ABSTRACT
For humans, gastric cancer (GC) is a common malignancy. Multiple circular RNAs (circRNAs) have been confirmed to be important cancer-promoting or tumor-suppressive factors. The present study discusses the roles and mechanisms of circ_0000423 in GC development. In this study, circ_0000423 expression in GC patient tissue samples and cell lines was detected via quantitative real-time polymerase chain reaction. Disheveled-Axin domain containing 1 (DIXDC1) expression in GC cells was examined via Western blot. Besides, cell counting kit-8 was utilized for detecting GC cell viability. GC cell migration and invasion were examined through Transwell assays. Bioinformatics and dual-luciferase reporter gene assays were employed to verify the regulatory relationships between microRNA-582-3p (miR-582-3p) and circ_0000423 or DIXDC1. In the present study, we demonstrated that circ_0000423 was highly expressed in GC. Circ_0000423 knockdown suppressed GC cell viability, migration and invasion. Moreover, miR-582-3p was confirmed as a direct target of circ_0000423, and an upstream regulator of DIXDC1. MiR-582-3p inhibition or DIXDC1 overexpression could reverse the above-mentioned effects of knocking down circ_0000423 on GC cells. In conclusion, circ_0000423 facilitates GC progression by modulating the miR-582-3p/DIXDC1 axis.
1. Introduction
Gastric cancer (GC), as the fifth most common carcinoma worldwide, ranks third among the causes of cancer-related death [Citation1]. It is estimated that more than 1 million patients are diagnosed with GC every year, and half of them are from China [Citation2,Citation3]. GC is associated with multiple pathological factors, and both environmental and genetic factors have a role in its etiology; despite dramatic advances in surgery and adjuvant therapy for GC, since over 80% of GC patients are diagnosed with advanced cancer, the mortality of GC still is very high: the 5-year overall survival rate is less than 30% due to frequent tumor recurrence and metastasis [Citation4–6]. In this context, seeking new diagnostic biomarkers and therapy targets for GC becomes urgent.
Non-coding RNA (ncRNA) has become a hot spot in cancer research. As a newly discovered class of ncRNA, circular RNAs (circRNAs) are widely expressed in eukaryotes; they are formed by non-canonical splicing and have a covalently closed loop with neither 3ʹ poly(A) tail nor 5ʹ end [Citation7,Citation8]. The special structure of circRNAs makes it relatively resistant to degradation mediated by exonuclease [Citation8,Citation9]. CircRNAs are strongly linked to the development of various diseases, especially cancer [Citation10]. For instance, circ_0007294 expression is markedly increased in triple-negative breast carcinoma, and its high expression is strongly associated with lymph node metastasis and advanced clinical stage [Citation11]; circ_0101432 inhibits hepatocellular cancer cell apoptosis through targeting miR-622 and miR-1258, and up-regulating mitogen-activated protein kinase 1 expression [Citation12]. In the present study, bioinformatics analysis suggested that circ_0000423 was highly expressed in GC tissues. Circ_0000423 is formed by reverse splicing of protein phosphatase 1 regulatory subunit 12A (PPP1R12A) mRNA. Reportedly, circ_0000423 expression is up-regulated in colon carcinoma tissues, and high circ_0000423 expression is strongly related to the short survival time of patients [Citation13]. Nonetheless, the specific role of circ_0000423 in GC is still not fully elucidated, and further exploration is needed.
We hypothesized that circ_0000423 played an important role in GC progression. The current study was aimed at exploring the role and mechanism of circ_0000423 in GC, providing a theoretical basis for clarifying the mechanism of the progression of this deadly disease.
2. Materials and methods
2.1. Clinical samples and ethics statement
A total of 43 GC patients (38–72 years old, 28 males and 15 females) who received gastrectomy at the First Affiliated Hospital of Zhengzhou University from 2016 to 2019 were enrolled in the study. Based on tumor, lymph node metastasis and tumor node metastasis (TNM) grading according to the International Union Against Cancer (7th edition), there were 14 cases at stage I, 10 cases at stage II, 19 cases at stage III. The tumorous and adjacent non-tumorous tissues (5 cm away from the tumor) were surgically removed and frozen with liquid nitrogen and then kept at −80°C. None of the participants had undergone chemotherapy or radiotherapy before tissue collection. The study was endorsed by the Ethics Committee of First Affiliated Hospital of Zhengzhou University (approval no. 201606032), and we obtained the written informed consent of each patient. The present study was carried out following the Declaration of Helsinki.
2.2. Cell culture
From the Cancer Institute and Hospital of the Chinese Academy of Medical Sciences (Beijing, China) and the American Type Culture Collection (Manassas, VA, USA), a human renal epithelial cell line (293 T), 4 kinds of GC cell lines (HGC-27, AGS, MGC-803 and MKN-45) and a human gastric epithelial cell line (GES-1). These cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM; Gibco, Grand Island, NY, USA) containing 100 U/ml penicillin, 0.1 mg/ml streptomycin and 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA) in 5% CO2 at 37°C.
2.3. CircRNA expression profile analysis
From the Gene Expression Omnibus (GEO) (http://www.ncbi.nlm.nih.gov/geo) database, we downloaded three datasets (GSE78092, GSE89143, and GSE141977) with GC circRNA expression profiles. To screen out differentially expressed circRNAs, the GEO2R online software was utilized to analyze them.
2.4. Cell transfection
From GenePharma (Shanghai, China), miR-582-3p inhibitors (anti-miR-582-3p: 5ʹ-GGUUCAGUUGUUCAACCAGUUA-3ʹ) and miR-582-3p mimics (5ʹ-UAACUGGUUGAACAACUGAACC-3ʹ), their negative controls (miR-con mimics: 5ʹ- UACGACUGAUACGAUCAUCAGA-3ʹ, and anti-miR-con: 5ʹ-GAAUGACUCACUGCUGAUUGUU-3ʹ), small interfering RNAs (siRNAs) targeting circ_0000423 (si-circ_0000423#1:5ʹ-ACCCAGCCCTGTAAGACCAGT-3ʹ and circ_0000423#2: 5ʹ-AAAAGGCCACCCAGCCCTGTA-3ʹ) and siRNA control (si-NC) were obtained. To construct overexpression vectors of Disheveled-Axin domain containing 1 (DIXDC1) (pcDNA3.1-DIXDC1) and circ_0000423 (pCD5-circ_0000423), the full-length cDNA of DIXDC1 sequence was amplified and cloned into the pcDNA3.1 vector (GenePharma, Shanghai, China); the full-length cDNA of circ_0000423 was amplified and inserted into the pCD5-ciR vector (Greenseed Biotech, Guangzhou, China), which contained a front and back circular frame to promote RNA circularization. They were transfected into AGS and MGC-803 cells employing Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s instructions.
2.5. Quantitative real-time polymerase chain reaction (qRT-PCR) and RNase R treatment
A TRIzol kit (Invitrogen, Carlsbad, CA, USA) was utilized for extracting the total RNA, and the PrimeScript™ RT Reagent Kit with gDNA Eraser (Takara Bio Inc., Shiga, Japan) was employed to reversely transcribe RNA into complementary DNA (cDNA). qRT-PCR was implemented with a SYBR® Premix Ex Taq™ II kit (Takara Bio Inc., Shiga, Japan) in Applied Biosystems 7500 Real-time PCR System (Applied Biosystems, Inc. Carlsbad, CA, USA). GAPDH and U6 were used as the internal reference for genes and miRNAs expression, respectively. For RNase R treatment, 1 μg of RNA was incubated with 1 U/μg RNase R (Epicenter, Madison, Wisconsin, USA) for 20 min at 37°C, and then reverse transcription and PCR was performed. Primer sequences are shown in .
Table 1. Primer sequences used in this study
2.6. Cell counting kit-8 (CCK-8) assay
After the transfection with the plasmids or oligonucleotides, MGC-803 and AGS cells (2 × 103 cells/well in a 96-well plate) were cultured for 0, 24, 48, and 72 h. Next, each well was added with CCK-8 reagent (10 μL; Dojindo, Tokyo, Japan), followed by the incubation for 2 h. After that, the absorbance of the cells in each well at 450 nm was detected with a microplate reader (Bio-Rad, Hercules, CA, USA) [Citation14].
2.7. Transwell assays
Using the Transwell chamber (24-well; Corning Incorporated, Corning, NY, USA), cell migration and invasion were measured via Transwell assays [Citation15]. The processes of migration assay and invasion assay were similar. For invasion assay, however, the filter of upper compartment was pre-coated with Matrigel (BD Bioscience, San Jose, CA, USA). GC cells (1 × 104 cells/well) cultured in FBS-free medium were transferred to the top compartment, and 10% FBS-containing DMEM was injected into the lower compartment. The cells were cultured for 24 h, and the cells on the top membrane surface were removed using cotton swabs, and the cells on the lower surface were fixed with methanol and stained with crystal violet (0.25%; Sigma, St. Louis, MO, USA). In three random fields under the microscope, the numbers of migrated or invaded cells were analyzed.
2.8. Luciferase reporter gene assay
The circ_0000423/DIXDC1 mRNA 3ʹUTR sequence containing binding sites to miR-582-3p were amplified and cloned into the pGL3 Basic reporter vector (Promega, Madison, WI, USA) to establish wild type (WT) circ_0000423-WT/DIXDC1-WT reporter vectors. Besides, the mutant type (MUT) circ_0000423/DIXDC1 mRNA 3ʹUTR sequence was inserted into pGL3 Basic reporter vectors to obtain the circ_0000423-MUT/DIXDC1-MUT1/2/1&2 reporter vector. Circ_0000423-WT/DIXDC1-WT or circ_0000423-MUT/DIXDC1-MUT1/2/1&2 and miR-582-3p mimics or miR-con mimics were co-transfected into 293 T cells, respectively. Ultimately, a Dual-Luciferase Assay System (Promega, Madison, WI, USA) was employed for determining the relative luciferase activity of the cells in each group.
2.9. Western blot
To obtain the total protein, protease inhibitors-containing radioimmunoprecipitation assay (RIPA) lysis buffer was applied to lyse cells. After being separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the protein was transferred to a polyvinylidene difluoride (PVDF) membrane (Amersham Bioscience, Piscataway, NJ). Subsequently, the membrane was blocked in 5% skim milk and then incubated with primary antibodies for DIXDC1 (ab226210, Abcam, Cambridge, MA, USA) and GAPDH (ab181602, Abcam, Cambridge, MA, USA) overnight at 4°C. After that, the membranes were incubated with secondary antibodies for 1 h at room temperature. A chemiluminescence detection system (Beyotime Biotechnology, Shanghai, China) was adopted to develop the protein bands.
2.10. Immunohistochemistry (IHC)
GC tissue and adjacent normal tissue were fixed in 4% paraformaldehyde for 24 h and embedded in paraffin. Serial sections of 4-μm thick were used to conducted IHC assay [Citation16]. Briefly, the sections were dewaxed, hydrated, and incubated with primary antibody at 4°C overnight in a wet box. The next day, the sections were incubated with secondary antibodies for 2 h. Then, the diaminobenzidine (DAB) system was used to develop the color, and the sections were counterstained with hematoxylin. Finally, the stained section images were captured using a microscope.
2.11. Statistical analysis
All experiments were conducted in triplicate. The tool for statistical analysis was GraphPad Prism 8.0 (GraphPad Software, Inc., San Diego, CA, USA). All data were represented by the ‘mean ± standard deviation’. Independent-sample t-test was conducted to compare the measurement data between the two groups. One-way analysis of variance (ANOVA) with Tukey’s post-hoc test was conducted to compare the data among more groups. Correlation analysis was conducted through Pearson’s correlation coefficient analysis. A difference was of statistical significance when P < 0.05.
3. Results
In this study, we investigated the expression pattern, function and molecular mechanism of circ_0000423 in GC progression through bioinformatics, function-gain and function-loss experiments. It was revealed that, circ_0000423, which is highly expressed in GC, promotes the GC cell proliferation, migration and invasion via regulating microR-582-3p (miR-582-3p)/Disheveled-Axin domain containing 1 (DIXDC1) axis.
3.1. Circ_0000423 is high-expressed in GC tissues and cell lines
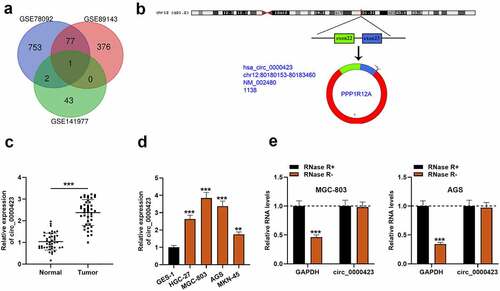
First of all, the microarray data of GSE78092, GSE89143 and GSE141977 downloaded from the GEO database were analyzed, and in each dataset, there were multiple up-regulated circRNAs. However, circ_0000423, was the only circRNA which was up-regulated in GC tissues in all of the three GEO datasets (). Hence, circ_0000423 was selected for further research. Circ_0000423 is formed by head-to-tail splicing of PPP1R12A transcript (). Then, qRT-PCR was employed to examine circ_0000423 expression in 43 pairs of GC and normal para-tumorous tissues, and it was found that circ_0000423 expression was elevated in GC tissues as against para-cancerous tissues (). Additionally, the amplification products of PCR were evaluated by Sanger sequencing, and the result validated the circular structure of circ_0000423 (Figure S1) . In comparison to GES-1 cells, circ_0000423 expression was markedly up-regulated in HGC-27, MGC-803, AGS and KN-45 cells (). Moreover, the stability of circ_0000423 in AGS and MGC-803 cells was further verified using RNase R exonuclease. RNase R treatment could degrade GAPDH, yet it did not affect circ_0000423 (). These findings suggested that up-regulated circ_0000423 expression might be implicated in GC development.
Figure 1. Circ_0000423 expression is significantly up-regulated in GC tissues and cell lines
3.2. Circ_0000423 knockdown inhibits AGS and MGC-803 cell multiplication, migration and invasion
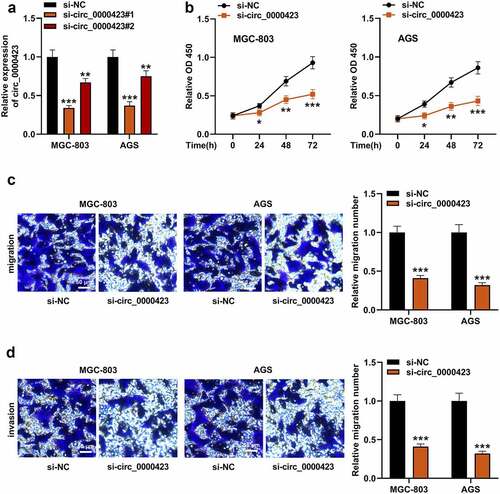
To ascertain circ_0000423’s role in GC development, AGS and MGC-803 cells with higher circ_0000423 expression, were transfected with si-circ_0000423#1 and si-circ_0000423#2 as well as si-NC to construct cell models with low circ_0000423 expression. Knockdown efficiency of si-circ_0000423#1 was higher, so it was used in subsequent experiments (). Subsequently, we investigated the impacts that circ_0000423 had on GC cell growth, migration and invasion. CCK-8 assay showed that knocking down circ_0000423 inhibited AGS and MGC-803 cell viability (). Transwell analysis suggested that circ_0000423 knockdown significantly inhibited the migration and invasion of MGC-803 and AGS cells ).
Figure 2. Circ_0000423 knockdown suppresses the proliferation, migration and invasion of MGC-803 and AGS cells
3.3. MiR-582-3p is the direct downstream target of circ_0000423
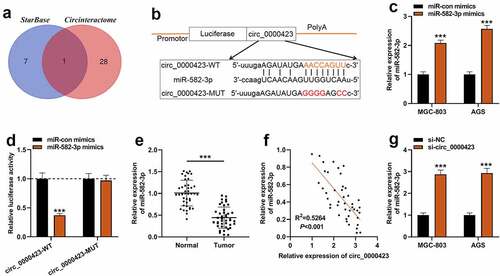
To dig deeper into the downstream mechanism of circ_0000423, bioinformatics tools were applied to predict the potential target miRNAs of circ_0000423. It was predicted that miR-582-3p was the only target of circ_0000423 predicted by both CircInteractome (https://circinteractome.nia.nih.gov/) and StarBase (https://starbase.sysu.edu.cn/) () [Citation17,Citation18]. qRT-PCR showed that AGS and MGC-803 cells were successfully transfected with miR-582-3p mimics (). Dual-luciferase reporter gene assay indicated that miR-582-3p could dramatically reduce circ_0000423-WT’s luciferase activity yet had no significant impact on circ_0000423-MUT’s luciferase activity, suggesting that circ_0000423 could directly bind to miR-582-3p (). Subsequently, qRT-PCR showed that miR-582-3p expression was markedly elevated in GC tissues as opposed to para-cancerous tissues (). Meanwhile, Pearson correlation analysis indicated that circ_0000423 expressions and miR-582-3p expressions were inversely correlated in GC samples (). Furthermore, in GC cells, knockdown of circ_0000423 enhanced miR-582-3p expression ().
Figure 3. Circ_0000423 directly targets miR-582-3p
3.4. The circ_0000423/miR-582-3p axis partakes in modulating the biological behaviors of GC cells
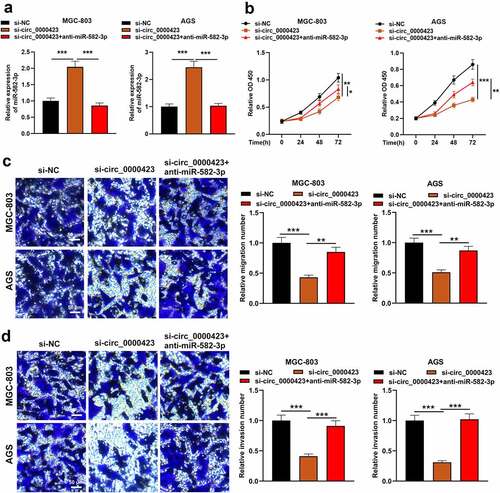
To pinpoint the role of circ_0000423/miR-582-3p axis in GC development, MGC-803 and AGS cells were transfected with si-NC, si-circ_0000423 or si-circ_0000423 + miR-582-3p inhibitor (). It was revealed miR-582-3p inhibition attenuated the inhibiting effects of knocking down circ_0000423 on the growth, migration and invasion of MGC-803 and AGS cells (). These data suggested that the circ_0000423/miR-582-3p axis participated in modulating the biological behaviors of GC cells.
Figure 4. MiR-582-3p inhibition weakens the inhibitory effects of silencing circ_0000423 expression on the malignant biological behaviors of GC cells
3.5. MiR-582-3p can target DIXDC1
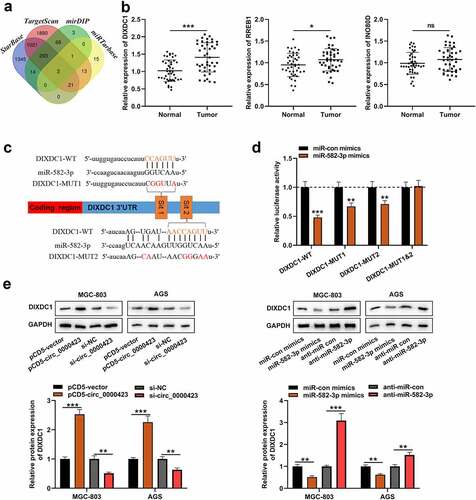
Subsequently, the downstream mRNAs of miR-582-3p were predicted, and three of them (DIXDC1, RREB1 and INO80D) were predicted by all of the four tools, StarBase (https://starbase.sysu.edu.cn/), TargetScan (http://www.targetscan.org/vert_72/), mirDIP (http://ophid.utoronto.ca/mirDIP/index.jsp) and miRTarbase (http://mirtarbase.mbc.nctu.edu.tw/php/index.php) databases (). qRT-PCR and IHC showed that the expression of DIXDC1 was significantly highly expressed in the GC tissues ( and Figure S2), while the changes of RREB1 expression and INO80D expression in GC tissues, compared with those in adjacent tissues, were not that significant (). The binding sites between DIXDC1 mRNA 3ʹUTR and miR-582-3p were presented by bioinformatics (). Subsequently, dual-luciferase reporter gene assay showed that miR-582-3p could reduce the luciferase activity of DIXDC1-WT reporter and DIXDC1-MUT1/2 reporters yet fail to significantly influence the luciferase activity of DIXDC1-MUT1&2 reporter (). Western blot assay showed that knocking down circ_0000423 or up-regulating miR-582-3p expression decreased DIXDC1 expression in GC cells, while circ_0000423 overexpression and miR-582-3p inhibition promoted DIXDC1 expression (). These results suggested that DIXDC1 was target gene of miR-582-3p, and could be positively regulated by circ_0000423.
Figure 5. MiR-582-3p targets DIXDC1
4. Discussion
GC is one of the most common malignancies of the digestive system; due to its insidious early symptom, high aggressiveness, and resistance to chemotherapy and radiotherapy, GC has a relatively high mortality [Citation19]. Therefore, exploring the biological mechanism underlying GC occurrence and development and identifying more biomarkers will help to develop better treatment strategies.
CircRNAs are vital in the tumorigenesis and progression of multiple cancers. For instance, in GC tissues, some circRNAs are differentially expressed and partake in regulating the malignancy of cancer cells [Citation20]. For example, circ_006100 expression is enhanced in GC tissues, and its high expression is closely associated with poor differentiation of tumor tissues, high TNM stage and lymph node metastasis [Citation21]. Circ-RPL15 regulates miR-502-3p/OLFM4/STAT3 axis to regulate the malignant phenotype of GC cells, thus facilitating GC progression; abnormal circ-RPL15 expression can predict the GC patient’s poor prognosis [Citation22]. Circ_0004872 expression is remarkably down-regulated in GC tissues, and circ_0004872 overexpression inhibits GC cell growth and metastasis in vitro and in vivo [Citation23]. Circ_0001546 modulates ATM/Chk2/p53 pathway via repressing miR-421, thereby inhibiting GC cell proliferation and resistance to oxaliplatin [Citation24]. Our study found that circ_0000423 was highly expressed in GC tissues and cells, and circ_0000423 knockdown restrained GC cell proliferation, migration and invasion. To the best of our knowledge, this is the first work to investigate the role of circ_0000423 in GC.
Emerging evidence shows that circRNA has multiple binding sites for miRNAs and may act as a miRNA sponge, thereby modulating the translation process of mRNA [Citation25,Citation26]. This competitive endogenous RNA (ceRNA) mechanism is reported to participate in regulating the pathogenesis of many human diseases, including GC [Citation27,Citation28]. MiR-582-3p has tumor-suppressive properties in several human malignancies. For example, it represses the growth and cell cycle progression of acute myeloid leukemia cells via inhibiting cyclin B2 [Citation29]. MiR-582-3p suppresses the malignancy of prostate cancer cells through regulating TGF-β pathway [Citation30]. Reportedly, in GC, miR-582-3p is low-expressed, and the transfection of miR-582-3p mimics inhibits GC cell growth, migration and invasion [Citation31], which is in line with the findings of our study. Moreover, herein, it was confirmed that circ_0000423 directly targeted miR-582-3p to repress its expression, which helps better understand the mechanism of miR-582-3p dysregulation in GC.
Disheveled-Axin domain containing 1 (DIXDC1) is a new type of Disheveled-Axin domain protein [Citation32]. There is growing evidence suggesting that DIXDC1 has carcinogenic effects in many cancers, for example, prostate cancer, glioma, non-small cell lung carcinoma and acute myeloid leukemia [Citation33–37]. Reportedly, DIXDC1 is high-expressed in GC and is linked to advanced TNM stage, lymph node metastasis and poor prognosis [Citation38]. By up-regulating MMPs expression, down-regulating E-cadherin expression and enhancing the nuclear accumulation of β-catenin to activate the WNT signaling, DIXDC1 participates in promoting GC cell invasion and metastasis [Citation38,Citation39]. Reportedly, DIXDC1 is a target gene of miR-154 in GC cells [Citation40]. In this work, DIXDC1 was also identified as a target gene of miR-582-3p. Additionally, it was revealed that circ_0000423 promoted DIXDC1 expression in GC cells. These findings suggest that circ_0000423 regulates DIXDC1 expression via serving as a ceRNA of miR-582-3p in GC cells.
Many circRNAs have been used as diagnostic markers for cancers to predict the prognosis of the patients [Citation41,Citation42]. Due to the limitation of the sample number in the present work, the potential of circ_0000423 as a prognostic biomarker for GC has not been evaluated. In the following work, more patients should be recruited to solve these issues. In addition, the biological function of circ_0000423/miR-582-3p/DIXDC1 axis in GC development needs to be further validated with in-vivo models in the following studies.
5. Conclusion
To sum up, our study reports that circ_0000423 expression is enhanced in GC, and circ_0000423 can promote GC cell proliferation, migration and invasion through modulating miR-582-3p/DIXDC1 axis. These findings provide a novel explanation of GC progression.
Ethics statement
Our study has been approved by the Ethics Review Board of the First Affiliated Hospital of Zhengzhou University.
Highlight
Circ_0000423 expression is up-regulated in gastric cancer tissues and cells.
Circ_0000423 promotes gastric cancer cell viability, migration and invasion.
Circ_0000423 acts as a competing endogenous RNA (ceRNA) of miR-582-3p to regulate DIXDC1 expression.
Supplemental Material
Download MS Word (1 MB)Acknowledgements
We kindly thank the colleagues from Department of General Surgery, the First Affiliated Hospital of Zhengzhou University, to recruit the patients.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data availability statement
The data used to support the findings of this study are available from the corresponding author upon request.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. 2020 Aug 29;396(10251):635–648.
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018 Nov;68(6):394–424.
- Li R, Liu B, Gao J. The application of nanoparticles in diagnosis and theranostics of gastric cancer. Cancer Lett. 2017;386:123–130.
- Rona KA, Schwameis K, Zehetner J, et al. Gastric cancer in the young: an advanced disease with poor prognostic features. J Surg Oncol. 2017;115(4):371–375.
- Zeng H, Zheng R, Guo Y, et al. Cancer survival in China, 2003-2005: a population-based study. Int J Cancer. 2015 Apr 15;136(8):1921–1930.
- Allemani C, Weir HK, Carreira H, Azevedo e Silva G, Chen WQ, Ogunbiyi OJ, Rachet B, Soeberg MJ, You H, Matsuda T, Bielska-Lasota M, Storm H, Tucker TC and Coleman MP; Concord Working Group. et al. Global surveillance of cancer survival 1995-2009: analysis of individual data for 25,676,887 patients from 279 population-based registries in 67 countries (Concord-2). Lancet. 2015 Mar 14;385(9972):977–1010.
- Chen BJ, Byrne FL, Takenaka K, et al. Analysis of the circular RNA transcriptome in endometrial cancer. Oncotarget. 2017 Dec 20;9(5):5786–5796.
- Chen LL, Yang L. Regulation of circRNA biogenesis. RNA Biol. 2015;12(4):381–388.
- Suzuki H, Tsukahara T. A view of pre-mRNA splicing from RNase R resistant RNAs. Int J Mol Sci. 2014;15(6):9331–9342.
- Patop IL, Kadener S. circRNAs in Cancer. Curr Opin Genet Dev. 2018 Feb;48:121–127.
- Zeng K, He B, Yang BB, et al. The pro-metastasis effect of circANKS1B in breast cancer. Mol Cancer. 2018 Nov 19;17(1):160.
- Zou H, Xu X, Luo L, et al. Hsa_circ_0101432 promotes the development of hepatocellular carcinoma (HCC) by adsorbing miR-1258 and miR-622. Cell Cycle. 2019 Oct;18(19):2398–2413.
- Zheng X, Chen L, Zhou Y, et al. A novel protein encoded by a circular RNA circPPP1R12A promotes tumor pathogenesis and metastasis of colon cancer via Hippo-YAP signaling. Mol Cancer. 2019 Mar 29;18(1):47.
- Cheng J, Lou Y, Jiang K. Downregulation of long non-coding RNA LINC00460 inhibits the proliferation, migration and invasion, and promotes apoptosis of pancreatic cancer cells via modulation of the miR-320b/ARF1 axis. Bioengineered. 2021;Dec;12(1):96–107.
- Xin H, Wang C, Chi Y, et al. MicroRNA-196b-5p promotes malignant progression of colorectal cancer by targeting ING5. Cancer Cell Int. 2020 Apr 10;20(1):119.
- Zhao L, Li C, Liu F, et al. A blockade of PD-L1 produced antitumor and antimetastatic effects in an orthotopic mouse pancreatic cancer model via the PI3K/Akt/mTOR signaling pathway. Onco Targets Ther. 2017 Apr 12;10:2115–2126.
- Dudekula DB, Panda AC, Grammatikakis I, et al. CircInteractome: a web tool for exploring circular RNAs and their interacting proteins and microRNAs. RNA Biol. 2016 Jan 2;13(1):34–42.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014 Jan;42( Database issue):D92–7.
- Sun TT, He J, Liang Q, et al. LncRNA GClnc1 promotes gastric carcinogenesis and may act as a modular scaffold of WDR5 and KAT2A complexes to specify the histone modification pattern. Cancer Discov. 2016;6(7):784–801.
- Shi P, Wan J, Song H, et al. The emerging role of circular RNAs in gastric cancer. Am J Cancer Res. 2018 Oct 1;8(10):1919–1932.
- Liang M, Huang G, Liu Z, et al. Elevated levels of hsa_circ_006100 in gastric cancer promote cell growth and metastasis via miR-195/GPRC5A signalling. Cell Prolif. 2019 Sep;52(5):e12661.
- Li Y, Gong Y, Ma J, et al. Overexpressed circ-RPL15 predicts poor survival and promotes the progression of gastric cancer via regulating miR-502-3p/OLFM4/STAT3 pathway. Biomed Pharmacother. 2020 Jul;127:110219.
- Ma C, Wang X, Yang F, et al. Circular RNA hsa_circ_0004872 inhibits gastric cancer progression via the miR-224/Smad4/ADAR1 successive regulatory circuit. Mol Cancer. 2020 Nov 10;19(1):157.
- Wu Q, Wang H, Liu L, et al. Hsa_circ_0001546 acts as a miRNA-421 sponge to inhibit the chemoresistance of gastric cancer cells via ATM/Chk2/p53-dependent pathway. Biochem Biophys Res Commun. 2020 Jan 8;521(2):303–309.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
- Fang J, Chen W, Meng X. Downregulating circRNA_0044516 inhibits cell proliferation in gastric cancer through miR-149/Wnt1/β-catenin pathway. J Gastrointest Surg. 2021 Jul;25(7):1696–1705.
- Qi X, Zhang DH, Wu N, et al. ceRNA in cancer: possible functions and clinical implications. J Med Genet. 2015 Oct;52(10):710–718.
- Kong X, Duan Y, Sang Y, et al. LncRNA-CDC6 promotes breast cancer progression and function as ceRNA to target CDC6 by sponging microRNA-215. J Cell Physiol. 2019 Jun;234(6):9105–9117.
- Li H, Tian X, Wang P, et al. MicroRNA-582-3p negatively regulates cell proliferation and cell cycle progression in acute myeloid leukemia by targeting cyclin B2. Cell Mol Biol Lett. 2019 Dec 4;24(1):66.
- Huang S, Zou C, Tang Y, et al. miR-582-3p and miR-582-5p suppress prostate cancer metastasis to bone by repressing TGF-beta signaling. Mol Ther Nucleic Acids. 2019 Jun 7;16:91–104.
- Xie M, Yu T, Jing X, et al. Exosomal circSHKBP1 promotes gastric cancer progression via regulating the miR-582-3p/HUR/VEGF axis and suppressing HSP90 degradation. Mol Cancer. 2020 Jun 29;19(1):112.
- Shiomi K, Uchida H, Keino-Masu K, et al. Ccd1, a novel protein with a DIX domain, is a positive regulator in the Wnt signaling during zebrafish neural patterning. Curr Biol. 2003;13(1):73–77.
- Zhong J, Liu Y, Xu Q, et al. Inhibition of DIXDC1 by microRNA-1271 suppresses the proliferation and invasion of prostate cancer cells. Biochem Biophys Res Commun. 2017;484(4):794–800.
- Xu Z, Liu D, Fan C, et al. DIXDC1 increases the invasion and migration ability of non-small-cell lung cancer cells via the PI3K-AKT/AP-1 pathway. Mol Carcinog. 2014;53(11):917–925.
- Chen J, Shen C, Shi J, et al. Knockdown of DIXDC1 inhibits the proliferation and migration of human glioma cells. Cell Mol Neurobiol. 2016;37(6):1009–1019.
- Xin H, Li C, Wang M. DIXDC1 promotes the growth of acute myeloid leukemia cells by upregulating the Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2018 Nov;107:1548–1555.
- Li X, Xiao Y, Fan S, et al. Overexpression of DIXDC1 correlates with enhanced cell growth and poor prognosis in human pancreatic ductal adenocarcinoma. Hum Pathol. 2016;57:182–192.
- Tan C, Qiao F, Wei P, et al. DIXDC1 activates the Wnt signaling pathway and promotes gastric cancer cell invasion and metastasis. Mol Carcinog. 2016 Apr;55(4):397–408.
- Wang L, Tan C, Qiao F, et al. Upregulated expression of DIXDC1 in intestinal-type gastric carcinoma: co-localization with β-catenin and correlation with poor prognosis. Cancer Cell Int. 2015 Dec 18;15(1):120.
- Song J, Guan Z, Li M, et al. MicroRNA-154 inhibits the growth and invasion of gastric cancer cells by targeting DIXDC1/WNT signaling. Oncol Res. 2018 Jul 5;26(6):847–856.
- Li XW, Yang WH, Xu J. Circular RNA in gastric cancer. Chin Med J (Engl). 2020 Aug 5;133(15):1868–1877.
- Wang HY, Wang YP, Zeng X, et al. Circular RNA is a popular molecule in tumors of the digestive system (Review). Int J Oncol. 2020 Jul;57(1):21–42.
