ABSTRACT
Bladder cancer (BC) is one of the most common malignant tumors in the urinary system. Our research aimed to explore the function and underlying mechanisms of long noncoding RNA (lncRNA) PSMA3-AS1 in BC. RT-qPCR was utilized to detect the levels of PSMA3-AS1, miR-214-5p, and PD-L1. ChIP assay was employed to confirm the transcription factor of PSMA3-AS1. Luciferase reporter assay was carried out to demonstrate the relationships between miR-214-5p and PSMA3-AS1 or PD-L1. The diagnostic value of PSMA3-AS1 was evaluated by the ROC curve. CCK-8, wound healing, transwell, and flow cytometry assays were applied to analyze cell viability, migration, invasion, and apoptosis. Western blotting was used to confirm the expression of cleaved caspase-3. The present study revealed that BC tissues and cells exhibited an increased expression in PSMA3-AS1. High expression of PSMA3-AS1 was related to poor prognosis in BC patients. Then, the area under the ROC curve for PSMA3-AS1 was up to 0.8954. Moreover, ChIP assay elaborated that YY1 could bind to the PSMA3-AS1 promoter region. Furthermore, it was found that that PSMA3-AS1 knockdown repressed BC cell viability and metastasis, and promoted apoptosis. In addition, miR-214-5p was inversely correlated with PSMA3-AS1 or PD-L1 levels. MiR-214-5p deletion reversed the impacts of PSMA3-AS1 deletion on BC progression, and PD-L1 inhibition also abrogated the influence of miR-214-5p deletion in BC development. In conclusion, YY1-induced PSMA3-AS1 exerted an oncogenic function in BC cells via targeting miR-214-5p and enhancing PD-L1, providing potential biomarkers for BC therapy.
Introduction
As the most common genitourinary cancer, bladder cancer (BC) is the 14th leading cause of cancer-associated death around the world [Citation1]. In 2021, approximately 83,730 new cases were diagnosed as BC, and 18,160 patients die from BC in United States [Citation2]. Many factors, including smoking, obesity, and bladder infections may lead to the occurrence of BC [Citation3]. Despite significant progress in clinical treatment, including radical cystectomy, radiotherapy and postoperative chemotherapy, or immunotherapy, the 5-year survival rate of BC patients is still unsatisfactory due to the high frequency of recurrence and metastasis [Citation4–6]. In addition, the prognosis of BC is closely associated with the tumor stage; however, patients do not exhibit specific symptoms at the early stage [Citation7]. Hence, early diagnosis of BC is quite beneficial for the treatment of BC [Citation8]. Therefore, exploring a regulatory network involved in BC development may help to find novel treatment strategies for BC.
Multiple lines of evidence have demonstrated that dysregulation of lncRNAs is implicated in the development of many human tumors, including BC. For example, Chen et al. indicated that KCNMB2-AS1 accelerated BC development via the miR-3194-3p/SAMD5 axis [Citation9]. Zhang et al. manifested that OIP5-AS1 facilitated BC progression via the miR-217/MTDH axis [Citation10]. Besides, Chen et al. revealed that lncRNA ROR1-AS1 promoted cell migration in BC via modulating miR-504 [Citation11]. LncRNA PSMA3 antisense RNA 1 (PSMA3-AS1) is located at chromosome 14q23.1 [Citation12], and was reported to exert a tumor-promoting effect in several malignant tumors, such as glioma, esophageal cancer and non-small cell lung cancer [Citation12–14]. Nevertheless, the function of PSMA3-AS1 in BC is unclear.
In the current research, we hypothesized that lncRNA PSMA3-AS1 was an oncogenic factor in BC. Our study aimed to investigate the biological function and mechanism of PSMA3-AS1 in the progression of BC. Our findings demonstrated that YY1-induced PSMA3-AS1 accelerated BC progression via the miR-214-5p/PD-L1 axis. These results may provide new insights for the understanding of PSMA3-AS1 in BC pathophysiology.
Materials and methods
Clinical samples
This study was approved by the Ethics Committee of the First Affiliated Hospital of Soochow University. Tissue specimens were obtained from 36 BC patients and 36 healthy subjects from May 2018 to June 2019. Tissue specimens were collected from patients with BC who underwent radical cystectomy without neoadjuvant chemotherapy. All tissues were gained and stored at −80°C until use. All participants have signed written informed consents.
Cell culture
Human BC cell lines (T24, 5637, J82, and UMUC3) and bladder epidermal cell line (HCV-29) were obtained from ATCC (Manassas, USA) and cultured in RPMI-1640 medium containing 10% FBS (Gibco) at 37°C under 5% CO2.
Cell transfection
MiR-214-5p mimics/inhibitor with their negative controls (NC mimics/inhibitor), shRNA targeting RNAs (shRNAs) including YY1(shYY1), shPSMA3-AS1, shPD-L1 with their negative control (shNC), and pcDNA3.1/YY1 and pcDNA3.1 were gained from GenePharma (Shanghai, China). The transfections were performed using Lipofectamine 2000 (Invitrogen).
Reverse transcription-quantitative PCR (RT-qPCR)
RT-qPCR was used to detect mRNA expression in tissues or cells [Citation15]. Total RNA was isolated using TRIzol Reagent (Invitrogen), followed by conversion into cDNA via a reverse transcription kit (Takara). RT-qPCR was conducted using an SYBR Green qPCR Mix (TaKaRa). The expression of RNAs was calculated using the 2−ΔΔCq method [Citation16]. The following primer sequences were used: PSMA3-AS1 forward, 5ʹ- TAGGAGAGTCAATCACTA-3ʹ and reverse, 5ʹ-TCGAGTTGACTCAATAGTGCT-3ʹ; miR-214-5p forward, 5ʹ-ATCCGTGAAGTTGTTCGTGG-3ʹ and reverse, 5ʹ-TATGGTTGTAGAGGACTCCTTGAC-3ʹ; PD-L1 forward, 5ʹ- GTCAGATTTTGTCCGTTCCACA-3ʹ and reverse, 5ʹ- CATGGACTTGACGTAGCTGTT-3ʹ; GAPDH forward, 5ʹ-CTCTGCTCCTCCTGTTCGAC-3ʹ and reverse, 5ʹ-GCGCCCAATACGACCAAATC-3ʹ; U6 forward, 5ʹ-GCTTCGGCAGCACATATACTAAAAT-3ʹ and reverse, 5ʹ-CGCTTCACGAATTTGCGTGTCAT-3ʹ.
CCK-8 assay
CCK-8 (Beyotime, China) was used to detect cell proliferation [Citation17]. The BC cell lines were plated into 96-well plates (6x103 cells/well), and incubated for 0, 24, 48 and 72 h. Then 10 μl of CCK-8 reagent (Beyotime) was added to the plate for another 2 h. The absorbance was recorded using a microplate reader (BioTek China) at 450 nm.
Wound-healing assay
Cell migration ability was evaluated using a wound healing assay [Citation18]. T24 and 5637 (2 x 106) cells were seeded into 6-well plates. Then, a wound in each group was scratched using a pipette tip. Images of migrated cells were acquired at 0 h and 24 h using a light microscope (Olympus).
Transwell assay
Transwell assay was applied to detect the invasive ability [Citation19]. Matrigel-coated membrane was used for transwell chamber assay. Cells (1x104) in serum-free medium were plated in the upper chamber. The lower chamber was filled with 500 μl of completed medium. Subsequently, cells underneath the membrane were stained with 0.1% crystal violet. At last, invaded cells were counted using a microscope (Olympus, Japan).
Flow cytometry assay
For cell apoptosis detection, cells were suspended in Annexin V binding buffer at a density of 1 × 105 cells/ml. After being stained with Annextin V-FITC reagent, cells were further stained with Propidium Iodide (Solarbio, Beijing, China) for 20 min. Cell apoptosis was assessed with flow cytometry (BD Biosciences) [Citation20].
Subcellular fractionation
Nuclear or cytoplasmic RNA in cells was separated using PARIS™ Kit (Ambion, USA). Expression of GAPDH, PSMA3-AS1, and U6 determined via RT-qPCR. U6 was used as the nuclear control, and GAPDH was used as the cytoplasmic control [Citation21].
RNA immunoprecipitation (RIP) assay
RIP was used to verify the binding relationship between miR-214-5p and PSMA3-AS1 or PD-L1 [Citation22]. RIP assay was conducted with an EZMagna RIP kit (Merck KGaA). Cell lysates were treated with magnetic beads conjugated to Ago2 antibody (Abcam) or control IgG antibody (Abcam) in RIP buffer. All precipitated RNAs were examined by RT-qPCR.
RNA pull‑down assay
RNA pull-down was used to verify the binding relationship between miR-214-5p and PSMA3-AS1 or PD-L1 [Citation23]. Cells were treated with biotin-labeled miR-214-5p (Bio-miR-214-5p-Wt or Bio-miR-214-5p-Mut) and their NC (Bio-NC) (RiboBio). Then, cell lysates were incubated with the streptavidin beads. The relative RNA enrichments were assayed.
Luciferase reporter assay
The PSMA3-AS1 promoter was sub-cloned into pGL3 vector to construct reporter and then was co-transfected with pcDNA3.1/YY1 or shYY1 in T24 and 5637 cells. PSMA3-AS1-wild-type (wt) or mutant (mut), PD-L1-wt or PD-L1-mut reporters were subcloned into pmirGLO vectors (Promega). BC cells were co-transfected with NC mimics or miR-214-5p mimics using Lipofectamine 3000 (Invitrogen).
Chromatin immunoprecipitation (ChIP) assay
ChIP assay was used to assess YY1 binding site at the promoter region of PSMA3-AS1. Briefly, ChIP assay was performed using the EZ ChIP™ Kit (Millipore). T24 and 5637 cells (1x104) were fixed with 1% formaldehyde and then were sonicated to generate DNA fragments (200–500 bp). Crosslinked chromatin DNA was collected and then immunoprecipitated with YY1 antibody (Abcam) or IgG control (Abcam). RT-qPCR was used to determine the enrichment of precipitated DNAs.
Western botting
Total proteins were isolated using RIPA buffer (Beyotime), separated on 10% SDS-PAGE, transferred to PVDF membranes and blocked with 5% skimmed milk for 2 h at 37°C. Primary antibodies against cleaved-caspase-3 (1:500; ab32042; Abcam) and GAPDH (1:5,000; ab8245; Abcam) were incubated with the membranes at 4°C overnight. Subsequently, the membranes were incubated with secondary antibody at 37°C for 1 h. The bands were detected using an ECL reagent (EMD Millipore).
Statistical analysis
SPSS 22.0 (SPSS Inc. IL, USA) was used for analyzing data, which are given as means ± SD. The results from different groups were assessed using Student’s t-test or one-way ANOVA. The diagnosed value of PSMA3-AS1 was evaluated by ROC curve. P < 0.05 was defined as statistically significant.
Results
In this study, a series of in vitro experiments were conducted to investigate role of PSMA3-AS1 in BC, and the data indicated that YY1-activated PSMA3-AS1 promoted the viability and metastasis of BC via regulating the miR-214-5p/PD-L1 axis. These findings may offer new insight into the pathogenesis of BC.
LncRNA PSMA3-AS1 expression is enhanced in BC tissues and cells
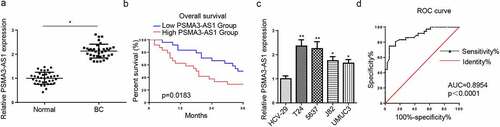
To explore the function of PSMA3-AS1 in BC progression, PSMA3-AS1 level was detected in BC tissues. As depicted in , PSMA3-AS1 was overexpressed in BC tissues. Then, BC patients with high expression of PSMA3-AS1 had a short survival time (). Moreover, it was determined that PSMA3-AS1 expression was higher in BC cell lines than that in bladder epidermal cells (HCV-29) (). T24 and 5637 cells were chosen for subsequent experiments due to the high expression of PSMA3-AS1. Furthermore, ROC curve was conducted to explore the potential diagnostic value of PSMA3-AS1 in BC. The result exhibited that the area under the curve (AUC) was 0.8954 (), implying its substantial value in BC diagnosis. Collectively, the above data demonstrated that PSMA3-AS1 exerted a pivotal role in BC tumorigenesis.
Figure 1. LncRNA PSMA3-AS1 level is enhanced in BC tissues and cells. (a) PSMA3-AS1 expression in BC tissues and normal tissues was detected by RT-qPCR. (b) kaplan–meier analysis was performed to investigate the relation between survival time of BC patients and PSMA3-AS1 expression. (c) PSMA3-AS1 expression in BC cells was detected by RT-qPCR. (d) ROC curve was used to evaluate the diagnostic value of PSMA3-AS1 in BC. *P < 0.05; **P < 0.01
PSMA3-AS1 deletion represses the viability and metastasis of BC cells
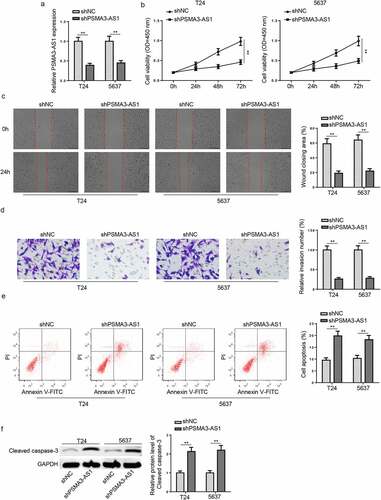
To further probe into PSMA3-AS1 functions in BC development, cells were transfected with shPSMA3-AS1 or shNC, and the knockdown efficiency was displayed in . Then, CCK-8 assay demonstrated that PSMA3-AS1 knockdown suppressed cell viability in BC cells (). Meanwhile, the migration and invasion ability were weakened by PSMA3-AS1 deficiency in BC cells (). Moreover, BC cells with PSMA3-AS1 deletion exhibited increased apoptosis rates (). Besides, western blotting revealed that PSMA3-AS1 knockdown increased the expression of the apoptosis-related protein cleaved caspase-3 (). These discoveries implied that interference of PSMA3-AS1 inhibited BC development.
Figure 2. PSMA3-AS1 deletion represses the viability and metastasis of BC cells. T24 and 5637 cells were transfected with shPSMA3-AS1 or shNC, respectively. (a) The efficiency of PSMA3-AS1 knockdown was detected by RT-qPCR. (b) CCK-8 assay was applied to assess the viability of T24 and 5637 cells. (c and d) Wound healing and transwell assays to detect migration and invasion ability. (e) cell apoptosis was assessed by flow cytometry. (f)western blotting showed the protein levels of cleaved caspase-3. **P < 0.01. n = 3
PSMA3-AS1 sponges miR-214-5p in BC
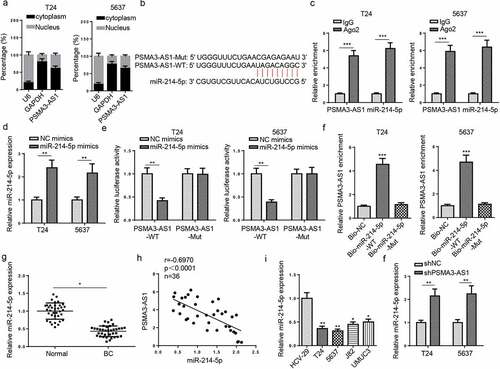
Analysis of PSMA3-AS1 regulation in BC began with the determination of its subcellular localization in BC cells and results elaborated that PSMA3-AS1 was mainly distributed in the cytoplasm of BC cells (). Then, we used starBase v2.0 to explore the miRNAs that targeted PSMA3-AS1. The data showed an underlying combination of PSMA3-AS1 and miR-214-5p (). Next, RIP experiment results manifested that PSMA3-AS1 interacted with miR-214-5p (). Moreover, miR-214-5p expression was enhanced in T24 and 5637 cells transfected with miR-214-5p mimics (). As indicated by luciferase reporter assay, miR-214-5p addition reduced the luciferase activity of PSMA3-AS1-wt rather than that of PSMA3-AS1-mut (). Meanwhile, RNA pull-down implied that PSMA3-AS1 could be pulled down when the cells were transfected with the biotin-labeled miR-214-5p mimics (). Besides, miR-214-5p expression was found to be reduced in BC tissues (). An inverse correlation was displayed between miR-214-5p and PSMA3-AS1 levels in BC tissues (). Similarly, miR-214-5p level was also reduced in BC cells (). Furthermore, T24 and 5637 cells showed an increase in miR-214-5p abundance after PSMA3-AS1 blocking (). Therefore, PSMA3-AS1 adversely regulated miR-214-5p in BC.
Figure 3. PSMA3-AS1 sponges miR-214-5p in BC. (a) subcellular localization of PSMA3-AS1 was determined in T24 and 5637 cells. (b) starbase software predicted the binding sites between miR-214-5p and PSMA3-AS1. (c) RIP assay was performed in T24 and 5637 cells to analyze the correlation between miR-214-5p and PSMA3-AS1. (d) T24 and 5637 cells were transfected with NC mimics or miR-214-5p mimics, respectively. the efficiency of miR-214-5p overexpression was detected by RT-qPCR. (e) THE association between miR-214-5p and PSMA3-AS1 was verified by luciferase reporter assay. (f) RNA pull-down assay was performed in T24 and 5637 cells. (g) MiR-214-5p expression in BC tissues and normal tissues was detected by RT-qPCR. (h) Pearson’s correlation analysis was used to analyze the correlation between miR-214-5p and PSMA3-AS1 levels in BC tissues. (i) MiR-214-5p expression in BC and normal cells was detected by RT-qPCR. (j) T24 and 5637 cells were transfected with shPSMA3-AS1 or shNC, respectively. MiR-214-5p expression in T24 and 5637 cells was detected by RT-qPCR. *P < 0.05; **P < 0.01, ***P < 0.001. n = 3
PSMA3-AS1 accelerates BC progression via modulating miR-214-5p
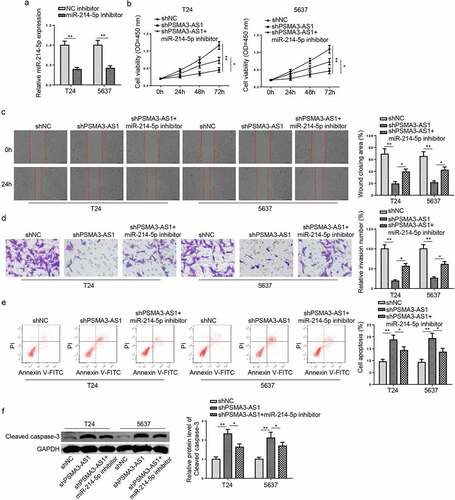
To assess the impacts of miR-214-5p on BC cell activities, a series of functional assays were performed. Firstly, the miR-214-5p level was reduced in T24 and 5637 cells (). It was manifested that PSMA3-AS1 deletion led to retard cell viability, migration, and invasion, as well as facilitated cell apoptosis, which was reversed by miR-214-5p inhibition (). Meanwhile, miR-214-5p inhibitor partially reversed the promotive effect of shPSMA3-AS1 on cleaved caspase-3 expression (). Therefore, PSMA3-AS1 exacerbated malignant phenotypes in BC cells via interaction with miR-214-5p.
Figure 4. PSMA3-AS1 accelerates BC progression via modulating miR-214-5p. T24 and 5637 cells were transfected with miR-214-5p inhibitor or NC inhibitor, respectively. (a) The efficiency of miR-214-5p knockdown was detected by RT-qPCR. (b) CCK-8 assay was applied to assess the viability of T24 and 5637 cells. (c and d) wound healing and transwell assays to detect migration and invasion ability. (e) cell apoptosis was assessed by flow cytometry. (f)Western blotting showed the protein levels of cleaved caspase-3. *P < 0.05; **P < 0.01. n = 3
PD-L1 binds to miR-214-5p in BC cells
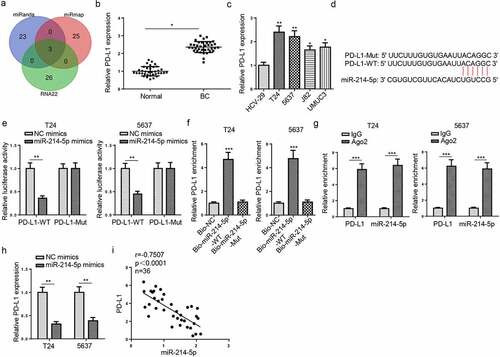
Venn diagram analysis was applied to predict target genes of miR-214-5p based on miRanda, miRmap and RNA22 databases (). Among the candidate genes, PD-L1 is a heatedly-discussed therapeutic target for the treatment of several cancers, including BC [Citation24]. Besides, it was also revealed by RT-qPCR that PD-L1 level was enhanced in BC tissues and cells (). Therefore, PD-L1 was chosen for further investigation in this work. As displayed in , the 3′-UTR of PD-L1 could bind to miR-214-5p. Then, luciferase reporter assay manifested that the activity of PD-L1-WT was inhibited following miR-214-5p addition, but no change in the PD-L1-Mut (). RNA pull-down determined that Bio-miR-214-5p-WT could bind to PD-L1 (). Moreover, RIP analysis manifested that PD-L1 and miR-214-5p were precipitated in abundance by Ago2 antibody (), verifying a direct association between PD-L1 and miR-214-5p. Then, overexpression of miR-214-5p led to a reduction in PD-L1 expression. (). Meanwhile, a negative correlation between PD-L1 and miR-214-5p was uncovered in BC tissues (). the above findings elaborated that miR-214-5p directly targeted PD-L1 in BC.
Figure 5. PD-L1 binds to miR-214-5p in BC cells. (a) Venn diagram of miR-214-5p targets predicted by miRanda, miRmap and RNA22 databases. (b and c) PD-L1 expression in BC and normal tissues and cells. (d) StarBase software predicted the binding sites between miR-214-5p and PD-L1. (e) The association between miR-214-5p and PD-L1 was verified by luciferase reporter assay. (f and g) RNA pull-down and RIP assays were detected in T24 and 5637 cells. (h) T24 and 5637 cells were transfected with NC mimics or miR-214-5p mimics. PD-L1 expression in T24 and 5637 cells was detected by RT-qPCR. (i) Pearson’s correlation analysis was used to analyze the correlation between miR-214-5p and PD-L1 levels in BC tissues. *P < 0.05; **P < 0.01, ***P < 0.001. n = 3
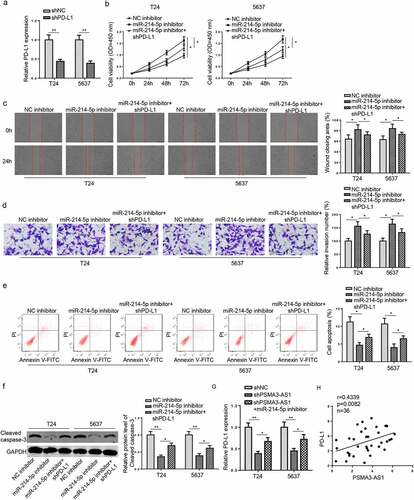
PSMA3-AS1 accelerates BC tumorigenesis in vitro via the miR-214-5p/PD-L1 axis
To further confirm whether PSMA3-AS1 exerted its functions in BC via regulating the miR-214-5p/PD-L1 axis, functional assays were conducted in T24 and 5637 cells. Firstly, RT-qPCR analysis manifested that PD-L1 was reduced by PD-L1 deficiency (). Then, miR-214-5p blocking promoted T24 and 5637 cell viability and metastasis, but restrained cell apoptosis, however, these influences were abrogated by PD-L1 inhibition (). Moreover, miR-214-5p inhibition decreased cleaved caspase-3 level, while PD-L1 deficiency reversed this effect (). Furthermore, RT-qPCR results implied that PSMA3-AS1 suppression downregulated PD-L1 abundance, while miR-214-5p deletion increased PD-L1 level (). Additionally, PD-L1 level was positively correlated to PSMA3-AS1 in BC tissues (). In sum, these observations revealed that PSMA3-AS1 contributed to BC tumorigenesis by targeting miR-214-5p and upregulating PD-L1.
Figure 6. PSMA3-AS1 accelerates BC tumorigenesis in vitro via the miR-214-5p/PD-L1 axis. T24 and 5637 cells were transfected with shPD-L1 or shNC, respectively. (a) The efficiency of PD-L1 knockdown was detected by RT-qPCR. (b-e) CCK-8, wound healing, transwell and flow cytometry assays were applied to assess the viability, migration, invasion and apoptosis of T24 and 5637 cells. (f)western blotting showed the protein levels of cleaved caspase-3. (g) RT-qPCR assay was applied to measure PD-L1 expression in T24 and 5637 cells transfected with shNC, shPSMA3-AS1, shPSMA3-AS1+ miR-214-5p. (h) Pearson’s correlation analysis was used to analyze the correlation between PD-L1 and PSMA3-AS1 levels in BC tissues. *P < 0.05; **P < 0.01. n = 3
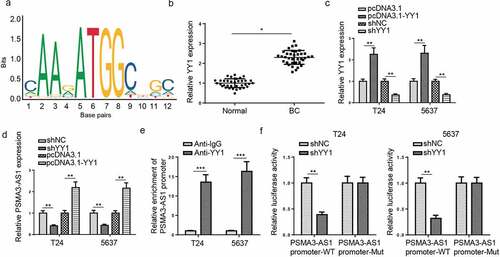
PSMA3-AS1 is activated by YY1 in BC
The transcription factor plays an important role in lncRNA [Citation25]. Through searching for UCSC (http://genom e.ucsc.edu/), YY1 was identified as the possible upstream factor of PSMA3-AS1. Next, JASPAR predicted that the transcription factor YY1 might combine with the PSMA3-AS1 promoter region (). Then, RT-qPCR assay manifested that YY1 expression was enhanced in BC tissues (). Moreover, the regulatory function of YY1 on PSMA3-AS1 level was assessed. It was demonstrated that YY1 was upregulated by YY1 addition, which was downregulated by YY1 deletion (). Moreover, RT-qPCR analysis determined that PSMA3-AS1 level was decreased by YY1 depletion and was elevated by YY1 addition in T24 and 5637 cells (). Furthermore, ChIP assay showed PSMA3-AS1 promoter was pulled down by anti-YY1 (). Subsequently, luciferase activity of PSMA3-AS1-wt promoter was impeded by YY1 deficiency while that of mutant PSMA3-AS1 had no change (). In sum, PSMA3-AS1 was transcriptionally activated by YY1 in BC.
Figure 7. PSMA3-AS1 is activated by YY1 in BC. (a) The binding motif of YY1 and PSMA3-AS1 promoter was presented according to the data from JASPAR database. (b) YY1 expression in BC tissues and normal tissues was detected by RT-qPCR. (c) RT-qPCR showed that YY1 expression in T24 and 5637 cells transfected with shYY1 or shNC and pcDNA3.1 or pcDNA3.1/YY1. (d) RT-qPCR analysis showed PSMA3-AS1 expression in T24 and 5637 cells transfected with shYY1 or shNC and pcDNA3.1 or pcDNA3.1/YY1 (e) ChIP assays showed YY1 could bind to the PSMA3-AS1 promoter. (f) Luciferase reporter assay further confirmed the correlation between miR-214-5p and PSMA3-AS1. *P < 0.05; **P < 0.01, ***P < 0.001. n = 3
Discussion
BC is the most common malignancy in the urinary system and poses threat to public health worldwide. Hence, it is important to understand the mechanisms of BC carcinogenesis. During this research, we uncovered that PSMA3-AS1 was enhanced in BC and PSMA3-AS1 deletion restrained the viability and metastasis of BC cells. These discoveries suggested that PSMA3-AS1, as an oncogene, played an essential function in BC development.
Mounting evidence has proved that dysregulation of lncRNAs participated in various biological processes, such as cell viability, apoptosis, and differentiation [Citation26,Citation27]. As reported in previous researches, lncRNA PSMA3-AS1 has been revealed to act as an oncogene involved in tumor development. To cite an instance, PSMA3-AS1 expedited colorectal cancer cell metastasis by modulating miR-4429 [Citation28]. Moreover, PSMA3-AS1 knockdown restrained lung cancer cell viability and metastasis via targeting miR-4504 [Citation29]. Furthermore, PSMA3-AS1 enhanced cell viability and metastasis via modulating miR-302a-3p/RAB22A in glioma [Citation12]. Here, our experiment results showed that PSMA3-AS1 was enhanced in BC cells and tissues. Moreover, functional assays elaborated that PSMA3-AS1 blocking repressed cell viability and metastasis, and induced cell apoptosis in BC cells. In sum, these findings highlighted the pivotal role of PSMA3-AS1 in BC progression in vitro.
Previous reports have revealed that transcription factors can lead to dysregulation of lncRNAs in cancers [Citation30,Citation31]. It was reported that YY1 was a transcription factor in many cancers [Citation32]. For example, YY1-mediated lncRNA ZFPM2-AS1 promoted cell viability in small cell lung cancer by increasing TRAF4 [Citation33]. YY1-induced lncRNA DSCR8 facilitated ovarian cancer development by sponging miR-3192-5p/YY1 axis [Citation34]. Hence, the interaction between YY1 and PSMA3-AS1 was assessed in this work. Based on the exploration, YY1 was uncovered to be possible factors participating in the upregulation of PSMA3-AS1 through JASPAR, and YY1 was confirmed to bind with PSMA3-AS1 promoter in BC cells. Overall, these essential discoveries suggested that YY1 activated PSMA3-AS1 translational levels to regulate PSMA3-AS1 in BC in vitro.
LncRNA acts as ceRNA or miRNA sponge by interacting with miRNAs and inhibits their expression [Citation35]. To explore the downstream mechanism of PSMA3-AS1 in BC cells, miR-214-5p was screened and verified to target 3′-UTR of PSMA3-AS1. Recent evidence indicated that miR-214-5p acted as an anti-tumor gene in multiple human cancers, such as osteosarcoma [Citation36], prostate cancer [Citation37], and cervical cancer [Citation38]. Herein, we illustrated that PSMA3-AS1 mainly accumulated in the cytoplasm of BC cells. Moreover, PSMA3-AS1 accelerated the viability and metastasis of BC by inhibiting miR-214-5p expression. Programmed death-ligand 1 (PD-L1, also called CD274, B7-H1), an immune checkpoint, has been applied in oncotherapy for multiple tumors as a therapeutic target. Roshani Asl et al. revealed that miR-124 addition restrained colorectal cancer cell viability and arrested cell cycle through downregulation of PD-L1 [Citation39]. In addition, Wei et al. implied that lncRNA MALAT1 expedited non-small cell lung cancer cell viability and mobility via the miR-200a-3p/PD-L1 axis [Citation40]. Results in our research illustrated that PD-L1 was inversely modulated by miR-214-5p and positively modulated by PSMA3-AS1 in BC. Also, the silencing of PD-L1 reversed the impacts of miR-214-5p deficiency on cell viability, metastasis, and apoptosis in BC cells.
Conclusion
This research elucidated that YY1-induced PSMA3-AS1 aggravated BC progression through the miR-214-5p/PD-L1 axis in vitro. The current study might provide new insights into the initiation and progression of BC, and PSMA3-AS1 might be a potential biomarker of BC diagnosis and treatment. In the future, in vivo experiments will be performed to further confirm the role and mechanism of PSMA3-AS1 in BC.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability
The datasets generated during and/or analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Mahdavifar N, Ghoncheh M, Pakzad R, et al. Epidemiology, incidence and mortality of bladder cancer and their relationship with the development index in the world. Asian Pac J Cancer Prev. 2016;17(1):381–386.
- Rl S, Kd M, He F, et al. Cancer Statistics, 2021. CA Cancer J Clin. 2021;71(1):7–33.
- Masaoka H, Matsuo K, Ito H, et al. Cigarette smoking and bladder cancer risk: an evaluation based on a systematic review of epidemiologic evidence in the Japanese population. Jpn J Clin Oncol. 2016;46(3):273–283.
- Humphrey PA, Moch H, Cubilla AL, et al. The 2016 WHO classification of tumours of the urinary system and male genital organs-part b: prostate and bladder tumours. Eur Urol. 2016;70(1):106–119.
- Alfred Witjes J, Lebret T, Comperat EM, et al. Updated 2016 EAU guidelines on muscle-invasive and metastatic bladder cancer. Eur Urol. 2017;71(3):462–475.
- Knowles MA, Cd H. Molecular biology of bladder cancer: new insights into pathogenesis and clinical diversity. Nat Rev Cancer. 2015;15(1):25–41.
- Babjuk M. Bladder cancer in the elderly. Eur Urol. 2018;73(1):51–52.
- Carneiro BA, Meeks JJ, Kuzel TM, et al. Emerging therapeutic targets in bladder cancer. Cancer Treat Rev. 2015;41(2):170–178.
- Chen YS, Xu YP, Liu WH, et al. Long noncoding RNA KCNMB2-AS1 promotes SMAD5 by targeting miR-3194-3p to induce bladder cancer progression. Front Oncol. 2021;11:649778.
- Zhang SF, Pang S, Wang FP, et al. Long noncoding RNA OIP5-AS1 exhibits oncogenic activity in bladder cancer through miR-217 and MTDH. Eur Rev Med Pharmacol Sci. 2021;25(8):3211–3220.
- Chen Q, Fu L. Upregulation of long non-coding RNA ROR1-AS1 promotes cell growth and migration in bladder cancer by regulation of miR-504. PLoS One. 2020;15(1):e0227568.
- Zhou LL, Zhang M, Zhang YZ, et al. Long non-coding RNA PSMA3-AS1 enhances cell proliferation, migration and invasion by regulating miR-302a-3p/RAB22A in glioma. Biosci Rep. 2020;40(9):9.
- Qiu BQ, Lin XH, Ye XD, et al. Long non-coding RNA PSMA3-AS1 promotes malignant phenotypes of esophageal cancer by modulating the miR-101/EZH2 axis as a ceRNA. Aging (Albany NY). 2020;12(2):1843–1856.
- Wang L, Wu L, Pang J. Long noncoding RNA PSMA3AS1 functions as a microRNA4093p sponge to promote the progression of nonsmall cell lung carcinoma by targeting spindlin 1. Oncol Rep. 2020;44(4):1550–1560.
- Zhou Q, Zhang L. MicroRNA-183-5p protects human derived cell line SH-SY5Y cells from mepivacaine-induced injury. Bioengineered. 2021;12(1):3177–3187.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Lai F, Zhang H, Xu B, et al. Long non-coding RNA NBR2 suppresses the progress of colorectal cancer in vitro and in vivo by regulating the polarization of TAM. Bioengineered. 2021;12(1):5462–5475.
- Liu Y, Zhang H, Wang H, et al. Long non-coding RNA DUXAP8 promotes the cell proliferation, migration, and invasion of papillary thyroid carcinoma via miR-223-3p mediated regulation of CXCR4. Bioengineered. 2021;12(1):496–506.
- Shi Z, Shen C, Yu C, et al. Long non-coding RNA LINC00997 silencing inhibits the progression and metastasis of colorectal cancer by sponging miR-512-3p. Bioengineered. 2021;12(1):627–639.
- Zan L, Chen Q, Zhang L, et al. Epigallocatechin gallate (EGCG) suppresses growth and tumorigenicity in breast cancer cells by downregulation of miR-25. Bioengineered. 2019;10(1):374–382.
- Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365.
- Zhou R, Miao S, Xu J, et al. Circular RNA circ_0000020 promotes osteogenic differentiation to reduce osteoporosis via sponging microRNA miR-142-5p to up-regulate bone morphogenetic protein BMP2. Bioengineered. 2021;12(1):3824–3836.
- An Q, Zhou Z, Xie Y, et al. Knockdown of long non-coding RNA NEAT1 relieves the inflammatory response of spinal cord injury through targeting miR-211-5p/MAPK1 axis. Bioengineered. 2021;12(1):2702–2712.
- Dermani FK, Samadi P, Rahmani G, et al. PD-1/PD-L1 immune checkpoint: potential target for cancer therapy. J Cell Physiol. 2019;234(2):1313–1325.
- Knauss JL, Miao N, Kim SN, et al. Long noncoding RNA Sox2ot and transcription factor YY1 co-regulate the differentiation of cortical neural progenitors by repressing Sox2. Cell Death Dis. 2018;9(8):799.
- Li XG, Liu SC, Qiao XF, et al. LncRNA MEG3 promotes proliferation and differentiation of osteoblasts through Wnt/beta-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2019;23(11):4521–4529.
- Zhang K, Zhao Z, Yu J, et al. LncRNA FLVCR1-AS1 acts as miR-513c sponge to modulate cancer cell proliferation, migration, and invasion in hepatocellular carcinoma. J Cell Biochem. 2018;119(7):6045–6056.
- Peng P, Wang Y, Wang BL, et al. LncRNA PSMA3-AS1 promotes colorectal cancer cell migration and invasion via regulating miR-4429. Eur Rev Med Pharmacol Sci. 2020;24(22):11594–11601.
- Li F, Yu L, Zhu J. LncRNA PSMA3-AS1 promotes lung cancer growth and invasion via sponging MiR-4504. Cancer Manag Res. 2020;12:5277–5283.
- Liu T, Han Z, Li H, et al. LncRNA DLEU1 contributes to colorectal cancer progression via activation of KPNA3. Mol Cancer. 2018;17(1):118.
- Zeng C, Liu S, Lu S, et al. The c-Myc-regulated lncRNA NEAT1 and paraspeckles modulate imatinib-induced apoptosis in CML cells. Mol Cancer. 2018;17(1):130.
- Agarwal N, Theodorescu D. The role of transcription factor YY1 in the biology of cancer. Crit Rev Oncog. 2017;22(1–2):13–21.
- Yan Z, Yang Q, Xue M, et al. YY1-induced lncRNA ZFPM2-AS1 facilitates cell proliferation and invasion in small cell lung cancer via upregulating of TRAF4. Cancer Cell Int. 2020;20(1):108.
- You Q, Yao Y, Wu J, et al. YY1-induced lncRNA DSCR8 promotes the progression of ovarian cancer via miR-3192-5p/YY1 axis. Biomed Pharmacother. 2020;129:110339.
- Wang LX, Wan C, Dong ZB, et al. Integrative analysis of long noncoding RNA (lncRNA), microRNA (miRNA) and mRNA expression and construction of a competing endogenous RNA (ceRNA) network in metastatic melanoma. Med Sci Monit. 2019;25:2896–2907.
- Zhang M, Wang D, Zhu T, et al. miR-214-5p Targets ROCK1 and suppresses proliferation and invasion of human osteosarcoma cells. Oncol Res. 2017;25(1):75–81.
- Zheng C, Guo K, Chen B, et al. miR-214-5p inhibits human prostate cancer proliferation and migration through regulating CRMP5. Cancer Biomark. 2019;26(2):193–202.
- Guo M, Lin B, Li G, et al. LncRNA TDRG1 promotes the proliferation, migration, and invasion of cervical cancer cells by sponging miR-214-5p to target SOX4. J Recept Signal Transduct Res. 2020;40(3):281–293.
- Roshani Asl E, Rasmi Y, Baradaran B. MicroRNA-124-3p suppresses PD-L1 expression and inhibits tumorigenesis of colorectal cancer cells via modulating STAT3 signaling. J Cell Physiol. 2021;236(10):7071–7087.
- Wei S, Wang K, Huang X, et al. LncRNA MALAT1 contributes to non-small cell lung cancer progression via modulating miR-200a-3p/programmed death-ligand 1 axis. Int J Immunopathol Pharmacol. 2019;33:2058738419859699.
