Since CSE1L is a direct target of miR-451a, its overexpression could eliminate miR-451a-dependent malignancy in 5–8 F and SUNE-1 cells.
Identification of miR-451a and CSE1L as the genes of interest in NPC
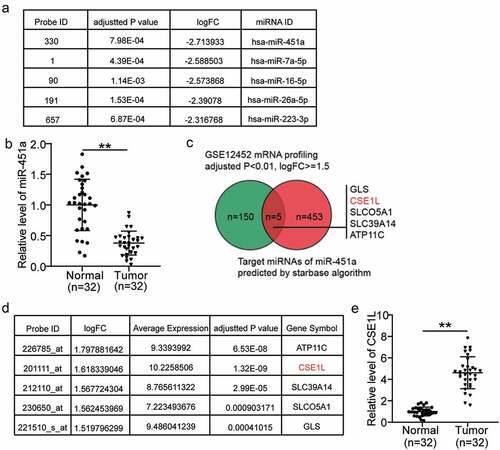
Upon analyzing the miRNA expression profiles in the GSE118613 dataset, we found that miR-451a was the most significantly down-regulated miRNA in NPC (). It has been reported to be a tumor suppressor in other human cancers, but not in NPC [Citation13,Citation33–38]. Remarkably, we found that miR-451a expression was significantly down-regulated in the NPC tumor tissues compared to that in the normal adjacent tissues (). Subsequently, we identified 458 unique downstream effectors of miR-451a using starbase algorithm. We also analyzed the mRNA expression profiles of NPC, retrieved from the GSE12452 dataset. We identified 155 mRNAs that were significantly up-regulated in NPC, among which 5 were also predicted to be the downstream effectors of miR-451a: GLS, CSE1L, SLCO5A1, SLC39A14, and ATP11C (). From the GSE12452 dataset, we also found that CSE1L had the highest up-regulated mRNA expression in NPC (average expression level = 10.22; ). In addition, our data suggested that the expression levels of ATP11 C, CSE1L, SLC39A14, SLCO5A1, and GLS were higher in tumor tissues than in normal tissue by a factor of 3.4, 4.7, 2.8, 1.7 and 3.6, respectively. The most significant up-regulation was observed in that of CSE1L expression; therefore, we focused on it in the subsequent experiments (Supplementary Figure 1a–d, and ). Studies have reported CSE1L to act as a tumor suppressor in colorectal cancer by functioning in ceRNA networks [Citation39,Citation40]. Nonetheless, the roles of miR-451a and CSE1L have not yet been investigated in NPC, and their interactions have not yet been studied in any human cancers.
Figure 1. The selection of miR-451a and CSE1L as our study objects in NPC. (a). The top 5 most downregulated miRNAs in NPC. Data obtained from GSE118613 analysis. Criteria: adjusted P < 0.05, logFC≤-1.5. (b). miR-451a expression in NPC tissues and non-cancerous tissues by RT-qPCR. (c). Venn diagram showing the intersection between the predicted target genes of miR-451a by starbase and the significantly upregulated genes from GSE12452 data analysis (adjusted P < 0.01, logFC≥1.5). (d). The expression of the five genes in GSE12452 data analysis. (e). CSE1L expression in NPC tissues and non-cancerous tissues by RT-qPCR
miR-451a expression attenuates cellular proliferation and promotes apoptosis in NPC
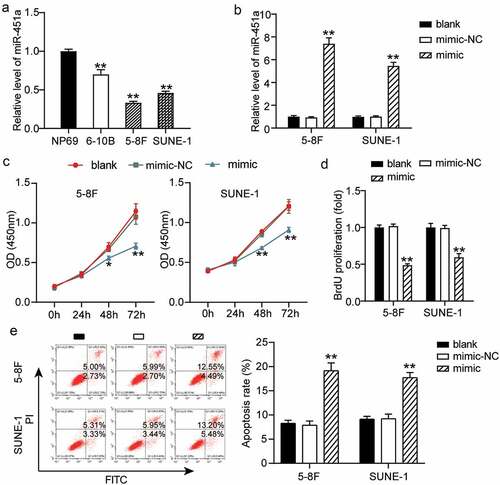
Previous observations have revealed that differential expression of miR-451a is associated with diverse types of cancers [Citation33,Citation37]. We investigated the clinical significance of miR-451a expression in NPC. To this end, we grouped NPC patients into miR-451a low expression (n = 16) and high expression (n = 16) groups based on the median value of the miR-451a expression levels. Subsequently, the correlation between the clinicopathological features of NPC patients and miR-451a expression levels was evaluated. As shown in , a low miR-451a expression was significantly associated with an advanced tumor, nodes, and metastasis (TNM) stage and with local or distant metastasis in NPC. The miR-451a expression profiles of the cell lines 6–10B, 5–8 F, and SUNE-1 revealed lower miR-451a expression in these cell lines than that in the NP69 cell line (). Hence, we chose 5–8 F and SUNE-1 cell lines, containing the lowest miR-451a expression levels, to examine whether synthetic miRNA analogs altered cellular processes in NPC. We transfected 5–8 F and SUNE-1 cells with synthetic miRNA mimics and NC and generated a cell transfection curve. Our results showed that in both the cell lines, expression levels of the mimic increased with prolonged transfection and stabilized at 48 h (Supplementary Figure S1); at this time poin,t a five-fold or greater increase in the miR-451a expression levels was observed (). Furthermore, we observed that upon over-expression of the miR-451a mimic in 5–8 F and SUNE-1 cells, cell viability was significantly impaired in both the cell lines, as assessed by the CCK-8 assay (). Moreover, we also observed that upon miR-451a over-expression, cell proliferation was impaired in these cell lines, as evaluated by the BrdU assay (). Additionally, we observed that NPC cells transfected with the synthetic miR-451a mimic () had a higher apoptotic rate than the ones transfected with the NC. These observations suggested that miR-451a over-expression suppressed cell proliferation and promoted apoptosis in the 5–8 F and SUNE-1 cell lines.
Figure 2. MiR-451a could inhibit the NPC cells proliferation. (a). mRNA levels of miR-451a in NP69, 6–10B, 5–8 F and SUNE-1 cells. **P < 0.001 vs.NP69. (b). The efficiency of transfected miR-451a mimic or negative control (NC) at 48 h was validated by RT-qPCR. (c). CCK8 assay revealing the suppression of miR-451a overexpression on the viability of 5–8 F and SUNE-1 cells. (d). BrdU assay demonstrated the inhibiting effect of miR-451a on the proliferation of 5–8 F and SUNE-1 cells. E. Flow cytometry assay showing the promoting effect of miR-451a on the apoptosis rate of 5–8 F and SUNE-1 cells. *P < 0.05, **P < 0.001 vs. blank
Over-expression of miR-451a suppresses cell migration and invasion in vitro and suppressed tumor growth in vivo in NPC
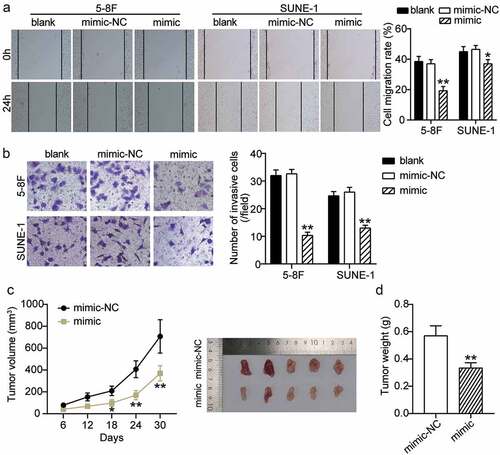
We assessed the effect of miR-451a over-expression on cell migration and invasion in the 5–8 F cand SUNE-1 cell lines. The wound healing assay indicated a significantly reduced cell migration rate in 5–8 F and SUNE-1 cells transfected with the synthetic miR-451a mimic compared to cells transfected with the blank control and NC (). Additionally, our transwell assay results indicated that cell invasiveness was reduced in cells over-expressing miR-451a (). Since miR-451a inhibited the survival of NPC cells in vitro, we examined whether over-expression of miR-451a could affect tumorigenesis in vivo. As shown in , over-expression of miR-451a in the tumor xenograft model significantly reduced the tumor growth and volume. In addition, the tumor weight in mice that over-expressed the synthetic miR-451a mimic was significantly lower than that in mice that expressed the NC (). These observations suggested that miR-451a inhibits malignant progression in NPC in vivo and in vitro.
Figure 3. MiR-451a negatively regulates the migration, invasion and xenograft tumor growth of NPC cells. 5–8 F and SUNE-1 cells were transfected with miR-451a synthetic miRNA analogs. The migratory and invasive capacities of both cells were evaluated by the wound-healing assay (a) and transwell assay (b). (c) Representative images of tumors formed and the growth curves of tumor volume. (d) Tumor weight. *P < 0.05, **P < 0.001 vs. blank or mimic-NC
CSE1L acts as a direct target of miR-451a
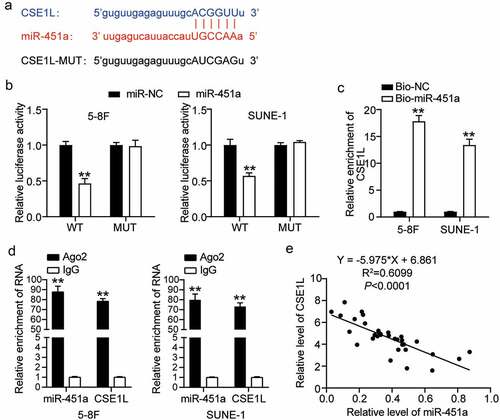
miRNAs are known to execute their functions via the miRNA-target gene regulatory network [Citation6]. We identified CSE1L among the putative target genes of miR-415a, which was reported to be associated with NPC tumorigenesis [Citation41]. Moreover, we demonstrated that the direct binding site of miR-451a was located on the 3΄ UTR region of the CSE1L mRNA (). For instance, we observed significantly impaired CSE1L-WT-mediated luciferase activity in cells that had been transfected with the synthetic miR-451a mimic; however, no such change was observed in CSE1L-MUT-mediated luciferase activity (). To validate the interaction between miR-451a and CSE1L, we measured the enrichment levels of CSE1L mRNA and biotin-labeled miR-451a by the RNA pull-down assay. As depicted in , CSE1L mRNA levels in both 5–8 F and SUNE-1 cells transfected with Bio-miR-451a were 12-fold higher than in the cells transfected with Bio-NC. Moreover, RIP results revealed that cells treated with anti-Ago2 molecules were more enriched in miR-451a and CSE1L than those treated with anti-IgG molecules (). In addition, Pearson’s correlation coefficient revealed a strong negative correlation between CSE1L expression and miR-451a in NPC tissues ().
Figure 4. CSE1L is a direct target of miR-451a. (a). starBase analysis predicted that 3′UTR sequence of CSE1L contained the complementary sequence of miR-451a. (b). Luciferase activity of CSE1L with wild type (WT) or mutant (MUT) 3ʹUTR in 5–8 F and SUNE-1 cells. **P < 0.001 vs. miR-NC. (c). RNA pull-down assay was performed to analysis the interaction between CSE1L and miR-451a in 5–8 F and SUNE-1 cells transfected with miR-451a mimic and miRNA-NC. **P < 0.001 vs. Bio-NC. (d). The expression of miR-451a and CSE1L extracted by RIP assay was evaluated in samples bound to Ago2 or IgG. **P < 0.001 vs. IgG. E. Pearson assay showed a strong negative correlation (R2 = 0.7123) between miR-451a level and CSE1L expression in NPC tissues
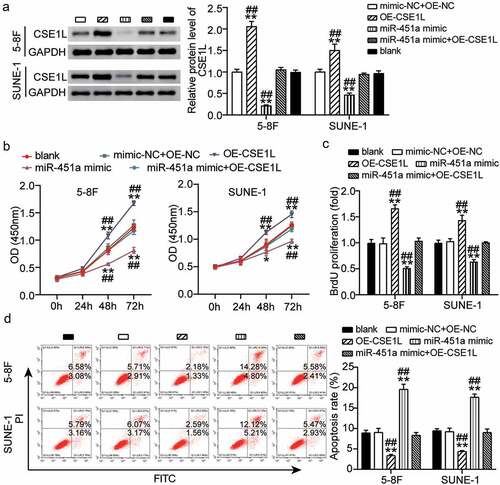
Overexpression of CSE1L alleviates miR-451a-mediated inhibition of proliferation and promotion of apoptosis.
CSE1L is recognized as an oncogene that is commonly amplified in NPC [Citation21] and is associated with proliferation, apoptosis, invasion, and metastasis [Citation22]. We observed that expression of CSE1L was rescued in NPC cells that were co-transfected with the synthetic miR-451a mimic and pcDNA 3.1-CSE1L, as evident by our western blot results (). Additionally, CCK-8 analysis indicated that 5–8 F and SUNE-1 cells transfected with OE-CSE1L had increased cell viability, partially eliminating the inhibitory effect that miR-451a overexpression had on cell viability (). Consistently, BrdU assay results indicated that ectopic expression of CSE1L significantly promoted cell proliferation in both 5–8 F and SUNE-1 cells, reversing the inhibitory effect of miR-451a overexpression on cell proliferation (). In addition, CSE1L up-regulation inhibited apoptosis in NPC cells, and to a certain extent, weakened the pro-apoptotic effect of the synthetic miR-451a mimic (). These observations implied that CSE1L over-expression could rescue the inhibition of cell proliferation mediated by miR-451a overexpression.
Figure 5. Ecotopic expression of CSE1L offsets the modulation of miR-451a on proliferation of NPC cells. (a). The expression of CSE1L in 5–8 F and SUNE-1 cells was detected by western blot. (b). CCK8 assay examining the viability of 5–8 F and SUNE-1 cells following transfection with pcDNA 3.1-CSE1L (OE), miR-451a mimic, OE-NC + mimic-NC, OE + mimic. (c). BrdU assay was performed to quantify the proliferation of 5–8 F and SUNE-1 cells following transfection with OE, miR-451a mimic, OE-NC + mimic-NC, OE, OE + mimic. (d). Flow cytometry assay was performed to quantify the apoptosis rate of 5–8 F and SUNE-1 cells following transfection with OE, miR-451a mimic, OE-NC + mimic-NC, OE + mimic. **P < 0.001 vs. blank; ##P < 0.001 vs. OE + mimic
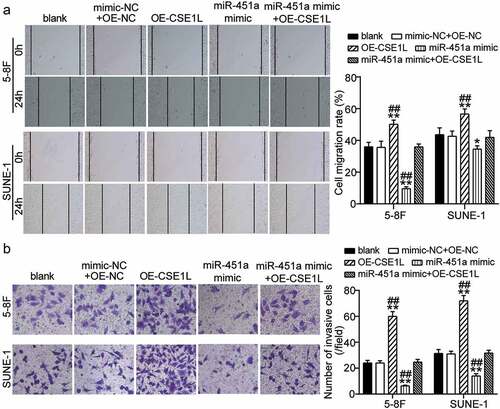
Ectopic expression of CSE1L alleviates miR-451a mimic-mediated inhibition of cell migration and invasion in NPC
Subsequently, we analyzed whether CSE1L overexpression affected cell migration and invasion in miR-451a-over-expressing 5–8 F and SUNE-1 cells. Remarkably, the area of the healed wound increased upon CSE1L over-expression in both cell lines and reduced post transfection with the synthetic miR-451a mimic. This indicated that wound healing was effectively rescued in cells over-expressing CSE1L (). Similarly, we also observed an increase in cell invasiveness upon CSE1L over-expression. Moreover, inhibition of cell invasion was rescued in cells co-transfected with miR-451a mimic and pcDNA 3.1-CSE1L in both the cell lines (). These findings suggested that CSE1L over-expression offsets the miR-451a-mediated suppression of cell migration and invasion in NPC in vitro.
Figure 6. CSE1L overexpression restores the migration and invasion impairment of NPC cells induced miR-451a mimic. (a). In vitro wound healing analysis of NPC cells migration after transfected with pcDNA 3.1-CSE1L (OE), miR-451a mimic, OE-NC + mimic-NC, OE + mimic at 0 and 48 h. (b). In vitro transwell assay was performed to examine the invasion of NPC cells following transfection with OE, miR-451a mimic, OE-NC + mimic-NC, OE + mimic. **P < 0.001 vs. blank; ##P < 0.001 vs. OE + mimic
Discussion
A new understanding of NPC etiopathological mechanisms offers novel insights into carcinogenesis. Thus, elucidating the function of miRNAs in cell proliferation, migration, and invasion can broaden our knowledge on NPC progression. In this study, we detected that miR-451a expression levels were reduced in NPC cell lines. Subsequent in vitro assays indicated that over-expression of miR-451a impaired cell proliferation, migration, and invasion in NPC. This impairment was at least partially controlled by CSE1L. Over-expression of CSE1L abrogated miR-451a-mediated inhibition of malignancy in NPC. In addition, the negative connection between miR-451a and CSE1L mRNA levels in NPC further validated our findings that CSE1L is a direct target of miR-451a. Collectively, our study revealed for the first time that miR-451a can inhibit cell proliferation, migration, and invasion and can promote apoptosis by targeting CSE1L; thus, it has an anticancer role in NPC.
Currently, pleotropic evidence has demonstrated that miR-451a abrogates malignant characteristics, such as cell proliferation, apoptosis, and epithelial-to-mesenchymal transition, that drive cancer progression and unfavorable treatment responses in patients [Citation12,Citation35,Citation42,Citation43]. For example, miR-451a suppresses disease aggressiveness in papillary thyroid cancer [Citation35]. Moreover, its down-regulated expression in multiple myeloma promoted cancer relapse in patients [Citation44]. Furthermore, exogenous miR-451a expression can inhibit drug resistance and restore drug sensitivity in patients with acute myeloid leukemia [Citation45]. However, thus far, the function of miR-451a in NPC remains unknown. The data presented here demonstrate that miR-451a is down-regulated in 5–8F and SUNE-1 cell lines compared to that in the NP69 cell line. Subsequent cell functional assays demonstrated that over-expression of miR-451a significantly impaired cell proliferation, migration, and invasion in both the cell lines. Therefore, our observations are congruent with those of previous studies that proposed that miR-451a is a tumor suppressor and that dysregulation of miR-451a may promote cancer progression.
Given the conserved regulatory functions of miRNA, miR-451a has been shown to post-transcriptionally modulate diverse genes implicated in tumor initiation and progression [Citation33,Citation34,Citation37,Citation46]. Therefore, we used microRNA target predictions starBase and identified gene targets associated with cell proliferation and apoptosis in NPC. One such predicted target, CSE1L, has been demonstrated to modulate tumor malignancy in various cellular models [Citation15,Citation18,Citation20,Citation47], and it is highly expressed in NPC cells [Citation21,Citation22,Citation41]. However, it is not clear whether interaction between miR-451a and CSE1L accounts for the tumor-suppressive effects observed in NPC. Here, we identified CSE1L as a direct target of miR-451a using a luciferase reporter assay. In addition, we found that over-expression of miR-451a in both 5–8 F and SUNE-1 cell lines significantly reduced the CSE1L mRNA level. We also observed that CSE1L and miR-451a exhibited contrasting cellular functions in NPC. Furthermore, we validated that over-expression of CSE1L in both 5–8 F and SUE-1 cells increased cell proliferation, migration, and invasion, and decreased apoptosis. However, the oncogenic role of CSE1L was offset by exogenous expression of the synthetic miR-451a mimic. Therefore, our results reiterate that miR-451a has anti-cancer properties in NPC, concurring with those of previous studies.
However, there were multiple limitations to this study. To this end, we will be validating our observations in in vivo models and in clinical experiments. Subsequently, as we have focused on only direct targets of miR-451a in the current study, we aim to elucidate the entire regulatory network of miR-451a in NPC in our future studies.
Conclusions
Collectively, our present data substantiate that miR-451a exerts tumor-suppressive functions by targeting CSE1L to hinder cell proliferation, migration, and invasion in NPC. Thus, we deduce that the miR-451a/CSE1L axis provides novel insights into targeted cancer therapeutics for NPC patients.
Consent for publication
Consent for publication was obtained from the participants.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of Affiliated Puren Hospital of Wuhan University of Science and Technology (Wuhan, China). The processing of clinical tissue samples is in strict compliance with the ethical standards of the Declaration of Helsinki. All patients signed written informed consent.
Authors’ contributions
XQ conducted the study, collected and analyzed the data. YL designed the study and methods and collected the funds. DK analyzed and interpreted the data. BLC collected materials and resources, conducted literature analysis and prepared manuscript. YL conducted literature analysis and prepared the manuscript. All authors read and approved the final manuscript.
Supplemental Material
Download ()Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
References
- Chen YP, Chan ATC, Le QT, et al. Nasopharyngeal carcinoma. Lancet. 2019;394(10192):64–80.
- Lee HM, Okuda KS, González FE, et al. Current perspectives on nasopharyngeal carcinoma. Adv Exp Med Biol. 2019;1164:11–34.
- Chua ML, Sun Y, Supiot S. Advances in nasopharyngeal carcinoma-”West meets East”. Br J Radiol. 2019;92(1102):20199004.
- Lam WKJ, Chan JYK. Recent advances in the management of nasopharyngeal carcinoma. F1000Res. 2018;7. DOI:10.12688/f1000research.15066.1
- Lee AWM, Ng WT, Chan JYW, et al. Management of locally recurrent nasopharyngeal carcinoma. Cancer Treat Rev. 2019;79:101890.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234(5):5451–5465.
- Lee YS, Dutta A. MicroRNAs in cancer. Annu Rev Pathol. 2009;4:199–227.
- Wang S, Claret FX, Wu W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma. Front Oncol. 2019;9:756.
- Cai L, Ye L, Hu X, et al. MicroRNA miR-330-3p suppresses the progression of ovarian cancer by targeting RIPK4. Bioengineered. 2021;12(1):440–449.
- Kang J, Huang X, Dong W, et al. MicroRNA-1269b inhibits gastric cancer development through regulating methyltransferase-like 3 (METTL3). Bioengineered. 2021;12(1):1150–1160.
- Zhao M, Luo R, Liu Y, et al. miR-3188 regulates nasopharyngeal carcinoma proliferation and chemosensitivity through a FOXO1-modulated positive feedback loop with mTOR-p-PI3K/AKT-c-JUN. Nat Commun. 2016;7:11309.
- Tao L, Shu-Ling W, Jing-Bo H, et al. MiR-451a attenuates doxorubicin resistance in lung cancer via suppressing epithelialmesenchymal transition (EMT) through targeting c-Myc. Biomed Pharmacothe. 2020;125:109962.
- Wang Q, Shang J, Zhang Y, et al. MiR-451a restrains the growth and metastatic phenotypes of papillary thyroid carcinoma cells via inhibiting ZEB1. Biomed Pharmacothe. 2020;127:109901.
- Zhao S, Li J, Zhang G, et al. Exosomal miR-451a functions as a tumor suppressor in hepatocellular carcinoma by targeting LPIN1. Cell Physiol Biochem. 2019;53(1):19–35.
- Jiang MC. CAS (CSE1L) signaling pathway in tumor progression and its potential as a biomarker and target for targeted therapy. Tumour Biol. 2016;37(10):13077–13090.
- Liu C, Wei J, Xu K, et al. CSE1L participates in regulating cell mitosis in human seminoma. Cell Prolif. 2019;52(2):e12549.
- Wang YS, Peng C, Guo Y, et al. CSE1L promotes proliferation and migration in oral cancer through positively regulating MITF. Eur Rev Med Pharmacol Sci. 2020;24(10):5429–5435.
- Pimiento JM, Neill KG, Henderson-Jackson E, et al. Knockdown of CSE1L gene in colorectal cancer reduces tumorigenesis in vitro. Am J Pathol. 2016;186(10):2761–2768.
- Yuksel UM, Turker I, Dilek G, et al. Does CSE1L overexpression affect distant metastasis development in breast cancer? Oncol Res Treat. 2015;38(9):431–434.
- Li Y, Yuan S, Liu J, et al. CSE1L silence inhibits the growth and metastasis in gastric cancer by repressing GPNMB via positively regulating transcription factor MITF. J Cell Physiol. 2020;235(3):2071–2079.
- Kim YJ, Go H, Wu HG, et al. Immunohistochemical study identifying prognostic biomolecular markers in nasopharyngeal carcinoma treated by radiotherapy. Head Neck. 2011;33(10):1458–1466.
- Hui AB, Lo KW, Teo PM, et al. Genome wide detection of oncogene amplifications in nasopharyngeal carcinoma by array based comparative genomic hybridization. Int J Oncol. 2002;20(3):467–473.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Hamano R, Miyata H, Yamasaki M, et al. Overexpression of miR-200c induces chemoresistance in esophageal cancers mediated through activation of the Akt signaling pathway. Clin Cancer Res off J Am Assoc Cancer Res. 2011;17(9):3029–3038.
- Shou J, Gao H, Cheng S, et al. LncRNA HOXA-AS2 promotes glioblastoma carcinogenesis by targeting miR-885-5p/RBBP4 axis. Cancer Cell Int. 2021;21(1):39.
- Li J, Kong F, Wu K, et al. miR-193b directly targets STMN1 and uPA genes and suppresses tumor growth and metastasis in pancreatic cancer. Mol Med Rep. 2014;10(5):2613–2620.
- Wang H, Jiao H, Jiang Z, et al. Propofol inhibits migration and induces apoptosis of pancreatic cancer PANC-1 cells through miR-34a-mediated E-cadherin and LOC285194 signals. Bioengineered. 2020;11(1):510–521.
- Wang X, Li T. Ropivacaine inhibits the proliferation and migration of colorectal cancer cells through ITGB1. Bioengineered. 2021;12(1):44–53.
- Li YQ, Lu JH, Bao XM, et al. MiR-24 functions as a tumor suppressor in nasopharyngeal carcinoma through targeting FSCN1. J Exp Clin Cancer Res. 2015;34:130.
- Chen L, Zhu Q, Lu L, et al. MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-β1. Bioengineered. 2020;11(1):91–102.
- Tian F, Tang P, Sun Z, et al. miR-210 in exosomes derived from macrophages under high glucose promotes mouse diabetic obesity pathogenesis by suppressing NDUFA4 expression. J Diabetes Res. 2020;2020:6894684.
- Li X, Liu F, Lin B, et al. miR‑150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int J Oncol. 2017;50(4):1097–1108.
- Liu Y, Yang HZ, Jiang YJ, et al. miR-451a is downregulated and targets PSMB8 in prostate cancer. Kaohsiung J Med Sci. 2020;36(7):494–500.
- Xu K, Han B, Bai Y, et al. MiR-451a suppressing BAP31 can inhibit proliferation and increase apoptosis through inducing ER stress in colorectal cancer. Cell Death Dis. 2019;10(3):152.
- Fan X, Zhao Y. miR-451a inhibits cancer growth, epithelial-mesenchymal transition and induces apoptosis in papillary thyroid cancer by targeting PSMB8. J Cell Mol Med. 2019;23(12):8067–8075.
- Uchida A, Seki N, Mizuno K, et al. Regulation of KIF2A by antitumor miR-451a inhibits cancer cell aggressiveness features in lung squamous cell carcinoma. Cancers (Basel). 2019;11:2.
- Zhang H, Chen P, Yang J. miR-451a suppresses the development of breast cancer via targeted inhibition of CCND2. Mol Cell Probes. 2020;54:101651.
- Dong Y, Wang G. Knockdown of lncRNA SNHG12 suppresses cell proliferation, migration and invasion in breast cancer by sponging miR-451a. Int J Clin Exp Pathol. 2020;13(3):393–402.
- Ma S, Yang D, Liu Y, et al. LncRNA BANCR promotes tumorigenesis and enhances adriamycin resistance in colorectal cancer. Aging (Albany NY). 2018;10(8):2062–2078.
- Wang X, Ren Y, Ma S, et al. Circular RNA 0060745, a novel circRNA, promotes colorectal cancer cell proliferation and metastasis through miR-4736 sponging. Onco Targets Ther. 2020;13:1941–1951.
- Fang WY, Liu TF, Xie WB, et al. Reexploring the possible roles of some genes associated with nasopharyngeal carcinoma using microarray-based detection. Acta Biochim Biophys Sin (Shanghai). 2005;37(8):541–546.
- Zong S, Liu X, Zhou N, et al. E2F7, EREG, miR-451a and miR-106b-5p are associated with the cervical cancer development. Arch Gynecol Obstet. 2019;299(4):1089–1098.
- Chang C, Liu J, He W, et al. A regulatory circuit HP1γ/miR-451a/c-Myc promotes prostate cancer progression. Oncogene. 2018;37(4):415–426.
- Zhong L, Jin X, Xu Z, et al. Circulating miR-451a levels as a potential biomarker to predict the prognosis of patients with multiple myeloma. Oncol Lett. 2020;20(5):263.
- Krakowsky RHE, Wurm AA, Gerloff D, et al. miR-451a abrogates treatment resistance in FLT3-ITD-positive acute myeloid leukemia. Blood Cancer J. 2018;8(3):36.
- Liu ZR, Song Y, Wan LH, et al. Over-expression of miR-451a can enhance the sensitivity of breast cancer cells to tamoxifen by regulating 14-3-3ζ, estrogen receptor α, and autophagy. Life Sci. 2016;149:104–113.
- Jiang K, Neill K, Cowden D, et al. Expression of CAS/CSE1L, the cellular apoptosis susceptibility protein, correlates with neoplastic progression in Barrett’s Esophagus. Appl Immunohistochem Mol Morphol. 2018;26(8):552–556.
