ABSTRACT
Oral squamous cell carcinoma (OSCC) poses a threat to public health worldwide. LncRNA APCDD1L-AS1 has been reported to participate in tumorigenesis and development of acquired chemoresistance. However, the role of APCDD1L-AS1 in 5-fluorouracil (5-FU) resistance regulation within OSCC is still obscure. In this study, 5-FU-resistant cell models were established with OSCC cell lines (HSC-3 and HN-4). Gene expressions and protein levels were detected by RT-qPCR and Western blotting, respectively. CCK-8, colony forming, and flow cytometry were utilized to measure IC50 value, cell viability, and cell apoptosis of 5-FU-resistant OSCC cells. Dual-luciferase reporter assay and RIP assay were applied to identify the associations between miR-1224-5p and APCDD1L-AS1 or NSD2. Herein, high APCDD1L-AS1 expression was shown in OSCC tissues and cells resistant to 5-FU and related to the worse prognosis of OSCC patients. APCDD1L-AS1 knockdown impaired 5-FU resistance in 5-FU-resistant OSCC cells by reducing IC50 value, suppressing cell viability, and accelerating cell apoptosis. Besides, extracellular APCDD1L-AS1 could be transferred to sensitive cells via exosome incorporation, thereby transmitting 5-FU resistance in OSCC cells. Besides, miR-1224-5p was a molecular target of APCDD1L-AS1 and directly targeted NSD2 in 5-FU-resistant cells. MiR-1224-5p exhibited a much lower level in 5-FU-resistant tissues and increased 5-FU sensitivity in 5-FU-resistant OSCC cells. Moreover, NSD2 upregulation neutralized the influence of blocking APCDD1L-AS1 in HSC-3/5-FU and HN-4/5-FU cells on 5-FU resistance. To sum up, our study demonstrated that exosomal APCDD1L-AS1 conferred resistance to 5-FU in HSC-3/5-FU and HN-4/5-FU cells via the miR-1224-5p/NSD2 axis, thus providing a novel target for OSCC chemoresistance.
Introduction
As a malignancy derived from oral mucosa, oral squamous cell carcinoma (OSCC) is the most frequently diagnosed subtype of head and neck squamous cell carcinoma (HNSC) [Citation1]. Since there are few typical symptoms for OSCC at early stages, many OSCC patients were diagnosed at advanced stages [Citation2]. Currently, surgery combined with adjuvant chemotherapy or radiotherapy is a major method for patients with advanced OSCC [Citation3]. In spite of progressions made in diagnosis and treatment, OSCC is still a type of human cancer characterized by high mortality [Citation4]. As indicated in multiple studies, OSCC patients may develop drug resistance which greatly impairs the efficacy of chemotherapy and constitutes a major cause for therapeutic failure of OSCC patients [Citation5]. 5-fluorouracil (5-FU) is a standard drug applied for OSCC chemotherapy [Citation6]. Nevertheless, 5-FU resistance brings great obstacles to its application in OSCC chemotherapy [Citation7]. Therefore, to overcome 5-FU resistance in OSCC therapies and assure 5-FU efficacy in OSCC chemotherapy, the underlying mechanism of 5-FU sensitivity should be further investigated in OSCC.
Long noncoding RNAs (lncRNAs) are non-coding RNAs (>200 nucleotides) possessing no protein-coding functions [Citation8]. The active involvement of lncRNAs in the development of human malignancies through gene expression and cellular function regulation has been fully evidenced by previous studies [Citation9]. What’s more, multiple studies have extensively investigated the development of lncRNA-modulated drug resistance in chemotherapy for human cancers. For example, Wang et al. found lncRNA NUTM2A-AS1 was upregulated and promoted tumor growth, metastasis, and chemoresistance in gastric cancer by elevating TET1 and HIF-1A levels through interaction with miR-376a [Citation10]. A report from Zhou et al. revealed that lncRNA PVT1 attenuated susceptibility to gemcitabine in pancreatic cancer through miR-619-5p/ATG14 and miR-619-5p/Pygo2 axes by Wnt/β-catenin and autophagy pathways [Citation11]. Besides, Lei et al. also demonstrated that lncRNA H19 conferred was packaged into exosomes, thus attenuating gefitinib susceptibility in non‑small cell lung cancer cells [Citation12]. Also, Ghafouri-Fard et al. disclosed that non-coding RNAs play critical roles in regulating the patients’ responses to 5-FU [Citation13]. A novel lncRNA, APCDD1L-AS1, showed higher expression in HNSC [Citation14]. Moreover, a report from Wu et al. also indicated that APCDD1L-AS1 impaired lung adenocarcinoma sensitivity to icotinib by upregulating SIRT5 level via miR-1322/miR-1972/miR-324-3p, thereby restraining EGFR autophagic degradation [Citation15], which indicates a promoting effect of APCDD1L-AS1 in the development of acquired chemoresistance during chemotherapy. Therefore, we supposed that APCDD1L-AS1 might promote chemoresistance in OSCC. However, the specific functions of APCDD1L-AS1 in regulating resistance of OSCC cells to 5-FU haven’t been studied yet.
LncRNAs may act as competing endogenous RNAs (ceRNAs) by specifically adsorbing miRNAs to regulate the levels of target genes, thus participating in the regulation of chemoresistance [Citation16,Citation17]. Therefore, we hypothesized that APCDD1L-AS1 increased the chemoresistance of OSCC by serving as a ceRNA to sponge miR-1224-5p and upregulate NSD2 expression. Through a series of functional assays, it was demonstrated that exosomal APCDD1L-AS1 induced 5-FU resistance in OSCC via the miR-1224-5p/NSD2 axis, thereby offering a novel treatment strategy for 5-FU-resistant OSCC.
Materials and methods
Tissue samples
Forty pairs of OSCC tissues and adjacent normal tissues were acquired from patients diagnosed and undergoing surgery at Henan Provincial People’s Hospital and accepted no radiotherapy or immunotherapy before surgery. The clinicopathologic details of the patients were as shown in . As 5-FU has a special effect on OSCC, short-term recurrence after 5-FU therapy means insensitivity to 5-FU [Citation18]. Therefore, the OSCC specimens were grouped into the 5-FU-sensitive group (from patients without recurrence or with recurrence after 6 months as of the completion of primary chemotherapy; n = 22) and the 5-FU-resistant group (from patients with recurrence within 6 months as of the completion of primary chemotherapy; n = 18). Prior to this study, each participant signed the informed consent. The current study was permitted by the Ethics Committee of Henan Provincial People’s Hospital.
Table 1. Clinicopathologic details of the patients
Cell culture and establishment of 5-FU-resistant cellular models
OSCC cell lines (SCC-4, HSC-3, TSCC1, SCC090, and HN-4) and normal human oral keratinocyte cell line (NHOK) were acquired from BeNa Culture Collection (Beijing, China). The cells were cultivated in DMEM (10% FBS) (Invitrogen, USA) in 5% CO2 at 37°C.
To establish OSCC cells resistant to 5-FU (HSC-3/5-FU and HN-4/5-FU) derived from parental HSC-3 and HN-4 cell lines, HSC-3 and HN-4 cells were treated with gradient 5-FU doses for more than 6 months. Then, HSC-3/5-FU and HN-4/5-FU cells were cultured in DMEM (10% FBS) containing 4 μg/mL 5-FU to maintain drug resistance.
Cell transfection
The short hairpin (sh)‑RNA against APCDD1L-AS1 (sh-APCDD1L-AS1: CCGGGAGAGUUAUGGCAUUTT), NSD2 (sh-NSD2: 5ʹ-CCAGCUAAGAAAGAGUCUUTT), and their negative control (sh-NC: UUCUCCGAACGUGUCACGUTT), pcDNA-APCDD1L-AS1 overexpression vector (oe-APCDD1L-AS1), pcDNA-NSD2 overexpression vector (oe-NSD2), and pcDNA 3.1 empty vector (Vector), as well as miR-1224-5p inhibitor (5ʹ-CCACCUCACCGUCCCCGAU-3ʹ), miR-1224-5p mimics (5ʹ-GUGAGGACUCGGGAGGUGG-3ʹ), and their corresponding negative controls (NC inhibitor, 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ; NC mimics: 5ʹ-UUCUCCGAACGUGUCACGUCC-3ʹ) were obtained from GenePharma (Shanghai, China). HSC-3/5-FU or HN-4/5-FU cells were transfected with sh-APCDD1L-AS1 (45 nM), sh‑NSD2 (45 nM), sh‑NC (15 nM), miR-1224-5p mimics (45 nM), miR-1224-5p inhibitor (45 nM), NC mimics (15 nM), NC inhibitor (15 nM), Vector (15 nM), oe-APCDD1L-AS1 (45 nM), or oe-NSD2 (45 nM) via Lipofectamine® 2000 reagent (Invitrogen). After 48 hours, the transfected cells were used for subsequent experiments.
RT-qPCR
Trizol reagent (Takara, China) was applied for total RNA extraction in tissues and cells. Then, the complementary DNA (cDNA) was synthesized by reversely transcribing RNA with TaqMan miRNA RT kit (Thermo Fisher Scientific). RT-qPCR was conducted on Applied Biosystems 7500 real-time PCR system with SYBR Green PCR Master Mix (Thermo Fisher Scientific). The relative enrichments of APCDD1L-AS1, NSD2, and miR-1224-5p were calculated by 2−ΔΔCt method. GAPDH or U6 was utilized as the internal reference. RT-qPCR primers were as follows: APCDD1L-AS1: 5ʹ-GAGCCTTGGAAAGGAGGACC-3ʹ (Forward (F)) and 5ʹ-GATCCATGCAGGTGGGAACA (Reverse (R)); NSD2: 5ʹ-AATATGACTCCTTGCTGGAGCAGG-3ʹ (F) and 5ʹ-ATTTCAACAGGTGGTCTTTGTCTC-3ʹ (R); GAPDH: 5ʹ-AACGGATTTGGTCGTATTGGG-3ʹ (F) and 5ʹ-CGCTCCTGGAAGATGGTGATG −3ʹ (R); miR-1224-5p: 5ʹ-ACACTCCAGCTGGGGTGAGGACTGGGG-3ʹ (F) and 5ʹ-TGGTGTCGTGGAGTCG-3ʹ (R); U6: 5ʹ- TGCGGGTGCTCGCTTCGGCAGC‑3ʹ (F) and 5ʹ‑CCAGTGCAGGGTCCGAGGT‑3ʹ (R).
CCK-8 assay
To evaluate 5-FU cytotoxicity, the IC50 (5-FU concentration causing 50% decrease in cell viability) value was assessed by CCK-8 assay. In brief, HSC-3/5-FU or HN-4/5-FU cells were seeded into 96-well plates (1 × 104 cells/well) after transfection. Then, the cells were incubated with 5-FU (1, 2, 4, 8, 16, 32, 64 μg/mL) for 24 hours. Thereafter, 10 μL CCK-8 reagent (Dojindo, Japan) was supplemented to each well. The cells were cultivated for 2 hours prior to the absorbance measurement performed at 450 nm with a microplate reader [Citation19].
Colony-forming assay
Briefly, transfected HSC-3/5-FU or HN-4/5-FU cells were firstly dispersed into single cells and then seeded into 6-well plates (500 cells/well). Then, the cells were cultivated for 2 weeks at 37°C with 5% CO2. Subsequently, the formed cell colonies were stained with crystal violet (0.1%) after being fixed with paraformaldehyde (4%). The cell colony number was measured under a microscope.
Flow cytometry assay
To detect apoptosis of OSCC cells, Annexin V-FITC Apoptosis Detection Kit (Yeasen, China) was utilized for flow cytometry assay [Citation20]. Transfected HSC-3/5-FU or HN-4/5-FU cells were washed in PBS and resuspended in binding buffer. Thereafter, the cell suspension was supplemented with Annexin V-FITC solution (5 μl) and PI solution (5 μl) followed by 15 minutes’ incubation in darkness. The cell apoptotic rate was analyzed with a flow cytometer (Agilent, China).
Dual-luciferase reporter assay
To build luciferase reporter vectors, the sequence of APCDD1L-AS1 or NSD2 3ʹ UTR with wild type (WT) or mutant type (MUT) binding sites to miR-1224-5p were cloned into pmirGLO dual-luciferase Vector (Promega, USA), respectively. Afterward, APCDD1L-AS1-WT (or APCDD1L-AS1-MUT) as well as NSD2-WT (or NSD2-MUT) were co-transfected into HSC-3/5-FU or HN-4/5-FU cells together with miR-1224-5p mimics or NC mimics via Lipofectamine 2000 (Invitrogen). To assess the luciferase activity, a dual luciferase assay kit (Promega) was employed after 48 hours of transfection [Citation21].
Western blotting
Extraction of total protein was performed with RIPA buffer. Thereafter, total protein was qualified with BCA kit (Solarbio). Then, proteins were separated by SDS-PAGE and transferred onto PVDF membranes. After blocked with nonfat milk for 1 hour, the membranes were cultivated with primary antibodies against CD63 (ab59479, 1:1000, Abcam, USA) or CD9 (ab233396, 1:1000, Abcam, USA) at 4°C overnight, respectively. After the secondary antibody (ab205718, 1:2000, Abcam, USA) was added, the membranes were incubated for another 2 hours at room temperature. Finally, the enhanced chemiluminescence detection kit (Pierce Biotechnology, USA) was employed to detect the blots [Citation22].
Exosome isolation
Exosome extraction from cell culture medium was performed with ExoQuick precipitation kit (System Biosciences, USA). Briefly, cells were collected when 80% confluency was achieved. After 10 minutes’ centrifugation at 3000 g, cells and cell debris were discarded. Thereafter, 250 μL supernatant was mixed with 63 μL ExoQuick precipitation kit and then incubated for 30 minutes at 4°C. After centrifugation at 1500 g for 30 minutes, the supernatant was discarded. Subsequently, the residual liquid was removed by centrifugation at 1500 g for 5 minutes to obtain the final exosome pellets.
Transmission electron microscopy (TEM)
The exosome pellets were re-suspended with 200 μL PBS. For TEM analysis, 10 μl exosome suspension was placed on a parafilm. Next, a carbon-coated copper grid was made floated on the drop of exosome suspension for 5 minutes. Thereafter, the grid was removed and the redundant liquid on the grid was blotted up with clean filter paper. Then, the grid was immersed in 2% phosphotungstic acid (pH = 7.0) for 30 seconds. Afterward, the redundant liquid was removed. Finally, the grid was observed under a transmission electron microscope (Tecnai G2 Spirit Bio TWIN, USA) after 5 minutes’ drying.
RNA immunoprecipitation (RIP) assay
RIP assay was performed with EZ-Magna Rip Kit (Millipore, USA) [Citation23]. HSC-3/5-FU or HN-4/5-FU cells were firstly lysed in RIP lysis buffer and then cultured with magnetic beads conjugated with human anti-Ago2 antibody. Thereafter, normal mouse IgG control was applied as the negative control. Trizol reagent was adopted to isolate RNAs from the precipitates. Thereafter, the relative RNA expression levels were assessed by RT-qPCR.
Statistical analysis
Each experiment in our study was done in triplicate, independently. All the data were shown as mean ± standard deviation. For statistical analysis, the GraphPad Prism 6.0 software was employed. Student’s t-test or one-way ANOVA was employed to analyze differences between two or more groups, respectively. Survival curves were plotted and analyzed by Kaplan-Meier method and log-rank test. P< 0.05 indicates a statistical significance of difference.
Results
High APCDD1L-AS1 is discovered in OSCC and correlative to the poor prognosis of OSCC patients
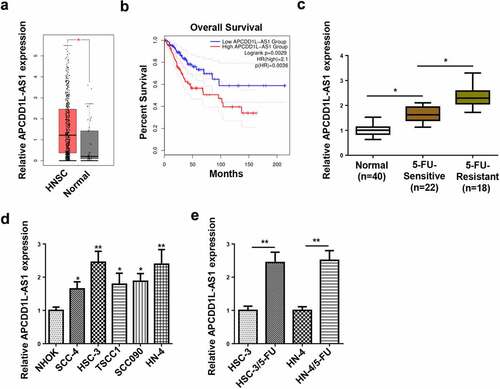
TCGA database revealed that APCDD1L-AS1 expression was apparently lifted in HNSC samples, relative to normal tissues (). Also, HNSC patients with high APCDD1L-AS1 levels exhibited shorter overall survival time than those exhibiting low APCDD1L-AS1 level, as indicated by the survival curve plotted by Kaplan-Meier analysis (). To further inquire into the specific regulatory effects of APCDD1L-AS1 on 5-FU resistance in OSCC, APCDD1L-AS1 abundance was assessed in OSCC tissues sensitive to 5-FU and OSCC tissues resistant to 5-FU. As shown by RT-qPCR, APCDD1L-AS1 level was mostly upregulated in 5-FU-resistant OSCC tissues and clearly increased in 5-FU-sensitive OSCC tissues, relative to normal tissues (). Furthermore, APCDD1L-AS1 level was determined in OSCC cell lines (SCC-4, HSC-3, TSCC1, SCC090, and HN-4) and normal human oral keratinocyte cell line (NHOK) by RT-qPCR, and OSCC cells exhibited an obvious increase in APCDD1L-AS1 expression, relative to normal cells (). Since APCDD1L-AS1 expression was much higher in HSC-3 and HN-4 cells, the two cell lines were chosen to establish corresponding 5-FU-resistant cellular models (HSC-3/5-FU and HN-4/5-FU) for subsequent experiments. According to RT-qPCR assay, APCDD1L-AS1 level was elevated in HSC-3/5-FU and HN-4/5-FU cells, relative to parental HSC-3 and HN-4 cells (). Therefore, it was indicated that APCDD1L-AS1 was remarkably overexpressed in 5-FU-resistant OSCC tissues and cells, suggesting its promoting role in regulating OSCC 5-FU resistance.
Figure 1. High APCDD1L-AS1 is discovered in OSCC and correlative to poor prognosis of OSCC patients
APCDD1L-AS1 confers resistance to 5-FU in OSCC cells
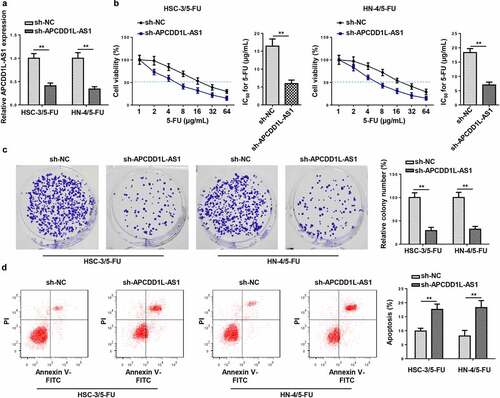
To confirm the effects of APCDD1L-AS1 on 5-FU resistance in OSCC cells, APCDD1L-AS1 silencing was firstly conducted in HSC-3/5-FU and HN-4/5-FU cells, and RT-qPCR was employed to evaluate the efficiency of APCDD1L-AS1 knockdown (). In addition, it was shown by CCK-8 that APCDD1L-AS1 inhibition significantly alleviated the resistance of HSC-3/5-FU and HN-4/5-FU cells to 5-FU, as indicated by the reduced IC50 values (). Thereafter, it was further demonstrated by colony formation and flow cytometry assays that APCDD1L-AS1 silencing visibly impaired the proliferation and accelerated the apoptosis of HSC-3/5-FU and HN-4/5-FU cells) ( and d). Taken together, APCDD1L-AS1 exacerbated 5-FU resistance in OSCC cells.
Figure 2. APCDD1L-AS1 confers resistance to 5-FU in OSCC cells
APCDD1L-AS1 is transferred extracellularly via exosome incorporation in 5-FU-Resistant OSCC Cells
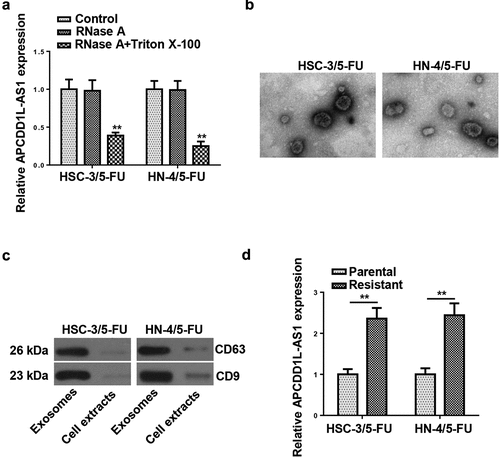
To explore whether APCDD1L-AS1 was exosome-encapsulated and transmitted through exosomes, we collected the culture medium in which 5-FU-resistant OSCC cells were cultivated. After treatment with RNase A or RNase A+ Triton X-100, APCDD1L-AS1 abundance in the medium was measured. The results exhibited that RNase A treatment exerted almost no effect on APCDD1L-AS1 level, whereas the combination of RNase A and Triton X-100 treatments caused an obvious decrease expression of APCDD1L-AS1 in HSC-3/5-FU and HN-4/5-FU cells, indicating extracellular APCDD1L-AS1 was encapsulated in membranes (). Thereafter, exosomes were isolated from the culture medium, and a typical lipid bilayer membrane was observed (). Western blotting results showed that the protein levels of exosome-associated proteins (CD63 and CD9) upregulated in exosomes (). Besides, it was revealed by RT-qPCR that APCDD1L-AS1 level was distinctly higher in exosomes extracted from 5-FU-resistant OSCC cells (). These data demonstrated that extracellular APCDD1L-AS1 was transferred via exosome incorporation in 5-FU-resistant OSCC cells.
Figure 3. APCDD1L-AS1 is transferred extracellularly via exosome incorporation in 5-FU-Resistant OSCC Cells
APCDD1L-AS1 inhibition attenuates 5-FU resistance mediated by exosomes
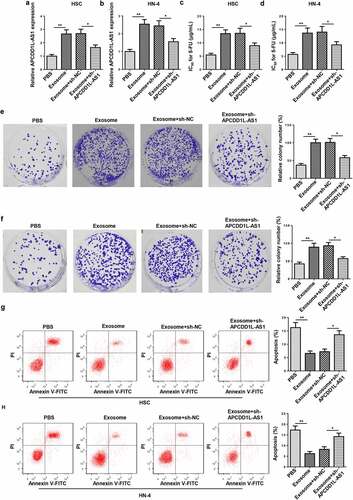
To further investigate the role of exosomes and their molecular mechanism, HSC and HN-4 cells were treated with exosomes isolated from the culture medium of HSC-3/5-FU and HN-4/5-FU cells and then transfected with sh-APCDD1L-AS1. The APCDD1L-AS1 enrichment was upregulated after treatment with exosomes and reduced by sh-APCDD1L-AS1 in HSC and HN-4 cells ( and b). CCK-8 and colony-forming assays manifested that 5-FU IC50 and cell viability were enhanced in HSC and HN-4 cells treated with exosomes and abrogated by APCDD1L-AS1 blocking (). Furthermore, exosome treatment reduced apoptosis in HSC and HN-4 cells, while APCDD1L-AS1 knockdown abolished this impact ( and h). To sum up, these results indicated that exosomes conferred 5-FU resistance by lifting APCDD1L-AS1 level in OSCC cells.
Figure 4. APCDD1L-AS1 inhibition attenuates 5-FU resistance mediated by exosomes
APCDD1L-AS1 serves as a sponge for miR-1224-5p in 5-FU-resistant OSCC cells
MiR-1224-5p was predicted as a target of APCDD1L-AS1 by Starbase software (). To confirm miR-1224-5p as a direct target of APCDD1L-AS1 in 5-FU-resistant OSCC cells (HSC-3/5-FU and HN-4/5-FU cells), a dual-luciferase reporter assay was performed. Firstly, miR-1224-5p was overexpressed in 5-FU-resistant OSCC cells, and the overexpression efficiency was evaluated (). It was shown that upregulation of miR-1224-5p level dramatically decreased the luciferase activity of 5-FU-resistant OSCC cells in the APCDD1L-AS1-WT group, relative to the APCDD1L-AS1-MUT group ( and d). In addition, it was also demonstrated by RIP assay that APCDD1L-AS1 directly targets miR-1224-5p in 5-FU-resistant OSCC cells ( and f). Furthermore, RT-qPCR assay exhibited that miR-1224-5p level was lower in OSCC tissues, relative to normal tissues (). Besides, it was discovered that miR-1224-5p abundance was much lower in 5-FU-resistant OSCC tissues, relative to 5-FU-sensitive OSCC tissues (). Thereafter, APCDD1L-AS1 expression was notably elevated in 5-FU-resistant OSCC cells after APCDD1L-AS1 overexpression transfection, as indicated in . As presented in and K, APCDD1L-AS1 knockdown elevated miR-1224-5p abundance, while APCDD1L-AS1 overexpression exerted an opposite effect in 5-FU-resistant OSCC cells. Then, miR-1224-5p knockdown efficiency was detected in HSC-3/5-FU and HN-4/5-FU cells (). As indicated in and N, APCDD1L-AS1 expression was significantly increased after miR-1224-5p inhibition and remarkably decreased after miR-1224-5p upregulation. These results demonstrated APCDD1L-AS1 was negatively correlative to miR-1224-5p in 5-FU-resistant OSCC cells.
Figure 5. APCDD1L-AS1 serves as a sponge for miR-1224-5p in 5-FU-resistant OSCC cells
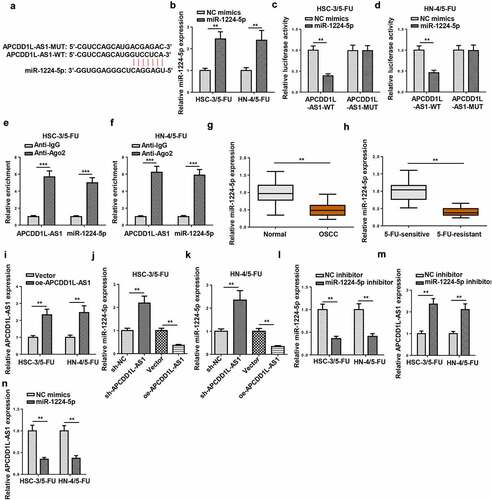
MiR-1224-5p overexpression overcomes 5-FU resistance in 5-FU-resistant OSCC cells
To explore the impact of miR-1224-5p on 5-FU resistance in OSCC cells resistant to 5-FU (HSC-3/5-FU and HN-4/5-FU), functional assays were conducted. As shown by CCK-8 assay, miR-1224-5p overexpression increased the sensitivity to 5-FU in HSC-3/5-FU and HN-4/5-FU cells (). Results of colony forming and flow cytometry assays revealed that increased miR-1224-5p abundance attenuate cell viability and increased the apoptotic rate of HSC-3/5-FU and HN-4/5-FU cells ( and c). To further verify whether APCDD1L-AS1 modulated the resistance to 5-FU in OSCC cells resistant to 5-FU by regulating miR-1224-5p level, a series of rescue assays were performed. According to the results of CCK-8, colony forming, and flow cytometry assays, downregulation of miR-1224-5p expression abrogated the alleviating effect on chemoresistance in 5-FU-resistant OSCC cells caused by APCDD1L-AS1 inhibition (). To sum up, miR-1224-5p overexpression attenuated 5-FU resistance in 5-FU-resistant OSCC cells, whereas APCDD1L-AS1 silencing led to increased miR-1224-5p enrichment, thus weakening 5-FU resistance in 5-FU-resistant OSCC cells.
Figure 6. MiR-1224-5p overexpression overcomes 5-FU resistance in 5-FU-resistant OSCC cells
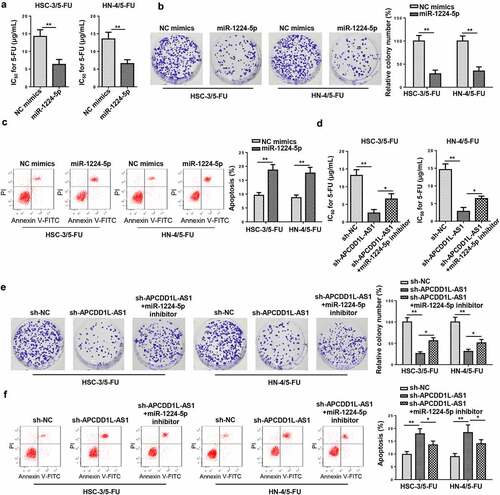
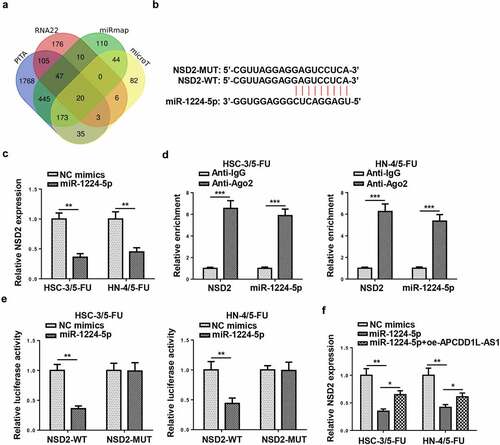
NSD2 is a direct target of miR-1224-5p in 5-FU-resistant OSCC cells
To investigate the downstream mechanism of miR-1224-5p, online tools, including PITA, RNA22, miRmap, and microT, were utilized to find target genes for miR-1224-5p via bioinformatic analysis, and there were 20 candidates (SEC14L1, ZNF107, DERL2, AIF1L, ZNF493, IKZF4, SH3RF1, ZNF91, ARID1A, NID1, NSD2, FYCO1, PCMT1, JAZF1, SIGMAR1, PRICKLE2, NAV1, RNF157, PSME3, and TAOK1) enrolled (). Among the 20 genes screened, nuclear receptor binding SET domain protein 2 (NSD2) has been identified to accelerate the progression of several malignancies and be positively correlated with chemoresistance [Citation24–26]. Hence, NSD2 was selected as the target for miR-1224-5p in this study. Starbase website predicted the latent-binding sites between miR-1224-5p and NSD2 (). After amplifying miR-1224-5p in 5-FU-resistant OSCC cells (HSC-3/5-FU and HN-4/5-FU cells), NSD2 expression was significantly decreased, as indicated by RT-qPCR (). RIP assay confirmed that NSD2 and miR-1224-5p coexisted in RNA-induced silencing complexes (). Thereafter, luciferase reporter assays were performed and the outcomes exhibited that the luciferase activity of HSC-3/5-FU and HN-4/5-FU cells transfected with NSD2-WT was declined by miR-1224-5p upregulation, while there was no obvious change found in the NSD2-MUT group (). As indicated by RT-qPCR assay, NSD2 expression in HSC-3/5-FU and HN-4/5-FU cells was markedly downregulated by miR-1224-5p overexpression, while APCDD1L-AS1 addition partly reversed such an inhibitory effect (). Taken together, APCDD1L-AS1 positively regulated NSD2 expression via interaction with miR-1224-5p.
Figure 7. NSD2 is a direct target of miR-1224-5p in 5-FU-resistant OSCC cells
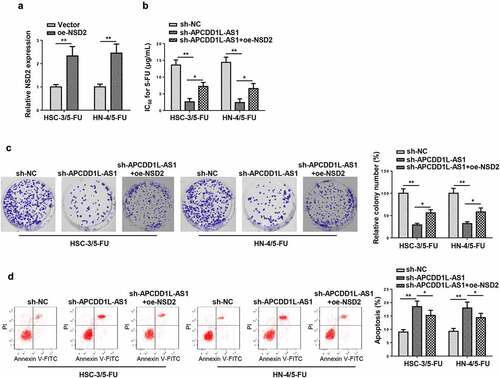
APCDD1L-AS1 knockdown enhances susceptibility to 5-FU in 5-FU-resistant OSCC cells by decreasing NSD2
To investigate how APCDD1L-AS1 knockdown affected 5-FU sensitivity of 5-FU-resistant OSCC cells (HSC-3/5-FU and HN-4/5-FU cells), NSD2 expression was firstly overexpressed in 5-FU-resistant OSCC cells, and it was revealed that NSD2 level was upregulated after NSD2 upregulation by RT-qPCR (). As shown in CCK-8 assay, APCDD1L-AS1 blocking increased the sensitivity of HSC-3/5-FU and HN-4/5-FU cells to 5-FU, while NSD2 overexpression abolished this phenomenon, as evidenced by the variation of IC50 value (). Colony forming and flow cytometry assays showed that APCDD1L-AS1 knockdown suppressed proliferation and increased apoptosis of HSC-3/5-FU and HN-4/5-FU cells; however, NSD2 overexpression abolished such impact on HSC-3/5-FU and HN-4/5-FU cells, indicating that APCDD1L-AS1 knockdown alleviated 5-FU resistance in OSCC cells resistant 5-FU via upregulating NSD2 ( and d). To sum up, APCDD1L-AS1 knockdown increased 5-FU sensitivity of OSCC cells by downregulating NSD2 level.
Figure 8. APCDD1L-AS1 knockdown enhances susceptibility to 5-FU in 5-FU-resistant OSCC cells by decreasing NSD2
Discussion
5-FU is frequently used in OSCC chemotherapy [Citation27]. However, OSCC cells may acquire resistance to 5-FU during continuous chemotherapy, which greatly impaired the clinical effectiveness of 5-FU in OSCC patients [Citation28]. In the current study, we mainly investigated the effect of APCDD1L-AS1 in regulating OSCC sensitivity to 5-FU.
Accumulated studies have demonstrated that abnormally expressed lncRNAs were heavily involved in the chemoresistance regulation of OSCC [Citation29]. For example, Fang et al. demonstrated that lncRNA UCA1 aggravated OSCC tumorigenesis and cisplatin resistance by repressing miR-184 expression [Citation30]. Wang et al. reported that lncRNA MALAT1 conferred OSCC resistance to cisplatin via the PI3K/AKT/m-TOR axis [Citation31]. Lian et al. discovered that higher APCDD1L-AS1 abundance was correlative to shorter survival of patients with bladder cancer [Citation32]. It was also demonstrated by Luo et al. that APCDD1L-AS1 was elevated in lung squamous cell carcinoma (LUSC) positively correlative to unsatisfactory prognosis of LUSC patients [Citation33]. In this study, APCDD1L-AS1 expression was apparently enhanced in OSCC tissues, especially in 5-FU-resistant OSCC tissues. In addition, APCDD1L-AS1 expression was significantly upregulated in OSCC cell lines, compared with normal cells. Moreover, APCDD1L-AS1 expression in 5-FU-resistant OSCC cell models was markedly higher than that in parental OSCC cells, indicating the positive relation between APCDD1L-AS1 expression and 5-FU resistance in OSCC. Functional analysis revealed that APCDD1L-AS1 inhibition decreased IC50 value of 5-FU, attenuated cell viability, and accelerated cell apoptosis in OSCC cells resistant to 5-FU. Therefore, it was assumed that APCDD1L-AS1 might play a promoting role in the development of 5-FU resistance in OSCC.
It has been widely recognized that exosomes participate in cellular communication and regulation of physiological processes, and exosomal transfer of lncRNAs was also correlative to drug resistance acquired in chemotherapy [Citation34–36]. Wang et al. discovered that exosome-mediated lncRNA HEIH regulated the miR-3619-5p/HDGF pathway to promote the chemoresistance of tongue squamous cell carcinoma to cisplatin [Citation37]. Kang et al. revealed that exosomal transmission of lncRNA PART1 conferred resistance to gefitinib in esophageal squamous cell carcinoma by sponging miR-129 [Citation38]. A report from Tong et al. demonstrated that exosomal lncRNA POU3F3 ESCC resistance to cisplatin by inducing differentiation in cancer-associated fibroblasts [Citation39]. These studies indicate that lncRNAs may exacerbate chemoresistance in HNSC via exosomal transmission. Herein, TEM, Western blotting, and RT-qPCR assays demonstrated that extracellular APCDD1L-AS1 was packaged into exosomes. Furthermore, exosomes conferred 5-FU resistance by lifting APCDD1L-AS1 level in OSCC cells. Therefore, our data proved that APCDD1L-AS1 could aggravate 5-FU resistance in OSCC in vitro via exosomal transfer.
Numerous microRNAs (miRNAs) have been identified as vital regulators in human cancers [Citation40]. MiR-1224-5p played an anti-cancer role in various cancers, such as rectal cancer [Citation41], pancreatic cancer [Citation42], ovarian cancer [Citation43], and glioma [Citation44]. In addition, a report from Shi et al. also elucidated that miR-1224-5p could impair the development of esophageal squamous cell carcinoma, which is another HNSC subtype [Citation45]. In the current study, miR-1224-5p was targeted and negatively regulated by APCDD1L-AS1 in OSCC cells resistant to 5-FU. We further discovered that miR-1224-5p was lowly expressed in OSCC tissues, particularly 5-FU-resistant OSCC tissues. In addition, miR-1224-5p overexpression attenuated 5-FU resistance in 5-FU-resistant OSCC cells, and miR-1224-5p downregulation offset the alleviative effect on 5-FU resistance in OSCC cells due to APCDD1L-AS1 depletion. These findings suggested that APCDD1L-AS1 promoted 5-FU resistance in OSCC via downregulating miR-1224-5p.
Massager RNAs (mRNAs) are deeply involved in cancer development and progression and play important roles in cancer detection and diagnosis [Citation46]. The oncogenic role of NSD2 has been widely studied in many malignancies, such as renal cancer [Citation47], colorectal cancer [Citation48], and prostate cancer [Citation49]. Herein, the binding ability between NSD2 and miR-1224-5p was also confirmed. Rescue assays further revealed that NSD2 abundance could partly neutralize the influence caused by APCDD1L-AS1 suppression on resistance to 5-FU in OSCC cells. Therefore, all these results indicated that APCDD1L-AS1 blocking enhanced 5-FU susceptibility in OSCC cells by reducing NSD2 abundance.
Conclusion
To sum up, our results suggested that exosomal-mediated transfer of APCDD1L-AS1 interacted with miR-1224-5p to positively regulate NSD2 level, thus promoting resistance to 5-FU in OSCC cells. The current study offers a novel clue for in-depth investigation of OSCC pathogenesis. The results in this paper will be further verified via in vivo experiments in the future.
Highlights
1. APCDD1L-AS1 induced 5-FU resistance in OSCC cells
2. APCDD1L-AS1 is transferred extracellularly via exosome incorporation in 5-FU-resistant OSCC
3. APCDD1L-AS1 increased chemoresistance of OSCC cells via miR-1224-5p/NSD2
References
- Chi AC, Day TA, Neville BW. Oral cavity and oropharyngeal squamous cell carcinoma–an update. CA Cancer J Clin. 2015;65:401–421.
- Zhang X, Guo B, Zhu Y, et al. Up-regulation of plasma lncRNA CACS15 distinguished early-stage oral squamous cell carcinoma patient. Oral Dis. 2020;26:1619–1624.
- Ren S, Cheng X, Chen M, et al. Hypotoxic and rapidly metabolic PEG-PCL-C3-ICG Nanoparticles for fluorescence-guided photothermal/photodynamic therapy against OSCC. ACS Appl Mater Interfaces. 2017;9:31509–31518.
- Rapidis AD, Gullane P, Langdon JD, et al. Major advances in the knowledge and understanding of the epidemiology, aetiopathogenesis, diagnosis, management and prognosis of oral cancer. Oral Oncol. 2009;45:299–300.
- Perez-Sayans M, Somoza-Martin JM, Barros-Angueira F, et al. Multidrug resistance in oral squamous cell carcinoma: the role of vacuolar ATPases. Cancer Lett. 2010;295:135–143.
- Yoshitomi I, Kawasaki G, Yanamoto S, et al. Orotate phosphoribosyl transferase mRNA expression in oral squamous cell carcinoma and its relationship with the dihydropyrimidine dehydrogenase expression and the clinical effect of 5-fluorouracil. Oral Oncol. 2006;42:880–887.
- Feng X, Luo Q, Zhang H, et al. The role of NLRP3 inflammasome in 5-fluorouracil resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res. 2017;36:81.
- Iyer MK, Niknafs YS, Malik R, et al. The landscape of long noncoding RNAs in the human transcriptome. Nat Genet. 2015;47:199–208.
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discov. 2011;1:391–407.
- Wang J, Yu Z, Wang J, et al. LncRNA NUTM2A-AS1 positively modulates TET1 and HIF-1A to enhance gastric cancer tumorigenesis and drug resistance by sponging miR-376a. Cancer Med. 2020;9:9499–9510.
- Zhou C, Yi C, Yi Y, et al. LncRNA PVT1 promotes gemcitabine resistance of pancreatic cancer via activating Wnt/beta-catenin and autophagy pathway through modulating the miR-619-5p/Pygo2 and miR-619-5p/ATG14 axes. Mol Cancer. 2020;19:118.
- Lei Y, Guo W, Chen B, et al. Tumorreleased lncRNA H19 promotes gefitinib resistance via packaging into exosomes in nonsmall cell lung cancer. Oncol Rep. 2018;40:3438–3446.
- Ghafouri-Fard S, Abak A, Tondro Anamag F, et al. 5-Fluorouracil: a narrative review on the role of regulatory mechanisms in driving resistance to this chemotherapeutic agent. Front Oncol. 2021;11:658636.
- Chaudhary R, Wang X, Cao B, et al. Long noncoding RNA, LINC00460, as a prognostic biomarker in head and neck squamous cell carcinoma (HNSCC). Am J Transl Res. 2020;12:684–696.
- Wu J, Zheng C, Wang Y, et al. LncRNA APCDD1L-AS1 induces icotinib resistance by inhibition of EGFR autophagic degradation via the miR-1322/miR-1972/miR-324-3p-SIRT5 axis in lung adenocarcinoma. Biomark Res. 2021;9:9.
- Wang X, Lan Z, He J, et al. LncRNA SNHG6 promotes chemoresistance through ULK1-induced autophagy by sponging miR-26a-5p in colorectal cancer cells. Cancer Cell Int. 2019;19:234.
- Han P, Li JW, Zhang BM, et al. The lncRNA CRNDE promotes colorectal cancer cell proliferation and chemoresistance via miR-181a-5p-mediated regulation of Wnt/β-catenin signaling. Mol Cancer. 2017;16:9.
- Kobayashi H, Koike T, Nakatsuka A, et al. Dihydropyrimidine dehydrogenase expression predicts survival outcome and chemosensitivity to 5-fluorouracil in patients with oral squamous cell carcinoma. Oral Oncol. 2005;41:38–47.
- Feng J, Li J, Qie P, et al. Long non-coding RNA (lncRNA) PGM5P4-AS1 inhibits lung cancer progression by up-regulating leucine zipper tumor suppressor (LZTS3) through sponging microRNA miR-1275. Bioengineered. 2021;12:196–207.
- Zhang L, Kang W, Lu X, et al. LncRNA CASC11 promoted gastric cancer cell proliferation, migration and invasion in vitro by regulating cell cycle pathway. Cell Cycle. 2018;17:1886–1900.
- Zhang CC, Li Y, Feng XZ, et al. Circular RNA circ_0001287 inhibits the proliferation, metastasis, and radiosensitivity of non-small cell lung cancer cells by sponging microRNA miR-21 and up-regulating phosphatase and tensin homolog expression. Bioengineered. 2021;12:414–425.
- Cai L, Ye L, Hu X, et al. MicroRNA miR-330-3p suppresses the progression of ovarian cancer by targeting RIPK4. Bioengineered. 2021;12:440–449.
- Li Z, Yu Z, Meng X, et al. LncRNA LINC00968 accelerates the proliferation and fibrosis of diabetic nephropathy by epigenetically repressing p21 via recruiting EZH2. Biochem Biophys Res Commun. 2018;504:499–504.
- Aytes A, Giacobbe A, Mitrofanova A, et al. NSD2 is a conserved driver of metastatic prostate cancer progression. Nat Commun. 2018;9:5201.
- Zhu L, Yu CL, Zheng Y. NSD2 inhibition suppresses metastasis in cervical cancer by promoting TGF-beta/TGF-betaRI/SMADs signaling. Biochem Biophys Res Commun. 2019;519:489–496.
- He C, Liu C, Wang L, et al. Histone methyltransferase NSD2 regulates apoptosis and chemosensitivity in osteosarcoma. Cell Death Dis. 2019;10:65.
- Longley DB, Harkin DP, Johnston PG. 5-fluorouracil: mechanisms of action and clinical strategies. Nat Rev Cancer. 2003;3:330–338.
- Nagata M, Nakayama H, Tanaka T, et al. Overexpression of cIAP2 contributes to 5-FU resistance and a poor prognosis in oral squamous cell carcinoma. Br J Cancer. 2011;105:1322–1330.
- Zhang L, Meng X, Zhu XW, et al. Long non-coding RNAs in oral squamous cell carcinoma: biologic function, mechanisms and clinical implications. Mol Cancer. 2019;18:102.
- Fang Z, Zhao J, Xie W, et al. LncRNA UCA1 promotes proliferation and cisplatin resistance of oral squamous cell carcinoma by sunppressing miR-184 expression. Cancer Med. 2017;6:2897–2908.
- Wang R, Lu X, Yu R. lncRNA MALAT1 Promotes EMT process and cisplatin resistance of oral squamous cell carcinoma via PI3K/AKT/m-TOR signal pathway. Onco Targets Ther. 2020;13:4049–4061.
- Lian P, Wang Q, Zhao Y, et al. An eight-long non-coding RNA signature as a candidate prognostic biomarker for bladder cancer. Aging (Albany NY). 2019;11:6930–6940.
- Luo Y, Xuan Z, Zhu X, et al. Long non-coding RNAs RP5-821D11.7, APCDD1L-AS1 and RP11-277P12.9 were associated with the prognosis of lung squamous cell carcinoma. Mol Med Rep. 2018;17:7238–7248.
- Qin X, Guo H, Wang X, et al. Exosomal miR-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting CDKN1B and ING5. Genome Biol. 2019;20:12.
- Qu L, Ding J, Chen C, et al. Exosome-transmitted lncARSR promotes sunitinib resistance in renal cancer by acting as a competing endogenous RNA. Cancer Cell. 2016;29:653–668.
- Dong H, Wang W, Chen R, et al. Exosome-mediated transfer of lncRNASNHG14 promotes trastuzumab chemoresistance in breast cancer. Int J Oncol. 2018;53:1013–1026.
- Wang X, Yu H, Yu Z, et al. Exosomal lncRNA HEIH promotes cisplatin resistance in tongue squamous cell carcinoma via targeting miR-3619-5p/HDGF axis. Acta Histochem. 2020;122:151647.
- Kang M, Ren M, Li Y, et al. Exosome-mediated transfer of lncRNA PART1 induces gefitinib resistance in esophageal squamous cell carcinoma via functioning as a competing endogenous RNA. J Exp Clin Cancer Res. 2018;37:171.
- Tong Y, Yang L, Yu C, et al. Tumor-secreted exosomal lncRNA POU3F3 promotes cisplatin resistance in ESCC by inducing fibroblast differentiation into CAFs. Mol Ther Oncolytics. 2020;18:1–13.
- Shin Low S, Pan Y, Ji D, et al. Smartphone-based portable electrochemical biosensing system for detection of circulating microRNA-21 in saliva as a proof-of-concept. Sens Actuators B Chem. 2020;308:127718.
- Song NS, Pei ZD, Fu G. MiR-1224-5p acts as a tumor suppressor via inhibiting the malignancy of rectal cancer through targeting SLC29A3. IUBMB Life. 2020;72:2204–2213.
- Kong L, Liu P, Zheng M, et al. The miR-1224-5p/ELF3 axis regulates malignant behaviors of pancreatic cancer via PI3K/AKT/Notch signaling pathways. Onco Targets Ther. 2020;13:3449–3466.
- Wang J, Hu Y, Ye C, et al. miR-1224-5p inhibits the proliferation and invasion of ovarian cancer via targeting SND1. Hum Cell. 2020;33:780–789.
- Qian J, Li R, Wang YY, et al. MiR-1224-5p acts as a tumor suppressor by targeting CREB1 in malignant gliomas. Mol Cell Biochem. 2015;403:33–41.
- Shi ZZ, Wang WJ, Chen YX, et al. The miR-1224-5p/TNS4/EGFR axis inhibits tumour progression in oesophageal squamous cell carcinoma. Cell Death Dis. 2020;11:597.
- Sudhakara Prasad K, Cao X, Gao N, et al. A low-cost nanomaterial-based electrochemical immunosensor on paper for high-sensitivity early detection of pancreatic cancer. Sens Actuators B Chem. 2020;305:127516.
- Han X, Piao L, Xu X, et al. NSD2 promotes renal cancer progression through stimulating Akt/Erk signaling. Cancer Manag Res. 2020;12:375–383.
- Chen LY, Zhi Z, Wang L, et al. NSD2 circular RNA promotes metastasis of colorectal cancer by targeting miR-199b-5p-mediated DDR1 and JAG1 signalling. J Pathol. 2019;248:103–115.
- Stangl-Kremser J, Lemberger U, Hassler MR, et al. The prognostic impact of tumour NSD2 expression in advanced prostate cancer. Biomarkers. 2020;25:268–273.
