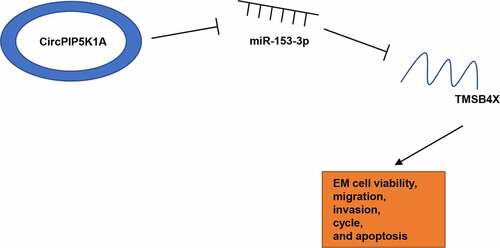
ABSTRACT
As a common gynecologic disease, endometriosis (EM) poses a threat to the reproductive health of about 10% women globally. Recent studies have revealed that circular RNAs (circRNAs) are deeply implicated in EM pathogenesis. However, the functions of circPIP5K1A in EM have not been studied yet. Our study intended to uncover the molecular mechanism of circPIP5K1A in EM. In this work, gene and protein expressions were determined by RT-qPCR or Western blotting. CCK-8, wound healing, transwell, and flow cytometry assays were conducted to analyze cell viability, migration, invasion, cell cycle, and apoptosis. Additionally, bioinformatics analysis, dual-luciferase reporter assay, as well as RIP assay were performed to investigate the combination between miR-153-3p and circPIP5K1A or TMSB4X. Herein, we found remarkable high circPIP5K1A expression in EM tissues and cells. Silencing of circPIP5K1A suppressed proliferation, restrained cell cycle, increased cell apoptosis, and decreased migration and invasion in EM cells. In addition, miR-153-3p inhibition could abrogate the impacts of circPIP5K1A knockdown on EM progression in vitro. Also, we found that circPIP5K1A regulated TMSB4X level via interaction with miR-153-3p in EM cells. Besides, circPIP5K1A promoted EM progression via TMSB4X. Moreover, TMSB4X could activate the TGF-β signaling in hEM15A cells. To sum up, our study elucidated that circPIP5K1A accelerated EM progression in vitro by activating the TGF-β signaling pathway via the miR-153-3p/TMSB4X axis, providing a potential clinical target for EM treatment.
KEYWORDS:
Introduction
As a common gynecological disorder, endometriosis (EM) is characterized by abnormal growth of functional endometrial tissues (glands and stroma) outside of uterus and closely associated with pain and/or infertility [Citation1,Citation2]. EM pathogenesis is related to endocrine, altered immune, environmental, and genetic factors [Citation3]. Although EM is a benign gynecologic disease, the biological behaviors of EM lesions are analogous to those of malignant tumors, including unbridled cell proliferation and metastasis caused by disrupted cell cycle control and enhanced epithelial-mesenchymal transition (EMT) [Citation4–6]. Besides, there is a high recurrence rate in EM patients despite complete lesion excision [Citation7]. Hence, it is widely believed that EM recurrence might be associated with its tumor-like biological characteristics [Citation8]. Unfortunately, due to diversified symptoms and complex pathogenesis of EM, there are still no efficient early diagnosis strategies or radical therapeutic methods for EM [Citation9]. Therefore, it is urgent to have an in-depth understanding of EM pathogenesis.
Circular RNAs (circRNAs) refer to transcriptionally or post-transcriptionally regulatory RNAs with covalently closed-loop structures [Citation10,Citation11]. Increasing evidence proves that circRNAs may participate in the pathophysiological progression of several gynecologic disorders, including EM [Citation12,Citation13]. To cite an instance, a report from Jiang et al. revealed that hsa_circ_0008433 was upregulated and induced cell-cycle arrest and apoptosis in EM via interaction with miRNAs [Citation14]. As demonstrated by He et al., through β-catenin signaling, circ_0004712/miR-148a-3p regulated estrogen-induced epithelial-to-mesenchymal transition (EMT) process in EM [Citation15]. Besides, it was elaborated by Dong et al. that circ_0007331 facilitated EM development by increasing the viability, proliferation, and invasive capacity of ectopic endometrial cells through the miR-200c-3p/HiF-1α pathway [Citation16]. In a previous report, Zhang et al. identified hsa_circ_0014130 (circPIP5K1A) as an upregulated circRNA in ovarian EM [Citation17]. However, the specific regulatory functions of circPIP5K1A in EM remain obscure.
In this work, we intended to investigate the role of circPIP5K1A in EM. We first demonstrated that circPIP5K1A was upregulated in EM and circPIP5K1A inhibition suppressed EM phenotypes through evaluation on EM cell viability, cell-cycle regulation, apoptosis, migration, and invasion via in vitro assays. Then, we further elaborated that circPIP5K1A exerted promoting effects on EM progression in vitro via the miR-153-3p/TMSB4X axis. Moreover, it was further disclosed that TMSB4X facilitated the activation of TGF-β signaling. Our findings might provide a novel direction for the development of effective EM diagnostic and therapeutic strategies.
Materials and methods
Clinical samples
All the tissue specimens were obtained from female patients (22 to 46 years old) who had regular menstruation cycles (22 to 35 days) and had not undergone any hormonotherapy at least half a year before sample collection. The clinical characteristics of the patients are as shown in . Among them, paired eutopic (euEM, n = 28) and ectopic (ecEM, n = 28) endometrial specimens were obtained from 28 EM patients during laparoscopy; normal endometrial specimens were randomly obtained from 28 women without EM in curettage. Specimen diagnoses were confirmed by histologic analysis. This study was permitted by the Ethics Committee of Maanshan Maternal and Child Health Care Hospital. Prior to specimen collection, each participant enrolled in the current study provided written informed consent. Patients with adenomyosis, pelvic inflammatory diseases, dysfunctional uterine bleeding, or endometrial cancer were excluded.
Table 1. Clinical and pathological characteristics of subjects
Cell culture and transfection
EM immortalized eutopic endometrial stromal cell line (hEM15A) and normal human endometrial stromal cell line (hESC) obtained from BeNa Culture Collection (Beijing, China) were cultivated in DMEM-H/F12 medium (Catalog No. 11,965,118; Gibco, USA) containing 10% FBS (Catalog No. 11,875–176; Thermo Fisher Gibco) with 5% CO2 at 37°C.
Small interfering RNA (siRNA) against circPIP5K1A (si-circPIP5K1A#1:5ʹ-AUUUUGCAUAGUCACAGAAAG-3ʹ and si-circPIP5K1A#2:5ʹ-GCAUAGUCAUUGAAAGUUACA-3ʹ), its negative control (si-NC: 5ʹ-AUCAUGUCUUACAUAAGAGAG-3ʹ), NC mimics(5ʹ-AUCUAAUGCAUAGUCAGAGUAC-3ʹ), miR-153-3p mimics(5ʹ-UUGCAUAGUCACAAAAGUGAUC-3ʹ), NC inhibitor (5ʹ-CUGAACUGCUAGGACGCGCUA-3ʹ), miR-153-3p inhibitor(5ʹ-GAUCACUUUUGUGACUAUGCAA-3ʹ), pcDNA3.1-circPIP5K1A overexpression vector (oe-circPIP5K1A), pcDNA3.1-TMSB4X overexpression vector (oe-TMSB4X) and pcDNA3.1 empty vector (Vector) were synthesized by GenePharma. These plasmids were, respectively, transfected into hEM15A cells via Lipofectamine 2000 reagent (Invitrogen) and harvested for later experiments after 24 hours transfection.
RT-qPCR
Trizol reagent (Catalog No. D312; Takara) was utilized to isolate total RNAs from tissues and cells. Then, a PrimeScript reverse transcriptase (RT) reagent kit was used to reversely transcribe total RNA into cDNA. Thereafter, PCR assay was conducted using an ABI PRISM 7500 real-time PCR System and an miScript SYBR Green PCR Kit (Qiagen), with GAPDH or U6 as the endogenous control. The primers used are listed in .
Table 2. List of RT-qPCR primers
CCK-8
To assess cell viability, CCK-8 assay was performed. In brief, 96-well plates were inoculated with hEM15A cells (2 × 103 cells/well). Then, CCK-8 solution (Catalog No. B34304; Bimake, USA) was added (10 µL/well) at indicated time points (0, 24, 48, 72 hours after inoculation). After cultivation for another 2 hours, a microplate reader (Bio-Rad, USA) was used to detect the optical density (OD) at 450 nm [Citation18].
Wound healing
The wound healing assay was utilized to evaluate migration capacity of hEM15A cells. The transfected hEM15A cells were inoculated into 6-well plates (2 × 105 cells/well) and cultivated until the cell convergence reached 90–100%. Then, a 20 μL micropipette tip was employed to create a straight wound in each well, and the plates were rinsed with cold phosphate buffer saline (PBS) (Catalog No. 10,010,031; Thermo Fisher Scientific) to remove the scraped cells. The cells left were cultivated for another 24 hours after fresh culture medium was supplemented to each well. Representative pictures were taken for each well with a microscope (Nikon, Japan) at 0 and 24 hours to measure wound widths [Citation19].
Transwell
A 24-well Transwell chamber with Matrigel-coated filters (8 μm pore size) was applied for cell invasion assessment [Citation20]. Briefly, 200 μL of serum-free medium (containing hEM15A cells; 2 × 105 cells/mL) was added into upper chambers; at the same time, 600 μL of complete culture medium containing 10% FBS (Catalog No. 11,875–176; Thermo Fisher Gibco) was added to lower chambers. After cultivation for 24 hours, Matrigel and cells in upper chambers were removed and cells below the filters were fixed with 4% paraformaldehyde and stained with 0.2% crystal violet. The invaded cells were counted under microscope (Nikon).
Flow cytometry
Transfected hEM15A cells were centrifuged for supernatant removal, washed with ice-cold PBS buffer, centrifuged again, and resuspended in binding buffer. The cells were mixed with 5 μL of Annexin V-FITC and 5 μL of propidium iodide (PI) (Catalog No. CA1020-20; Solarbio, China) for 10 minutes in the dark. Then, the cell apoptosis was analyzed by the flow cytometer (BD Biosciences, USA) [Citation21].
Regarding cell-cycle analysis, treated cells were washed three times with PBS and fixed with ethanol at 4°C overnight. Thereafter, the cells were permeabilized and stained with eBioscience™ Permeabilization Buffer (Catalog No. 88–8823-88; Thermo Fisher Scientific, Inc.) and PI solution for 30 min. Thereafter, cell cycle was measured and analyzed using an FACScan flow cytometer (BD Biosciences, CA) and FlowJo (TreeStar, CA) [Citation22].
Western blotting
The hEM15A cells were lysed in RIPA buffer (Sigma, USA) containing protease inhibitor cocktail (Sigma). Then, proteins were isolated using 10% SDS-PAGE gels, transferred onto PVDF membranes (Millipore, USA), and blocked with nonfat milk (5%) at room temperature for 2 h. Afterward, the membranes were incubated with specific primary antibodies against CDK2 (ab32147, 1:1000; Abcam, USA), CDK4 (ab137675, 1:1000), CDK6 (ab124821, 1:1000), Cyclin D1 (ab16663, 1:1000), Bcl-2 (ab32124, 1:1000), Bax (ab32124, 1:1000), cleaved Caspase-3 (ab4051, 1:1000), N-cadherin (ab18203, 1:1000), vimentin (ab137321, 1:1000), E-cadherin (ab15148, 1:1000), TMSB4X (ab167650, 1:1000), or TGF-β2 (ab113670, 1:1000) at 4°C overnight and incubated with corresponding HRP-conjugated secondary antibodies (ab131368 and ab191866; both 1:5,000) for 2 h at room temperature. Eventually, the proteins were visualized via ECL (Thermo Pierce), with GAPDH as the loading control.
Bioinformatics analysis
StarBase website (http://starbase.sysu.edu.cn) was used to find candidate microRNAs (miRNAs) for circPIP5K1A, and miRmap (https://mirmap.ezlab.org), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html), and PicTar (http://www.pictar.mdc-berlin.de) databases were used to predict the candidate target genes for miR-153-3p.
Dual-luciferase reporter assay
To generate wild-type or mutant luciferase reporter plasmids of circPIP5K1A (circPIP5K1A-WT or circPIP5K1A-MUT) and TMSB4X (TMSB4X-WT or TMSB4X-MUT), gene fragments of circPIP5K1A and 3′-UTR of TMSB4X with wild-type or mutant miR-153-3p binding sites were amplified and inserted into pmirGLO luciferase reporter vectors (Promega) and then transfected into hEM15A cells with miR-153-3p mimics or NC mimics, via Lipofectamine 2000. The cells were lysed after 48 hours of cultivation. Finally, Double-Luciferase Reporter Assay Kit Luminometer LB960 (Berthold Technologies, Germany) was utilized to detect luciferase activity [Citation23].
RNA immunoprecipitation (RIP) assay
For RIP assay, an EZ-Magna RIP kit (Millipore, MA) was used. In brief, cell lysates were cultured in RIP buffer with magnetic beads in conjugation with anti-Argonaute2 (Ago2) and Immunoglobulin G (IgG) antibodies (Catalog No. 03–110; Millipore). Immunoprecipitated RNA was isolated after the specimens were cultivated with Proteinase K buffer. Afterward, purified RNAs were extracted. Eventually, RT-qPCR was performed to analyze relative circPIP5K1A, miR-153-3p, and TMSB4X expressions.
Statistical analysis
Every experiment was repeated three times. All data were expressed as mean ± standard deviation (SD). Student’s t-test (2 groups) or one-way ANOVA (≥ 3 groups) was conducted to analyze group differences. Statistical analysis was conducted via GraphPad Prism 6.0. Any difference with p-value ≤0.05 was deemed statistically significant.
Results
In this study, we aimed to explore the regulatory effects of circPIP5K1A in regulating EM progression. Through in vitro assays on EM cell viability, cell-cycle regulation, apoptosis, migration, and invasion, we demonstrated that circPIP5K1A promoted EM progression in vitro by activating the TGF-β signaling via the miR-153-3p/TMSB4X axis, providing a novel biomarker for EM detection and diagnosis.
CircPIP5K1A knockdown inhibits EM progression in vitro
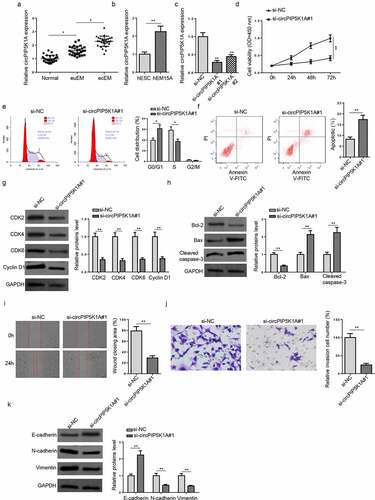
We firstly detected circPIP5K1A expression in ecEM, euEM, and normal tissues. RT-qPCR showed that circPIP5K1A level was prominently higher in ecEM tissues than that in euEM and normal tissues (). Besides, circPIP5K1A expression enrichment was also obviously upregulated in EM immortalized eutopic endometrial stromal cell line (hEM15A), compared with normal human endometrial stromal cell line (hESC) (). Hence, circPIP5K1A expression was elevated in EM, suggesting that it might play a vital role in regulating EM progression. To explore the potential biological function of circPIP5K1A in EM, a series of functional assays were performed. First of all, circPIP5K1A knockdown was performed in hEM15A cells (). As si-circPIP5K1A#1 showed a better transfection effect, si-circPIP5K1A#1 was adopted for the following experiments. CCK-8 and flow cytometry assays exhibited that circPIP5K1A inhibition suppressed the cell proliferative capability, induced G0/G1 cell cycle arrest, and increased cell apoptosis in hEM15A cells (). Western blotting analysis showed that the expression levels of cell cycle-related proteins (CDK2, CDK4, CDK6, and Cyclin D1) were decreased after circPIP5K1A knockdown (). Besides, circPIP5K1A inhibition suppressed Bcl-2 expression but stimulated Bax and cleaved Caspase-3 levels (). Wound healing and transwell assays disclosed that the migrative and invasive abilities of hEM15A cells were remarkably restrained by circPIP5K1A depletion (Fig. I and J). Consistently, circPIP5K1A silencing down-regulated N-cadherin and vimentin levels but up-regulated E-cadherin level in hEM15A cells (). To sum up, circPIP5K1A played a crucial role in expediting EM cellular activities.
Figure 1. CircPIP5K1A knockdown inhibits EM progression in vitro. (a) Relative expression levels of circPIP5K1A in normal, euEM and ecEM tissues by RT-qPCR (n = 28). (b) Relative expression levels of circPIP5K1A in hESC cells and hEM15A cells by RT-qPCR. (c) The knockdown efficiency of si-circPIP5K1A#1 and si-circPIP5K1A#2 was assessed by RT-qPCR in hEM15A cells. (d) CCK-8 assay was performed to identify the effect of circPIP5K1A inhibition on proliferative ability of hEM15A cells. (e and f) flow cytometry assays were performed to analyze cell-cycle regulation and cell apoptosis in hEM15A cells. (g) The expression levels of cell cycle-related proteins (CDK2, CDK4, CDK6, and cyclin D1) were detected by western blotting. (h) The expression levels of apoptosis-associated proteins (Bcl-2, Bax, and cleaved Caspase-3) were detected by western blotting. (i and j) wound healing and transwell assays were performed to identify the migrative and invasive ability of hEM15A cells. (k) The expression levels of EMT-related proteins (N-cadherin, vimentin, and E-cadherin) were detected by western blotting. Data are shown as mean ± SD; *P < 0.05, **P < 0.01
CircPIP5K1A sponges miR-153-3p in EM cells
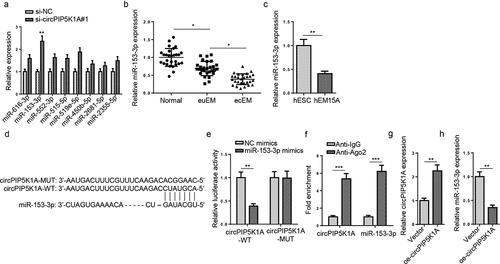
Next, we probed into the specific molecular mechanism of circPIP5K1A in EM. StarBase website predicted potential miRNAs with binding sites with circPIP5K1A. Eight candidate miRNAs (miR-616-3p, miR-153-3p, miR-552-3p, miR-515-5p, miR-519e-5p, miR-450b-5p, miR-2681-5p, and miR-2355-5p) were predicted under the condition of strict stringency (≥5) in CLIP Data. Then, the abundances of the 8 candidate miRNAs in hEM15A cells transfected with si-circPIP5K1A#1 or si-NC were detected. RT-qPCR results exhibited that circPIP5K1A silencing markedly upregulated miR-153-3p enrichment in hEM15A cells (). MiR-153-3p expression was distinctly lower in ecEM tissues, relative to euEM and normal tissues (). Moreover, miR-153-3p enrichment was also obviously downregulated in hEM15A cells, compared with hESC cells (). shows the binding site of miR-153-3p to circPIP5K1A. Dual-luciferase reporter assay manifested that the luciferase activity of circPIP5K1A-WT was remarkably reduced by miR-153-3p overexpression, whereas miR-153-3p upregulation had no obvious effects on the luciferase activity of circPIP5K1A-MUT (), implying a direct binding between circPIP5K1A and miR-153-3p. Additionally, RIP assay exhibited higher circPIP5K1A and miR-153-3p expressions in Ago2 group than IgG group (), thus further verifying the association between circPIP5K1A and miR-153-3p. Then, circPIP5K1A was overexpressed in hEM15A cells (). RT-qPCR revealed that miR-153-3p expression was substantially downregulated after circPIP5K1A upregulation (). The above findings suggested that circPIP5K1A acted as a sponge for miR-153-3p in EM.
Figure 2. CircPIP5K1A sponges miR-153-3p in EM cells. (a) Relative expression of 8 miRNAs in hEM15A cells after circPIP5K1A knockdown. (b) Relative expression levels of miR-153-3p in normal, euEM and ecEM tissues by RT-qPCR (n = 28). (c) Relative expression levels of miR-153-3p in hESC cells and hEM15A cells by RT-qPCR. (d) The molecular binding site between miR-153-3p and circPIP5K1A-WT or circPIP5K1A-MUT. (e) Dual-luciferase reporter assays confirmed the combination between circPIP5K1A and miR-153-3p. (f) The interaction between circPIP5K1A and miR-153-3p was further confirmed by RIP assay. (g) CircPIP5K1A overexpression efficiency in hEM15A cells was assessed by RT-qPCR. (h) miR-153-3p expression in hEM15A cells transfected with vector or oe-circPIP5K1A was detected by RT-qPCR. Data are shown as mean ± SD; *P < 0.05, **P < 0.01, ***P < 0.001
MiR-153-3p inhibition reverses EM phenotypes induced by circPIP5K1A silencing in vitro
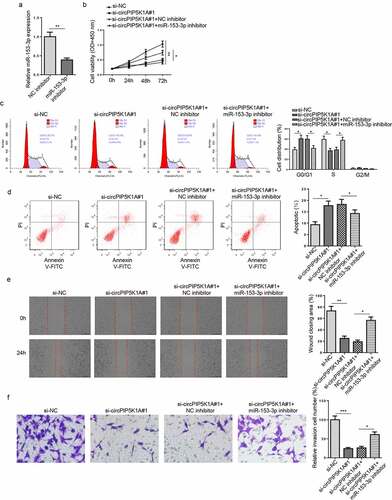
To investigate the role of miR-153-3p in EM progression via interaction with circPIP5K1A in vitro, hEM15A cells were transfected with si-NC, si-circPIP5K1A#1, si-circPIP5K1A#1+ NC inhibitor, or si-circPIP5K1A#1+ miR-153-3p inhibitor. First, the efficiency of miR-153-3p inhibition was confirmed by RT-qPCR (). As indicated by CCK8 assay and flow cytometry assays, miR-153-3p inhibition significantly enhanced hEM15A cell proliferation capability, accelerated cell cycle process, and reduced apoptosis (), thereby abrogating the effects on hEM15A cells induced by circPIP5K1A depletion. Wound healing and transwell assays also showed that circPIP5K1A blocking suppressed hEM15A cell migration and invasion, while miR-153-3p downregulation manifestly eliminated such impacts (). In summary, circPIP5K1A might expedite EM progression in vitro via interaction with miR-153-3p.
Figure 3. miR-153-3p inhibition reverses EM phenotypes induced by circPIP5K1A silencing in vitro. (a) The knockdown efficiency of miR-153-3p was assessed by RT-qPCR in hEM15A cells. (b) hEM15A cells were transfected with si-NC, si-circPIP5K1A#1, si-circPIP5K1A#1+ NC inhibitor, or si-circPIP5K1A#1+ miR-153-3p inhibitor. Then, CCK-8 assay was performed to analyze the proliferative ability of hEM15A cells. (c and d) flow cytometry assays were performed to analyze cell-cycle regulation and cell apoptosis in hEM15A cells. (e and f) wound healing and transwell assays were performed to analyze the migrative and invasive ability of hEM15A cells. Data are shown as mean ± SD; *P < 0.05, **P < 0.01
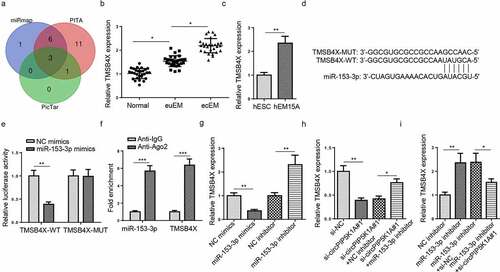
CircPIP5K1A regulates TMSB4X expression in EM cells as a sponge for miR-153-3p
In order to find the downstream target of miR-153-3p, 3 databases (miRmap, PITA, and PicTar) were adopted to predict latent target genes of miR-153-3p. As indicated in candidate target genes (AHR, ORC2, and TMSB4X) were screened under certain conditions (Degradome Data: high stringency (≥3); CLIP Data: strict stringency (≥5)). Since TMSB4X has been verified to promote cell proliferation and metastasis [Citation24], TMSB4X was selected as the target for miR-153-3p in this work. It was found that TMSB4X enrichment was clearly higher in ecEM tissues than euEM and normal tissues () and upregulated in hEM15A cells, relative to hESC cells (). StarBase website predicted a binding site between miR-153-3p and TMSB4X (). Subsequently, a dual-luciferase reporter assay was conducted to analyze the binding condition between miR-153-3p and TMSB4X. The results revealed that the luciferase activity was visibly reduced in the TMSB4X-WT groups, relative to the TMSB4X-MUT groups after miR-153-3p overexpression (). RIP assay further confirmed that both miR-153-3p and TMSB4X were enriched in the anti-Ago2 group (). Afterward, RT-qPCR showed that miR-153-3p upregulation reduced TMSB4X abundance in hEM15A cells, whereas miR-153-3p inhibition had an opposite effect (). Therefore, TMSB4X was a target for miR-153-3p in EM. Additionally, circPIP5K1A knockdown significantly decreased TMSB4X level in hEM15A cells, whereas miR-153-3p inhibition counteracted this inhibitory effect on TMSB4X level (). Also, miR-153-3p depletion remarkably increased TMSB4X level in hEM15A cells, while circPIP5K1A silencing partly offset such a promoting effect on TMSB4X expression (). Therefore, it could be concluded that circPIP5K1A regulated TMSB4X via directly binding to miR-153-3p.
Figure 4. CircPIP5K1A regulates TMSB4X expression in EM cells as a sponge for miR-153-3p. (a) The target genes of miR-153-3p were predicted by 3 databases (miRmap, PITA, and PicTar). (b) Relative mRNA levels of TMSB4X in normal, euEM and ecEM tissues by RT-qPCR (n = 28). (c) Relative mRNA levels of TMSB4X in hESC cells and hEM15A cells by RT-qPCR. (d) The molecular binding site of miR-153-3p and TMSB4X-WT or TMSB4X-MUT. (e) Dual-luciferase reporter assays confirmed the combination between miR-153-3p and TMSB4X. (f) The interaction between miR-153-3p and TMSB4X was further confirmed by RIP assay. (g) MSB4X mRNA expression was evaluated by RT-qPCR in hEM15A cells respectively transfected with NC mimics, miR-153-3p mimics, NC inhibitor, miR-153-3p inhibitor. (h) TMSB4X mRNA expression was evaluated by RT-qPCR in hEM15A cells respectively transfected with si-NC, si-circPIP5K1A#1, si-circPIP5K1A#1+ NC inhibitor, or si-circPIP5K1A#1+ miR-153-3p inhibitor. (i) TMSB4X mRNA expression was evaluated by RT-qPCR in hEM15A cells respectively transfected with NC inhibitor, miR-153-3p inhibitor, miR-153-3p inhibitor+si-NC, or miR-153-3p inhibitor+si-circPIP5K1A#1. Data are shown as mean ± SD; *P < 0.05, **P < 0.01
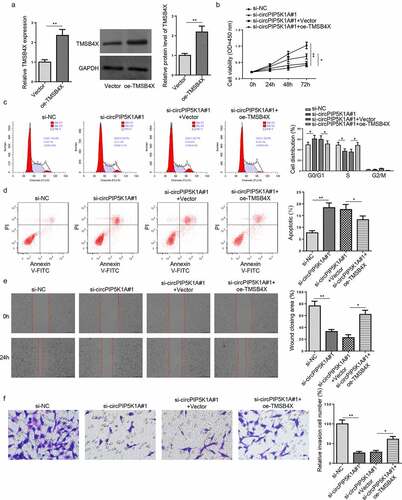
TMSB4X overexpression abrogates the effects of circPIP5K1A deletion on EM progression in vitro
To inquire into whether TMSB4X is involved in the regulation of EM progression mediated by circPIP5K1A, rescue assays were performed. First, TMSB4X overexpression efficiency was confirmed in hEM15A cells via RT-qPCR and western blotting (). Then, hEM15A cells were, respectively, transfected with si-NC, si-circPIP5K1A#1, si-circPIP5K1A#1+ Vector, and si-circPIP5K1A#1+ oe-TMSB4X. According to CCK-8 and flow cytometry assays, TMSB4X overexpression partly reversed the inhibitory effect on hEM15A cell proliferation and the promoting impact on cell-cycle arrest and apoptosis induced by circPIP5K1A knockdown (). Besides, wound healing and transwell assays showed that the impacts of circPIP5K1A knockdown on hEM15A cell migration and invasion could be abrogated by TMSB4X overexpression (). These results implied that circPIP5K1A might accelerate EM development in vitro via regulating TMSB4X.
Figure 5. TMSB4X overexpression abrogates the effects of circPIP5K1A deletion on EM progression in vitro. (a) The overexpression efficiency of oe-TMSB4X was assessed by RT-qPCR and Western blotting in hEM15A cells. (b) hEM15A cells were respectively transfected with si-NC, si-circPIP5K1A#1, si-circPIP5K1A#1+ vector, si-circPIP5K1A#1+ oe-TMSB4X. Then, CCK-8 assay was performed to analyze proliferative ability of hEM15A cells. (c and d) Flow cytometry assays were performed to analyze cell-cycle regulation and cell apoptosis in hEM15A cells. (e and f) wound healing and transwell assays were performed to analyze the migrative and invasive ability of hEM15A cells. Data are shown as mean ± SD; *P < 0.05, **P < 0.01
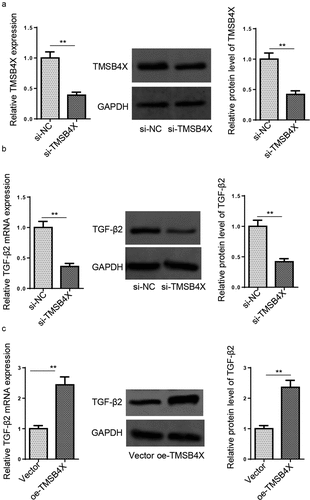
TMSB4X activates TGF-β signaling pathway in hEM15A cells
TGF-β signaling is critical in regulating cell invasion and migration [Citation25]. TGF-β2 is an isoform of the TGF-β cytokine [Citation26]. First of all, the efficiency of TMSB4X knockdown in hEM15A cells was verified by RT-qPCR and western blotting (). As shown in and C, TMSB4X depletion decreased TGF-β2 expression in hEM15A cells, while TMSB4X overexpression increased TGF-β2 expression. The above data indicated that TMSB4X could activate the TGF-β signaling pathway in hEM15A cells.
Figure 6. TMSB4X activates TGF-β signaling pathway in hEM15A cells. (a) The knockdown efficiency of si-TMSB4X was assessed by RT-qPCR and Western blotting in hEM15A cells. (b and c) RT-qPCR and western blotting showed the expression levels of TGF-β2 in hEM15A cells transfected with si-NC, si-TMSB4X, vector, or oe-TMSB4X. Data are shown as mean ± SD; **P < 0.01
Discussion
EM is a kind of gynecological disorder threatening millions of women of childbearing age [Citation27]. Although there are various hypotheses about EM pathogenesis, none of them can solely explain EM occurrence in an appropriate manner [Citation28]. Therefore, it is very important to find novel biomarkers for EM detection and treatment. As evidenced by multiple studies, circRNAs play essential roles in EM development and progression [Citation29]. As a circRNA, circPIP5K1A has been proven to facilitate the progression of multiple human tumors, including hepatocellular carcinoma [Citation30], gastric cancer [Citation31], glioma [Citation32], colon cancer [Citation33], and ovarian cancer [Citation34], through promoting cell proliferation, migration, and invasion. In this work, we verified that circPIP5K1A expression was upregulated in EM tissues and cells. A series of loss-of-function assays manifested that circPIP5K1A promoted cell proliferation, accelerated cell-cycle progression, reduced cell apoptosis, and increased cell migration and invasion in EM, indicating that circPIP5K1A facilitated EM progression in vitro.
As competing endogenous RNAs (ceRNAs) for miRNAs, circRNAs could regulate mRNA expression post-transcriptionally [Citation35]. Besides, it has been demonstrated that circRNA-regulated ceRNA networks are deeply implicated [Citation36]. Herein, miR-153-3p was verified as a target of circPIP5K1A by bioinformatics analyses, dual-luciferase reporter assay, and RIP assay. In this work, we detected that miR-153-3p abundance in hEM15A cells was distinctly increased after circPIP5K1A knockdown and markedly decreased after circPIP5K1A overexpression. In addition, miR-153-3p expression was lowly expressed in EM tissues and cells. Previous studies reported that many miRNAs have been identified as potential biomarkers for EM and participate in EM development and progression [Citation37,Citation38]. As a widely recognized anti-cancer gene, miR-153-3p has been demonstrated as a tumor suppressor in retinoblastoma [Citation39], melanoma [Citation40], and gastric cancer [Citation41], indicating its function of suppressing cell proliferation, migration, and invasion. Herein, it was observed from rescue assays that miR-153-3p inhibition could increase cell proliferation, expedite cell cycle process, decrease cell apoptosis, and promote cell migration and invasion, thereby partially, though not completely, abrogating the suppressive effects of circPIP5K1A knockdown on the EM cellular process. Therefore, it was suggested that circPIP5K1A promoted EM development in vitro via negatively regulating miR-153-3p.
Next, Thymosin Beta-4 X-Linked (TMSB4X) was identified as the target gene of miR-153-3p in this work. Chu et al. elaborated that TMSB4X upregulation in mesenchymal stem cells promoted tumor growth in ovarian cancer [Citation24]. Besides, An et al. disclosed that TMSB4X promoted diffuse-type gastric cancer metastasis [Citation42]. In addition, Makowiecka et al. uncovered that TMSB4X facilitated melanoma migration and invasion [Citation43]. To sum up, TMSB4X might play a promotive role in regulating cell proliferation, migration, and invasion. Herein, it was found that TMSB4X enrichment was lifted in EM tissues and cells. Besides, circPIP5K1A positively regulated TMSB4X level via interaction with miR-153-3p in hEM15A cells. Rescue assays further exhibited that TMSB4X overexpression partly offset the inhibitory effects of circPIP5K1A silencing on EM progression in vitro, suggesting that circPIP5K1A accelerated EM development in vitro via the miR-153-3p/TMSB4X axis.
TGF-beta signaling plays a vital role in a variety of cellular processes, including cell growth, differentiation, cell-cycle regulation, apoptosis, and EMT [Citation44,Citation45]. Besides, TGF-β signaling is also involved in EM pathophysiology [Citation46]. Chen et al. demonstrated that HES5-induced FBXW7 suppression alleviated EM progression by inhibiting the activation of the TGF-β signaling, as indicated by the enhanced apoptosis and inhibited proliferation and invasion of hESC cells [Citation47]. A previous study also elaborated that Syndecan-1 also modulated endometrioma invasion via the TGF-β signaling in an EM subgroup [Citation48]. Herein, it was shown that TGF-β2 expression in hEM15A cells was decreased by TMSB4X knockdown and increased by TMSB4X upregulation, indicating that TMSB4X activated TGF-β signaling pathway in EM.
Conclusion
This study is the first to probe into the biological functions of circPIP5K1A and its regulatory mechanism in EM. Our findings indicated that circPIP5K1A might expedite EM development in vitro by accelerating biological activities of EM cells through activation of the TGF-β signaling pathway via miR-153-3p/TMSB4X axis. However, our work is still limited because of the small sample size and the lack of in vivo experiments. In the future, additional in vitro studies on more EM cell lines, in vivo studies, and larger sample sizes will be applied to further evaluate the role and molecular mechanisms of circPIP5K1A in EM.
Research highlights
CircPIP5K1A depletion suppressed EM progression in vitro.
CircPIP5K1A promoted EM phenotypes in vitro via interaction with miR-153-3p.
CircPIP5K1A regulated TMSB4X expression in EM cells via miR-153-3p.
CircPIP5K1A accelerated EM progression in vitro via regulating TMSB4X expression.
Availability of data and material
All data generated or analyzed during this study are included in this published article or are available from the corresponding author on reasonable request.
Authors’ contributions
LS and YW designed and carried out the study. LS and JW participated in the experiments and statistical analysis. LS, YW and JW wrote the manuscript. JW revised the manuscript. All authors read and approved the final manuscript.
Consent for publication
All of the authors have consented to publication of this research.
Ethics approval and consent to participate
This study was permitted by the Ethics Committee of Maanshan Maternal and Child Health Care Hospital.
Disclosure statement
The authors declare that they have no competing interests.
Additional information
Funding
References
- Yuan Z, Wang L, Wang Y, et al. Tubal origin of ovarian endometriosis. Mod Pathol. 2014;27(8):1154–1162.
- Vercellini P, Vigano P, Somigliana E, et al. Endometriosis: pathogenesis and treatment. Nat Rev Endocrinol. 2014;10(5):261–275.
- Lai ZZ, Yang HL, Ha SY, et al. Cyclooxygenase-2 in Endometriosis. Int J Biol Sci. 2019;15(13):2783–2797.
- Baranov V, Malysheva O, Yarmolinskaya M. Pathogenomics of Endometriosis Development. Int J Mol Sci. 2018;19(7):1852.
- Patel BG, Rudnicki M, Yu J, et al. Progesterone resistance in endometriosis: origins, consequences and interventions. Acta Obstet Gynecol Scand. 2017;96(6):623–632.
- Zondervan KT, Becker CM, Koga K, et al. Endometriosis. Nat Rev Dis Primers. 2018;4:9.
- Li H, Ma X, Yang D, et al. PCAT-1 contributes to cisplatin resistance in gastric cancer through epigenetically silencing PTEN via recruiting EZH2. J Cell Biochem. 2020;121(2):1353–1361.
- Zhang J, Wang H, Meng Q, et al. Expression of MTA1 in endometriosis and its relationship to the recurrence. Medicine (Baltimore). 2018;97(35):e12115.
- Cui L, Chen S, Wang D, et al. LINC01116 promotes proliferation and migration of endometrial stromal cells by targeting FOXP1 via sponging miR-9-5p in endometriosis. J Cell Mol Med. 2021;25(4):2000–2012.
- Qu S, Yang X, Li X, et al. Circular RNA: a new star of noncoding RNAs. Cancer Lett. 2015;365(2):141–148.
- Zaiou M. circRNAs signature as potential diagnostic and prognostic biomarker for diabetes mellitus and related cardiovascular complications. Cells. 2020;9(3):659.
- Xu X, Jia SZ, Dai Y, et al. The relationship of circular RNAs With ovarian endometriosis. Reprod Sci. 2018;25(8):1292–1300.
- Sheng R, Li X, Wang Z, et al. As and their emerging roles as diagnostic and prognostic biomarkers in ovarian cancer. Cancer Lett. 2020;473:139–147.
- Jiang N, Pan W, Li J, et al. Upregulated circular RNA hsa_circ_0008433 regulates pathogenesis in endometriosis via miRNA. Reprod Sci. 2020;27(11):2002–2017.
- He X, Liu N, Mu T, et al. Oestrogen induces epithelial-mesenchymal transition in endometriosis via circ_0004712/miR-148a-3p sponge function. J Cell Mol Med. 2020;24(17):9658–9666.
- Dong L, Zhang L, Liu H, et al. Circ_0007331 knock-down suppresses the progression of endometriosis via miR-200c-3p/HiF-1alpha axis. J Cell Mol Med. 2020;24(21):12656–12666.
- Zhang M, Ren C, Xiao Y, et al. Expression profile analysis of circular RNAs in ovarian endometriosis by microarray and bioinformatics. Med Sci Monit. 2018;24:9240–9250.
- Chen L, Zhu Q, Lu L, et al. MiR-132 inhibits migration and invasion and increases chemosensitivity of cisplatin-resistant oral squamous cell carcinoma cells via targeting TGF-β1. Bioengineered. 2020;11:91–102.
- Shi G, Yang F. Krüppel-like factor 1 (KLF1) promoted the proliferation, migration and invasion of human lens epithelial cells by enhancing the expression of Zinc Finger and BTB domain containing 7A (ZBTB7A) and activating Wnt/β-catenin pathway. Bioengineered. 2021;12(1):4374–4384.
- Yang CY, Wang J, Zhang JQ, et al. Human circular RNA hsa_circRNA_101705 (circTXNDC11) regulates renal cancer progression by regulating MAPK/ERK pathway. Bioengineered. 2021;12(1):4432–4441.
- Cheng Q, Zhang M, Zhang M, et al. Long non-coding RNA LOC285194 regulates vascular smooth muscle cell apoptosis in atherosclerosis. Bioengineered. 2020;11(1):53–60.
- Liang Z, Xu J, Ma Z, et al. MiR-187 suppresses non-small-cell lung cancer cell proliferation by targeting FGF9. Bioengineered. 2020;11(1):70–80.
- Li H, Xuan J, Zhang W, et al. Long non-coding RNA SNHG5 regulates ulcerative colitis via microRNA-375/Janus kinase-2 axis. Bioengineered. 2021;12(1):4150–4158.
- Chu Y, You M, Zhang J, et al. Adipose-Derived mesenchymal stem cells enhance ovarian cancer growth and metastasis by increasing thymosin Beta 4X-Linked expression. Stem Cells Int. 2019;2019:9037197.
- Prud’homme GJ. Pathobiology of transforming growth factor beta in cancer, fibrosis and immunologic disease, and therapeutic considerations. Lab Invest. 2007;87(11):1077–1091.
- Munger JS, Sheppard D. Cross talk among TGF-beta signaling pathways, integrins, and the extracellular matrix. Cold Spring Harb Perspect Biol. 2011;3(11):a005017.
- Ahn SH, Singh V, Tayade C. Biomarkers in endometriosis: challenges and opportunities. Fertil Steril. 2017;107(3):523–532.
- Lagana AS, Garzon S, Gotte M, et al. The pathogenesis of endometriosis: molecular and cell biology insights. Int J Mol Sci. 2019;20(22):5615.
- Dana PM, Taghavipour M, Mirzaei H, et al. Circular RNA as a potential diagnostic and/or therapeutic target for endometriosis. Biomark Med. 2020;14(13):1277–1287.
- Zhang T, Jing B, Bai Y, et al. Circular RNA circTMEM45A acts as the sponge of microRNA-665 to promote hepatocellular carcinoma progression. Mol Ther Nucleic Acids. 2020;22:285–297.
- Ma Y, Cong X, Zhang Y, et al. CircPIP5K1A facilitates gastric cancer progression via miR-376c-3p/ZNF146 axis. Cancer Cell Int. 2020;20(1):81.
- Zheng K, Xie H, Wu W, et al. CircRNA PIP5K1A promotes the progression of glioma through upregulation of the TCF12/PI3K/AKT pathway by sponging miR-515-5p. Cancer Cell Int. 2021;21(1):27.
- Zhang Q, Zhang C, Ma JX, et al. Circular RNA PIP5K1A promotes colon cancer development through inhibiting miR-1273a. World J Gastroenterol. 2019;25(35):5300–5309.
- Sun Q, Li Q, Xie F. LncRNA-MALAT1 regulates proliferation and apoptosis of ovarian cancer cells by targeting miR-503-5p. Onco Targets Ther. 2019;12:6297–6307.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Shen L, Zhang Y, Zhou W, et al. Circular RNA expression in ovarian endometriosis. Epigenomics. 2018;10(5):559–572.
- Bjorkman S, Taylor HS. MicroRNAs in endometriosis: biological function and emerging biomarker candidatesdagger. Biol Reprod. 2019;100:1135–1146.
- Bjorkman S, Taylor HS. Erratum: microRNAs in endometriosis biological function and emerging biomarker candidates. Biol Reprod. 2019;101(6):1179.
- Wang Y, Wang J, Hao H, et al. lncRNA KCNQ1OT1 promotes the proliferation, migration and invasion of retinoblastoma cells by upregulating HIF-1alpha via sponging miR-153-3p. J Investig Med. 2020;68(8):1349–1356.
- Luan W, Shi Y, Zhou Z, et al. circRNA_0084043 promote malignant melanoma progression via miR-153-3p/Snail axis. Biochem Biophys Res Commun. 2018;502(1):22–29.
- Gao Y, Xie M, Guo Y, et al. Long non-coding RNA FGD5-AS1 regulates cancer cell proliferation and chemoresistance in gastric cancer through miR-153-3p/CITED2 axis. Front Genet. 2020;11:715.
- An HW, Kim SY, Kwon JW, et al. In vivo CRISPR-Cas9 knockout screening using quantitative PCR identifies thymosin beta-4 X-linked that promotes diffuse-type gastric cancer metastasis. Mol Carcinog. 2021;60(9):597–606.
- Makowiecka A, Malek N, Mazurkiewicz E, et al. Thymosin beta4 regulates focal adhesion formation in human melanoma cells and affects their migration and invasion. Front Cell Dev Biol. 2019;7:304.
- Moustakas A, Pardali K, Gaal A, et al. Mechanisms of TGF-beta signaling in regulation of cell growth and differentiation. Immunol Lett. 2002;82(1–2):85–91.
- Heldin CH, Landstrom M, Moustakas A. Mechanism of TGF-beta signaling to growth arrest, apoptosis, and epithelial-mesenchymal transition. Curr Opin Cell Biol. 2009;21(2):166–176.
- Young VJ, Ahmad SF, Duncan WC, et al. The role of TGF-beta in the pathophysiology of peritoneal endometriosis. Hum Reprod Update. 2017;23(5):548–559.
- Chen LJ, Hu B, Han ZQ, et al. Repression of FBXW7 by HES5 contributes to inactivation of the TGF-beta signaling pathway and alleviation of endometriosis. FASEB J. 2021;35:e20938.
- Ponandai-Srinivasan S, Saare M, Boggavarapu NR, et al. Syndecan-1 modulates the invasive potential of endometrioma via TGF-beta signalling in a subgroup of women with endometriosis. Hum Reprod. 2020;35(10):2280–2293.