ABSTRACT
Circular RNAs (CircRNAs) were reported to play vital roles in the progression of DN. Herein, the action of circular RNA_0037128 (circ_0037128) was investigated in DN. The level of circ_0037128, microRNA-497-5p (miR-497-5p) and nuclear factor of activated T cells 5 (NFAT5) was determined using quantitative real-time polymerase chain reaction (qRT-PCR). The feature of circ_0037128 was tested by RNase R and Actinomycin D treatment assays. Cell Counting Kit-8 (CCK-8) and 5-ethynyl-2ʹ-deoxyuridine (EdU) staining assays were conducted to evaluate the proliferation ability. The relative protein expression was determined via Western blot analysis. Levels of the inflammatory cytokines, like tumor necrosis factor α (TNF-α), interleukin-1β (IL-1β) and interleukin-6 (IL-6), were assessed by enzyme-linked immunosorbent assay (ELISA). Reactive oxygen species (ROS) production, lactate dehydrogenase (LDH) and superoxide dismutase (SOD) activity were determined by the matched kits. Dual-luciferase reporter and RNA immunoprecipitation (RIP) assays were conducted for evaluating the correlation between miR-497-5p and circ_0037128 or NFAT5. Circ_0037128 and NFAT5 were enhanced, while miR-497-5p was weakened in kidney tissues of DN patients and high glucose (HG)-cultured HK-2 cells. Circ_0037128 inhibition bated HG-caused inhibition effect on cell proliferation and promotion effects on oxidative stress, inflammation and fibrosis in HK-2 cells. Moreover, circ_0037128 knockdown alleviated HG-caused cell damage via regulating miR-497-5p. In addition, NFAT5 overexpression could reverse the influence of miR-497-5p on HG-induced injury in HK-2 cells. Mechanically, circ_0037128 sponged miR-497-5p to modulate NFAT5. Circ_0037128 downregulation could mitigate HG-stimulated cell damage via regulating the miR-497-5p/NFAT5 axis in HK-2 cells in vitro, providing a possible therapy target for DN.
Introduction
Diabetic nephropathy (DN) is a frequently lethal complication of patients diagnosed with diabetes accompanying with renal impairment and accounts for 30–47% of end-stage renal disease and chronic renal failure [Citation1,Citation2]. It was reported that hyperglycemia-caused metabolic disorder is vital to the pathogenesis of DN, and long duration of diabetes, glucose metabolism disorder, dyslipidemia, obesity, and high blood pressure were the risk factors in DN progression [Citation3,Citation4]. Recently, the therapy strategies for DN, including glycemic control, blood pressure control, cardiovascular reduction, and renin–angiotensin system inhibitors, did not achieve the effective progress [Citation5]. High concentration glucose treatment-induced renal tubular cell damage is considered as a main feature of DN, and tubular cells are verified to be the main targets of DN [Citation6]. Hence, investigating the potential mechanism of tubular cell injury under high glucose condition might develop an efficient therapeutic target for DN.
In recent years, non-coding RNAs have attracted intensive attention due to its function in mammalian development [Citation7,Citation8]. Circular RNAs (circRNAs) are a novel type of endogenous non-coding RNAs with the covalently closed loop and obtain a high degree of sequence conservation and stability due to lack of the 3ʹ end of the polyadenylated tail and the 5ʹ cap [Citation9–11]. MicroRNAs (MiRNAs), the short, small, non-coding RNAs containing only 20–22 nucleotides, are capable of degrading target mRNAs or inhibiting its translation [Citation12]. Increasing evidence indicated that circRNAs could canonically act as miRNA sponges and play essential roles in the regulation of various human diseases pathogenesis by sponging miRNA [Citation13,Citation14]. Multiple studies demonstrated that dysregulation of some circRNAs and miRNAs was found in human diseases and largely related to their pathological progression, including DN [Citation15–17]. For example, upregulation of circHIPK3 in Ang II-triggered cardiac fibroblasts accelerated the metastasis of cardiac fibroblasts via regulating miR-29b-3p, indicating the promotion action of circHIPK3 on fibrosis via functioning as miR-29b-3p sponge [Citation18]. A recent report revealed that highly expressed circRNA_15698 was validated in both mouse mesangial cells under high glucose (HG) treatment in vitro and db/db mice in vivo, exacerbating the accumulation of extracellular matrix and aggravating DN through inhibiting miR-185 expression [Citation19]. Of note, circular RNA_0037128 (circ_0037128; Position: chr16:142,593–150,507) originates from the 9–12 exons of nitrogen permease regulator like protein 3 (NPRL3) and was identified to be overexpressed in mouse DN model and HG-stimulated mesangial cells, suggesting that circ_0037128 might be involved in DN pathogenesis [Citation20]. Moreover, miR-497 was proved to be lowly expressed in the renal tissues from DN patients [Citation21]. However, the precise role, biological function and the underlying detail mechanism of circ_0037128 are completely unclear in DN development. Whether circ_0037128 could serve as a microRNA-497-5p (miR-497-5p) sponge in DN needs further investigation.
Nuclear factor of activated T cells (NFAT) is the zymolyte for calcineurin, and nuclear factor of activated T cells (NFAT5) belongs to NFAT protein family [Citation22]. Increasing researches showed that NFAT participated in various physiological processes, such as glomerulosclerosis and renal tubular cell apoptosis, and suppression of NFAT-related pathway could alleviate early-stage DN [Citation23,Citation24]. Thus, exploration of the role of NFAT5 in DN progression is of value.
Here, we hypothesis that circ_0037128 might play an important role in the development of DN. The aim and goal of this research was to investigate the biological function of circ_0037128, miR-497-5p and NFAT5 in DN pathogenesis.
Material and methods
Tissue samples
Seventy-five fresh samples of kidney tissues including DN kidney tissues (n = 45) and the normal kidney tissues (n = 30) were obtained from the First Affiliated Hospital of Zhengzhou University. Forty-five patients, who diagnosed with DN by renal biopsy examination according to the diagnostic criteria for diabetes in the World Health Organization [Citation25], provided the DN kidney tissues. Thirty patients with renal trauma were recruited to collect the normal kidney tissues during nephrectomy, which served as controls. Renal damage caused by other primary and secondary factors or complications was not found in these patients, and all participants did not receive any treatment before admission. Written informed consent was provided by each patient prior to taking part in this research. All experimental procedures were ratified by the Ethics Committee of The First Affiliated Hospital of Zhengzhou University.
Cell culture and treatment
HK-2 cells (a renal tubular epithelial cell line) were provided by the American Type Culture Collection (Manassas, VA, USA). The culture medium for HK-2 cells was the Dulbecco’s Modified Eagle Medium (DMEM) (Thermo Fisher Scientific, Rockford, IL, USA) containing penicillin (100 U/mL), streptomycin (100 µg/mL) and 10% FBS (Gibco, Carlsbad, CA, USA), and the culture condition was a 5% CO2 atmosphere incubator at 37°C. HK-2 cells treated with 30 mM glucose were included in the HG group, and the cells treated with 5 mM glucose were served as the normal-glucose (NG) group as previously described [Citation26].
Cell transfection
The oligonucleotides, including small interfering RNA against circ_0037128 (si-circ_0037128: 5ʹ-GGUCGAACGTCGAGU-3ʹ), miR-497-5p mimic (5ʹ-CAGCAGCACACUGUGGUUUGU-3ʹ) and inhibitor (5ʹ-ACAAACCACAGUGUGCUGCUG-3ʹ), and the matched negative controls (si-NC: 5ʹ-CACUGAUUUCAAAUGGUGCUAUU-3ʹ, miRNA NC: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ, and inhibitor NC: 5ʹ-CAGUACUUUUGUGUAGUACAA-3ʹ), and overexpression-NFAT5 vector (pc-NFAT5) and the empty pcDNA3.1 vector (pc-NC) were procured from Genepharma (Shanghai, China). HK-2 cells were transfected with the oligonucleotides and aforementioned plasmids using Lipofectamine 3000 (Invitrogen, Carlsbad, CA, USA) [Citation27].
Quantitative real-time polymerase chain reaction (qRT-PCR)
Total RNA was isolated using TRIzol kit (Invitrogen) and purified in line with the instruction of the RNeasy Maxi kit (Qiagen, Dusseldorf, Germany). The cDNA synthesis was obtained using the PrimeScript RT Reagent Kit (Takara, Shiga, Japan). Then, qRT-PCR analysis was conducted utilizing the SYBR® Green PCR Master Mix kit (TaKaRa, Dalian, China). The level of NPRL3, circ_0037128, miR-497-5p and NFAT5 was quantified using the 2−∆∆Ct method, and U6 and β-actin were used for standardization [Citation26]. The primers of the relative genes were listed as below: circ_0037128, F 5ʹ-CCAGTTCCCATCTCATGACC-3ʹ, R 5ʹ-GATGTGAAGCCGAACTACGCC-3ʹ; miR-497-5p, F 5ʹ-GCCGAGCAGCAGCACACTGTG-3ʹ, R 5ʹ-GTGCAGGGTCCGAGGT-3ʹ; NFAT5, F 5ʹ-CAGGCCAACCACAAAACGAG-3ʹ, R 5ʹ-TCATCTTGTGAGAAAGCCACA-3ʹ; NPRL3, F 5ʹ-ACGGCGATTCCAGGTTTTCA-3ʹ, R 5ʹ-GCATGCTGTAGCAGTGTTGG-3ʹ; β-actin, F 5ʹ-CTTCGCGGGCGACGAT-3ʹ, R 5ʹ-CCACATAGGAATCCTTCTGACC-3ʹ; U6, F 5ʹ-CTCGCTTCGGCAGCACATA-3ʹ, R 5ʹ-CGAATTTGCGTGTCATCCT-3ʹ.
CircRNA identification
For RNase R treatment, the extracted total RNA (3 µg) was treated with RNase R (10 U) for 30 min at 37°C [Citation28]. In actinomycin D test, HK-2 cells were treated with actinomycin D (2 mg/ml; R&D Systems, Shanghai, China) to block the new RNA synthesis for 0, 6, 12, 18 and 24 h. Finally, the relative level of circ_0037128 and NPRL3 was examined using qRT-PCR [Citation29].
Cell Counting Kit-8 (CCK-8) assay
A total of 5000 HK-2 cells suspended in 100 μL medium were seeded in the 96-well plates. Ten microliters of CCK-8 solution (Beyotime, Shanghai, China) was added and incubated with the cells for 4 h. Cell viability was analyzed by determining the optical density with a microscope reader (Thermo Labsystems, Waltham, MA, USA) at 450 nm [Citation30].
5-Ethynyl-2ʹ-deoxyuridine (EdU) staining assay
EdU staining assay was conducted through the usage of the BeyoClick™ EdU Cell Proliferation Kit with Alexa Fluor 488 (Beyotime). Briefly, the treated HK-2 cells were incubated with EdU reagent (10 μM) for 2 h and dyed with 4,6-diamidino-2-phenylindole (DAPI; Beyotime). Finally, an inverted fluorescence microscope (Nikon Microsystems, Shanghai, China) was utilized to detect the fluorescence. The EdU-positive cells were counted and quantified with ImageJ software [Citation30].
Western blot
Radio immunoprecipitation assay (RIPA) lysis buffer (Beyotime) containing proteinase inhibitor was utilized to lyse the transfected HK-2 cells to extract the total protein. Twenty micrograms of proteins were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The proteins were transferred onto the polyvinylidene fluoride membrane and then blocked with 5% nonfat milk. The antibodies were proliferating cell nuclear antigen (PCNA; 1:1000, ab18197, Abcam, Cambridge, UK), fibronectin (FN; 1:1000, ab45688, Abcam), transforming growth factor-β1 (TGF-β1; 1:1000, ab215715, Abcam), collagen type I (Col. I; 1:1000, ab34710, Abcam), NFAT5 (1:1000, ab3446, Abcam), β-actin (1:2000, ab8227, Abcam), as well as the secondary antibody (goat anti-rabbit; 1:10,000, ab205718, Abcam). Finally, the protein signals were obtained using the enhanced chemiluminescence kit (Amersham Pharmacia Biotech, Little Chalfont, UK) [Citation28].
Enzyme-linked immunosorbent assay (ELISA)
The production of the inflammatory cytokines, including tumor necrosis factor alpha (TNF-α), interleukin-1β (IL-1β) and interleukin-6 (IL-6), was assessed using ELISA kits (BD Biosciences, Franklin Lakes, NJ, USA) based on the manufacturer's direction [Citation28].
Determination of reactive oxygen species (ROS) production, superoxide dismutase (SOD) activity, and lactate dehydrogenase (LDH) activity
To detect ROS production, HK-2 cells were harvested and incubated with 5 μM 2ʹ,7ʹ-dichlorodihydrofluorescein diacetate (DCF-DA; Sigma-Aldrich) for 1 h at 37°C, followed by determining the fluorescence intensity at the wavelength 488/525 nm. Then, ROS generation was quantified using FACS-Calibur (BD Biosciences). To measure SOD and LDH activity, the cells were collected, lysed, and then cell extract was obtained. Then, the examination of SOD and LDH activity in cell extract was performed with a SOD kit-WST (Dojindo, Kumamoto, Japan) and LDH kit (Beyotime) based on the manufacturer’s specification [Citation28].
Dual-luciferase reporter assay
Circ_0037128 and NFAT5 containing the wild-type (WT) or mutant (MUT) complementary sites of miR-497-5p were synthesized and fused in pGL3 vector (Promega, Madison, WI, USA) to form the WT- or MUT-circ_0037128 and NFAT5-3ʹUTR vectors. These WT/MUT vectors and the indicated miRNAs were co-transfected into cells using Lipofectamine 3000 (Invitrogen). Lastly, the luciferase activity was estimated [Citation28].
RNA immunoprecipitation (RIP) assay
In brief, the magnetic beads combined with the anti-Ago2 or IgG antibody were added in cell supernatant and incubated for 6 h at 4°C. Then, protease K was utilized to remove the proteins. Finally, circ_0037128, miR-497-5p and NFAT5 enrichment were determined via qRT-PCR [Citation30].
Statistical analysis
Data were obtained from three independent experiments. Measurement data analyzed by GraphPad Prism version 6.0 (GraphPad Software, La Jolla, CA, USA) were represented as the mean ± standard deviation (SD). Difference in groups was estimated by one-way analysis of variance (ANOVA) followed by Tukey’s test or Student’s t test. P < 0.05 was deemed as significantly statistical difference.
Results
We hypothesized that circ_0037128 might play a vital role in the development of DN. Thus, this study explored the functional role of circ_0037128 in DN progression and investigated the underlying molecular mechanism. Here, a variety of experiments were performed to explore the effect of circ_0037128 knockdown on cell proliferation, inflammation, oxidative stress and fibrosis in HG-induced HK-2 cells.
Circ_0037128 was increased in kidney tissues from DN patients and HG-induced HK-2 cells in vitro
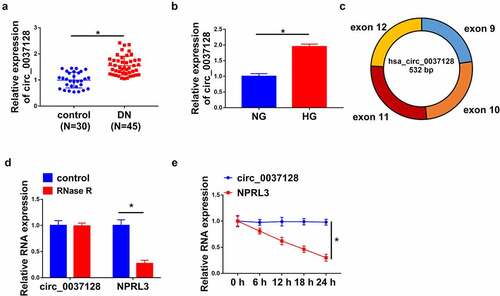
As represented in , circ_0037128 was highly expressed in DN tissues (n = 45) compared with the normal kidney tissues (n = 30). Moreover, the data in qRT-PCR analysis suggested that circ_0037128 was facilitated in HG-stimulated HK-2 cells (). The circRNA circ_0037128 originates from exon 9–12 of NPRL3 gene (). Our results demonstrated that the level of circ_0037128 was not changed, while linear NPRL3 level was significantly depressed after RNase R digestion treatment or Actinomycin D treatment ( e), suggesting that circ_0037128 is indeed a circRNA.
Figure 1. Circ_0037128 was increased in kidney tissues from DN patients and HG-stimulated HK-2 cells in vitro. (a) Circ_0037128 level in DN kidney tissues (n = 45) and normal kidney tissues (n = 30) was measured using qRT-PCR. (b) Circ_0037128 level in HG-stimulated HK-2 cells and NG-cultured HK-2 cells was evaluated via qRT-PCR. (c) The genomic loci and structure of circ_0037128 were described. RNase R digestion (d) and actinomycin D assays (e) were performed to evaluate the stability of circ_0037128, respectively. *P < 0.05
Circ_0037128 knockdown alleviated HG-caused cell damage in HK-2 cells
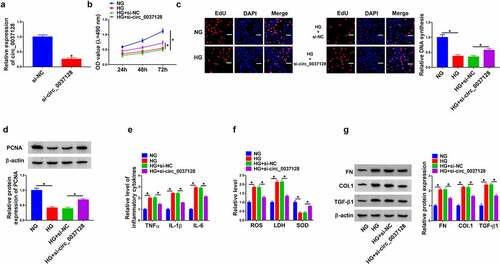
Circ_0037128 was knocked down in treated-HK-2 cells to find out the action of circ_0037128 in the progression of DN in vitro. The data of qRT-PCR showed the successful knockdown efficiency of circ_0037128 (). CCK-8 assay demonstrated that cell viability was suppressed by HG treatment, which was significantly reversed by circ_0037128 repression in HK-2 cells (). Consistently, HG-caused reduction of EdU-positive HK-2 cells was alleviated by circ_0037128 silencing (). The protein expression of PCNA, a proliferation indicator, was dramatically decreased in HK-2 cells under HG condition, which was rescued by circ_0037128 downregulation (). Moreover, we detected the levels of inflammatory cytokines including TNF-α, IL-1β and IL-6 in HK-2 cells. As described in , the level of TNF-α, IL-1β, and IL-6 was significantly elevated in the HG treatment group, which was suppressed by circ_0037128 knockdown in HK-2 cells under HG treatment, suggesting that circ_0037128 played a repression role in the inflammatory response in HG-stimulated HK-2 cells. Moreover, the decline of SOD level, and the increase of ROS and LDH levels caused by HG treatment were overturned in circ_0037128 silencing group, indicating that circ_0037128 absence could significantly weaken HG-induced oxidative stress (). Also, our data showed that the augment expression of FN, Col. I and TGF-β1 caused by HG treatment was dramatically abrogated by the absence of circ_0037128 in HK-2 cells ().
Figure 2. Circ_0037128 knockdown alleviated HG-induced cell damage in HK-2 cells. (a) The transfection efficiency of si-circ_0037128 was assessed. (b–i) HK-2 cells were treated with NG, HG, HG + si-NC, or HG + si-circ_0037128. (b) Cell viability was detected using CCK-8 assay. (c) The proliferative ability was examined by EdU staining assay. (d) The protein expression of PCNA was determined by Western blot. (e) Level of TNF-α, IL-1β and IL-6 was detected in treated HK-2 cells via ELISA assay. (f) ROS production, LDH and SOD activity in treated HK-2 cells were assessed. (g) Western blot was employed for detection of protein levels of FN, Col. I and TGF-β1. *P < 0.05
Circ_0037128 directly interacted with miR-497-5p
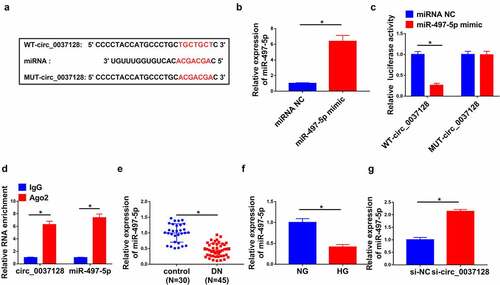
CircBank predicted that circ_0037128 had the complementary bases pairing with miR-497-5p (). The data in qRT-PCR analysis displayed that the expression of miR-497-5p was significantly elevated by miR-497-5p mimic (). As indicated in , the luciferase activity was depressed by miR-497-5p mimic in WT-circ_0037128 group, while there was no remarkable change in MUT-circ_0037128 group. RIP assay revealed that circ_0037128 and miR-497-5p were dramatically enriched by anti-Ago2, not anti-IgG, in HK-2 cells (). Furthermore, miR-497-5p was decreased in DN kidney tissues (n = 45) compared with the normal kidney tissues (n = 30) (). Moreover, the decrease of miR-497-5p was detected in HK-2 cells under HG treatment (). Our data represented that circ_0037128 knockdown significantly boosted miR-497-5p level ().
Figure 3. Circ_0037128 directly interacted with miR-497-5p. (a) The complementary binding sites of circ_0037128 and miR-497-5p were predicted by circBank. (b) The expression of miR-497-5p in HK-2 cells transfected with miRNA NC or miR-497-5p mimic was detected by qRT-PCR. (c) The relative luciferase activity in HK-2 cells co-transfected with WT-circ_0037128, MUT-circ_0037128, miR-497-5p mimic or miRNA NC was determined. (d) RIP assay verified the enrichment of circ_0037128 and miR-497-5p. (e) MiR-497-5p level in DN kidney tissues (n = 45) and the normal kidney tissues (n = 30) was assessed by qRT-PCR. (f) MiR-497-5p level in HG or NG-treated HK-2 cells was evaluated by qRT-PCR. (g) The level of miR-497-5p in HK-2 cells transfected with si-NC or si-circ_0037128 was measured by qRT-PCR. *P < 0.05
Circ_0037128 downregulation mitigated HG-caused damage in HK-2 cells through sponging miR-497-5p
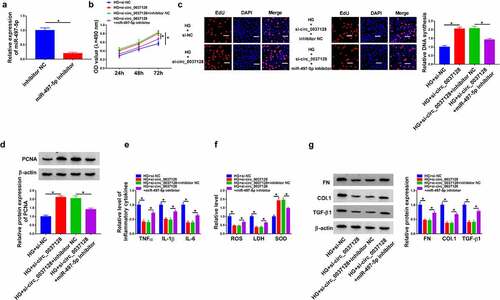
In view of the regulatory effect of circ_0037128 on miR-497-5p expression, whether miR-497-5p was involved in the influence of circ_0037128 on HG-induced damage in HK-2 cells was investigated. The data displayed that miR-497-5p was significantly repressed by miR-497-5p inhibitor (). CCK-8 and EdU staining assays demonstrated that miR-497-5p inhibition abated circ_0037128 knockdown-caused promotion action on cell proliferation in HG-treated HK-2 cells ( and 4 C), accompanied by decreased PCNA expression (). In addition, our results revealed that circ_0037128 inhibition-induced decrease of TNFα, IL-1β, and IL-6 was mitigated by depression of miR-497-5p in HG-treated HK-2 cells (). Moreover, the augment of SOD activity and the decline of ROS production and LDH activity caused by circ_0037128 depletion were hindered by miR-497-5p inhibitor in HG-stimulated HK-2 cells (). Additionally, the suppression effect of circ_0037128 deletion on the expression on FN, Col. I and TGF-β1 was blocked by repression of miR-497-5p in HG-stimulated HK-2 cells ().
Figure 4. Circ_0037128 downregulation mitigated HG-caused damage in HK-2 cells through sponging miR-497-5p. (a) miR-497-5p expression in HK-2 cells transfected with inhibitor NC or miR-497-5p inhibitor was determined by qRT-PCR. (b-i) HK-2 cells were treated with HG + si-circ_0037128, HG + si-NC, HG + si-circ_0037128 + inhibitor NC, or HG + si-circ_0037128 + miR-497-5p inhibitor. (b-d) CCK-8 assay (b), EdU staining assay (c) and Western blot analysis (d) were employed for proliferation assessment. (e) ELISA was utilized for TNF-α, IL-1β and IL-6 determination. (f) ROS generation, LDH and SOD activity were evaluated. (g) FN, Col. I and TGF-β1 expression were determined by Western blot. *P < 0.05
NFAT5 was a target of miR-497-5p
StarBase predicted that NFAT5 harbored the binding sites of miR-497-5p (). The interaction between miR-497-5p and NFAT5 was attested by dual-luciferase reporter and RIP assays ( and c). Of note, the mRNA and protein expression of NFAT5 were elevated in DN kidney tissues (n = 45) in comparison to the normal kidney tissues (n = 30) ( and e). Also, the protein expression of NFAT5 was highly expressed in HG-treated HK-2 cells (). Furthermore, miR-497-5p significantly suppressed NFAT5 expression ().
Figure 5. NFAT5 was a target of miR-497-5p. (a) StarBase predicted the target sites of mir-497-5p and NFAT5. (b) Dual-luciferase reporter assay was conducted. (c) The enrichment of miR-497-5p and NFAT5 was measured by RIP assay. (d–e) The mRNA and protein expression of NFAT5 in DN tissues (n = 45) and control tissues (n = 30) were determined. (f) Western blot was utilized to detect NFAT5 expression in HG- or NG-treated HK-2 cells. (g) NFAT5 expression in HK-2 cells with transfection of miRNA NC or miR-497-5p mimic was measured by Western blot. *P < 0.05
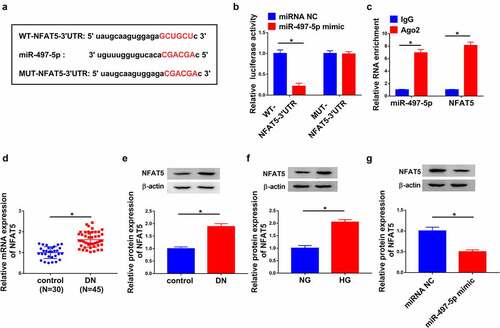
MiR-497-5p weakened HG-induced injury in HK-2 cells through regulating NFAT5 expression
Next, the detailed mechanism of miR-497-5p and NFAT5 in the progression of DN was investigated. As shown in , the successful overexpression efficiency of pc-NFAT5 was observed. Functionally, elevated cell proliferation caused by miR-497-5p overexpression was impeded by NFAT5 upregulation in HG-treated HK-2 cells, as depicted by reduced PCNA expression (). Synchronously, the level of TNF-α, IL-1β and IL-6 suppressed by miR-497-5p was reversed by NFAT5 overexpression in HG-stimulated HK-2 cells, suggesting that miR-497-5p caused inhibition action on inflammatory response was overturned by NFAT5 overexpression (). Additionally, the suppressive influence of miR-497-5p on oxidative stress was impaired by upregulation of NFAT5, as proved by increased LDH activity and ROS production, and decreased SOD activity in HG-stimulated HK-2 cells (). As exhibited in , the expression of FN, Col. I and TGF-β1 was significantly repressed by miR-497-5p, which was evidently rescued by NFAT5 overexpression in HG-stimulated HK-2 cells, implying that miR-497-5p weakened HG-induced fibrosis in HK-2 cells by targeting NFAT5. In addition, our data showed that the repression action of circ_0037128 knockdown on NFAT5 expression was hindered by miR-497-5p inhibitor ().
Figure 6. MiR-497-5p alleviated HG-induced injury in HK-2 cells through regulating NFAT5 expression. (a) NFAT5 protein expression was detected using Western blot. (b–i) HK-2 cells were treated with HG + miR-497-5p mimic, HG + miRNA NC, HG + miR-497-5p mimic + pc-NC, or HG + miR-497-5p mimic + pc-NFAT5. (b-d) CCK-8 (b), EdU (c) and Western blot analysis (d) were performed to evaluate cell proliferation. (e) The level of TNF-α, IL-1β and IL-6 was assessed by ELISA assay. (f) The measurement of ROS generation, LDH and SOD activity via the corresponding kits. (g) The protein expression of FN, Col. I and TGF-β1 was determined by Western blot. *P < 0.05
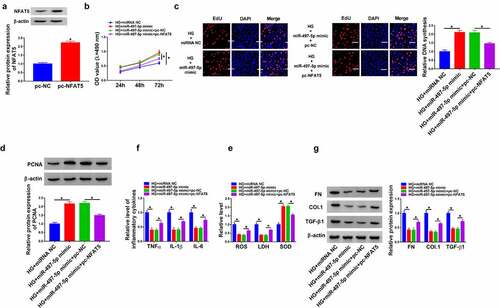
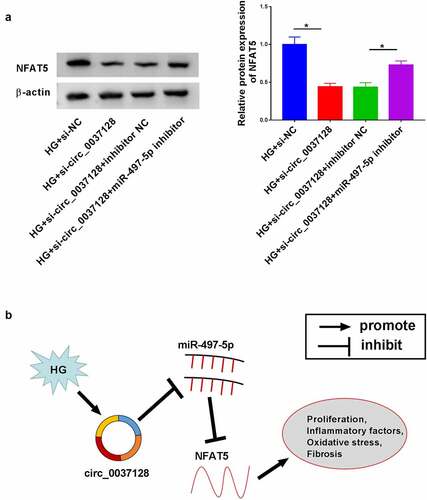
Figure 7. Circ_0037128 downregulation mitigated HG-caused damage in HK-2 cells through targeting miR-497-5p to regulate NFAT5 expression. (a) HK-2 cells were treated with HG + si-circ_0037128, HG + si-NC, HG + si-circ_0037128 + inhibitor NC, or HG + si-circ_0037128 + miR-497-5p inhibitor, and NFAT5 expression was detected using Western blot. (b) Schematic model of the circ_0037128/miR-497-5p/NFAT5 regulatory axis in HG-induced cell damage in HK-2 cells. *P < 0.05
Discussion
DN is a microangiopathic complication of diabetes mellitus with high death rate and severely threatens the health of patients [Citation31]. Accumulating researches suggested that DN progression was accompanied by renal tubular damage, and the death of renal tubular epithelial cells participated in DN [Citation32]. Moreover, tubulointerstitial fibrosis was one of the main features of DN pathogenesis [Citation33]. TGF-β1, an important inducer of renal fibrosis, could facilitate extracellular matrix proteins expression, predominantly FN and collagen [Citation34]. Importantly, oxidative stress, apoptosis, and inflammatory response were regarded as inseparable factors that were related to the pathogenesis of DN [Citation35,Citation36]. Therefore, exploration of effective therapeutic agents that definitely repressed inflammatory injury, oxidative stress and fibrosis might be useful for DN therapy.
A convergence of researches illustrated that circRNAs functioned as regulators by sponging miRNA in DN progression through participating in renal damage [Citation37]. For instance, increased circ_0123996 was observed in Type 2 diabetes patients with DN, DN mice model, and mice mesangial cells under HG condition, which played a promoting role in DN development, as evidenced by elevating mice mesangial cell growth and fibrosis [Citation38]. Also, the effects of circ_0000712 on cell apoptosis, fibrosis, oxidative stress and inflammation were verified in mesangial cells, and circ_0000712 knockdown downregulated SOX6 expression to assuage cell injury caused by HG treatment in SV40-MES13 cells via sponging miR-879-5p, implying the mainly regulatory role of circRNA in DN progression [Citation28]. In HG-stimulated HK-2 cells, cell fibrosis and inflammatory responses were aggravated by circ_WBACR17 through the regulation of SOX6 via targeting miR-185-5p [Citation39]. Moreover, circ_0037128 was previously revealed to be increased in kidney tissues from DN mice and DN patients, as well as the HG-stimulated human mesangial cells [Citation20]. Consistent with this result, we found that circ_0037128 was significantly upregulated in kidney tissues from DN patients and HK-2 cells under HG treatment. Furthermore, our results clarified that HG-caused cell injury including fibrosis, inflammation and oxidative stress was dampened by downregulating circ_0037128 in HK-2 cells, indicating the possible inhibition action of circ_0037128 downregulation in HG-stimulated HK-2 cell damage.
Previous studies documented that circRNAs were involved in regulating diverse biological processes via functioning as competing endogenous RNAs for miRNAs [Citation40]. Accumulating evidence discovered that miR-497-5p was involved in cancer development, like laryngeal squamous cell carcinoma [Citation41], papillary thyroid cancer [Citation42], and non-small cell lung cancer [Citation43]. Moreover, the vital role of miR-497 in inflammation has also been reported. By targeting NF-κB pathway, the inflammatory responses in human bronchial epithelial cells were dramatically repressed by miR-497 [Citation44]. A previous study discovered that miR-497 was reduced in HG-cultured HK-2 cells and promoted cell pyroptosis and caspases-1 activation [Citation21]. Consistently, our data represented the lowly expressed miR-497-5p in HK-2 cells under HG condition. Circ_0037128 sponged miR-497-5p, thereby modulating miR-497-5p expression. Additionally, circ_0037128 silencing weakened HG treatment-induced cell damage in HK-2 cells via sponging miR-497-5p.
MiRNAs directly bind to their target mRNA to degrade or inhibit its expression, thereby exerting function of miRNAs [Citation45]. In this research, we found that miR-497-5p directly targeted NFAT5. NFAT5 was implicated in cellular stress response, inflammatory processes, and cell differentiation and proliferation in a tonicity-independent manner [Citation46]. Moreover, the abnormal expression of NFAT5 was found in some cancers and NFAT5 exerted functions in cancer progression, like the promotion effect on cell growth and metastasis in melanoma and renal carcinoma [Citation47,Citation48]. More importantly, a recent research reported that NFAT5 was increased in DN mouse kidney tissues, HG-stimulated mesangial cells and DN patients [Citation49]. In our study, we also found the upregulation expression of NFAT5 in DN, consistent with the previous data [Citation49]. In terms of mechanism, the repression effect of miR-497-5p on cell damage in HK-2 cells under HG was reversed by NFAT5 overexpression. Our results disclosed that circ_0037128 upregulated NFAT5 expression through sponging miR-497-5p, supporting that the circ_0037128/miR-497-5p/NFAT5 regulatory axis was involved in HG-caused cell injury in HK-2 cells ().
Conclusion
Our findings revealed the augment of miR-497-5p, as well as the reduction of circ_0037128 and NFAT5 in DN, and testified the functional roles of circ_0037128, miR-497-5p and NFAT5 in DN progression, indicating that circ_0037128 could elevate NFAT5 expression by targeting miR-497-5p to aggravate cell damage in HG-treated HK-2 cells. Our study provided a promising target for the therapy of DN.
Authors’ contributions
Tao Feng carried out the experiments. Weifang Li and Tianyi Li are responsible for data collection and analysis. Wenjun Jiao wrote the manuscript and provide some comments. Sufang Chen conceived and designed the concept. All authors read and approved the final version.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Magee C, Grieve DJ, Watson CJ, et al. Diabetic nephropathy: a tangled web to unweave. Cardiovasc Drugs Ther. 2017;31(5–6):579–592.
- Lin YC, Chang YH, Yang SY, et al. Update of pathophysiology and management of diabetic kidney disease. J Formos Med Assoc. 2018;117(8):662–675.
- Flyvbjerg A. The role of the complement system in diabetic nephropathy. Nat Rev Nephrol. 2017;13(5):311–318.
- Zhuang L, Wang Z, Hu X, et al. CircHIPK3 alleviates high glucose toxicity to human renal tubular epithelial HK-2 cells through regulation of miR-326/miR-487a-3p/SIRT1. Diabetes Metab Syndr Obes. 2021;14:729–740.
- Long B, Wan Y, Zhang S, et al. LncRNA XIST protects podocyte from high glucose-induced cell injury in diabetic nephropathy by sponging miR-30 and regulating AVEN expression. Arch Physiol Biochem. 2020;17:1–8.
- Gilbert RE, Cooper ME. The tubulointerstitium in progressive diabetic kidney disease: more than an aftermath of glomerular injury? Kidney Int. 1999;56(5):1627-1637.
- Carninci P, Kasukawa T, Katayama S, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309(5740):1559–1563.
- Djebali S, Davis CA, Merkel A, et al. Landscape of transcription in human cells. Nature. 2012;489:101–108.
- Chen LL. The biogenesis and emerging roles of circular RNAs. Nat Rev Mol Cell Biol. 2016;17:205–211.
- Lasda E, Parker R. Circular RNAs: diversity of form and function. RNA. 2014;20:1829–1842.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. RNA. 2013;19:141–157.
- Assmann TS, Recamonde-Mendoza M, de Souza BM, et al. MicroRNAs and diabetic kidney disease: systematic review and bioinformatic analysis. Mol Cell Endocrinol. 2018;477:90–102.
- Jeck WR, Sharpless NE. Detecting and characterizing circular RNAs. Nat Biotechnol. 2014;32:453–461.
- Rong D, Sun H, Li Z, et al. An emerging function of circRNA-miRNAs-mRNA axis in human diseases. Oncotarget. 2017;8:73271–73281.
- Jin J, Sun H, Shi C, et al. Circular RNA in renal diseases. J Cell Mol Med. 2020;24:6523–6533.
- Zhang JR, Sun HJ. Roles of circular RNAs in diabetic complications: from molecular mechanisms to therapeutic potential. Gene. 2020;763:145066.
- Li X, Yang L, Chen -L-L. The biogenesis, functions, and challenges of circular RNAs. Mol Cell. 2018;71(3):428–442.
- Ni H, Li W, Zhuge Y, et al. Inhibition of circHIPK3 prevents angiotensin II-induced cardiac fibrosis by sponging miR-29b-3p. Int J Cardiol. 2019;292:188–196.
- Hu W, Han Q, Zhao L, et al. Circular RNA circRNA_15698 aggravates the extracellular matrix of diabetic nephropathy mesangial cells via miR-185/TGF-beta1. J Cell Physiol. 2019;234:1469–1476.
- Wang Q, Cang Z, Shen L, et al. circ_0037128/miR-17-3p/AKT3 axis promotes the development of diabetic nephropathy. Gene. 2021;765:145076.
- Wang J, Zhao SM. LncRNA-antisense non-coding RNA in the INK4 locus promotes pyroptosis via miR-497/thioredoxin-interacting protein axis in diabetic nephropathy. Life Sci. 2021;264:118728.
- Muller MR, Rao A. NFAT, immunity and cancer: a transcription factor comes of age. Nat Rev Immunol. 2010;10:645–656.
- Barzegar-Fallah A, Alimoradi H, Razmi A, et al. Inhibition of calcineurin/NFAT pathway plays an essential role in renoprotective effect of tropisetron in early stage of diabetic nephropathy. Eur J Pharmacol. 2015;767:152–159.
- Zhang L, Li R, Shi W, et al. NFAT2 inhibitor ameliorates diabetic nephropathy and podocyte injury in db/db mice. Br J Pharmacol. 2013;170:426–439.
- Chi C, Loy SL, Chan SY, et al. Impact of adopting the 2013 World Health Organization criteria for diagnosis of gestational diabetes in a multi-ethnic Asian cohort: a prospective study. BMC Pregnancy Childbirth. 2018;18:69.
- An L, Ji D, Hu W, et al. Interference of Hsa_circ_0003928 alleviates high glucose-induced cell apoptosis and inflammation in HK-2 cells via miR-151-3p/Anxa2. Diabetes Metab Syndr Obes. 2020;13:3157–3168.
- Zou J, Liu KC, Wang WP, et al. Circular RNA COL1A2 promotes angiogenesis via regulating miR-29b/VEGF axis in diabetic retinopathy. Life Sci. 2020;256:117888.
- Zhao L, Chen H, Zeng Y, et al. Circular RNA circ_0000712 regulates high glucose-induced apoptosis, inflammation, oxidative stress, and fibrosis in (DN) by targeting the miR-879-5p/SOX6 axis. Endocr J. 2021;68:1155–1164.
- Zhao X, Dong W, Luo G, et al. Silencing of hsa_circ_0009035 suppresses cervical cancer progression and enhances radiosensitivity through microRNA 889-3p-dependent regulation of HOXB7. Mol Cell Biol. 2021;41:e0063120.
- Zhu Y, Ma C, Lv A, et al. Circular RNA circ_0010235 sponges miR-338-3p to play oncogenic role in proliferation, migration and invasion of non-small-cell lung cancer cells through modulating KIF2A. Ann Med. 2021;53:693–706.
- Alebiosu CO, Ayodele OE. The increasing prevalence of diabetic nephropathy as a cause of end stage renal disease in Nigeria. Trop Doct. 2006;36:218–219.
- Morii T, Fujita H, Narita T, et al. Association of monocyte chemoattractant protein-1 with renal tubular damage in diabetic nephropathy. J Diabetes Complications. 2003;17:11–15.
- Fioretto P, Mauer M. Histopathology of diabetic nephropathy. Semin Nephrol. 2007;27:195–207.
- Yu L, Border WA, Huang Y, et al. TGF-beta isoforms in renal fibrogenesis. Kidney Int. 2003;64:844–856.
- Brezniceanu ML, et al. Reactive oxygen species promote caspase-12 expression and tubular apoptosis in diabetic nephropathy. J Am Soc Nephrol. 2010;21(6):943–954.
- Okamura DM, Himmelfarb J. Tipping the redox balance of oxidative stress in fibrogenic pathways in chronic kidney disease. Pediatr Nephrol. 2009;24:2309–2319.
- Ren GL, Zhu J, Li J, et al. Noncoding RNAs in acute kidney injury. J Cell Physiol. 2019;234:2266–2276.
- Wang W, Feng J, Zhou H, et al. Circ_0123996 promotes cell proliferation and fibrosisin mouse mesangial cells through sponging miR-149-5p and inducing Bach1 expression. Gene. 2020;761:144971.
- Li G, Qin Y, Qin S, et al. Circ_WBSCR17 aggravates inflammatory responses and fibrosis by targeting miR-185-5p/SOX6 regulatory axis in high glucose-induced human kidney tubular cells. Life Sci. 2020;259:118269.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495:384–388.
- Song K, Yu P, Zhang C, et al. The LncRNA FGD5-AS1/miR-497-5p axis regulates septin 2 (SEPT2) to accelerate cancer progression and increase cisplatin-resistance in laryngeal squamous cell carcinoma. Mol Carcinog. 2021;60:469–480.
- Ren J, Zhang FJ, Wang JH, et al. LINC01315 promotes the aggressive phenotypes of papillary thyroid cancer cells by sponging miR-497-5p. Kaohsiung J Med Sci. 2021;37:459–467.
- Xu SH, Bo YH, Ma HC, et al. lncRNA LINC00473 promotes proliferation, migration, invasion and inhibition of apoptosis of non-small cell lung cancer cells by acting as a sponge of miR-497-5p. Oncol Lett. 2021;21:429.
- Jia R, Zhao XF. MicroRNA-497 functions as an inflammatory suppressor via targeting DDX3Y and modulating toll-like receptor 4/NF-kappaB in cigarette smoke extract-stimulated human bronchial epithelial cells. J Gene Med. 2020;22:e3137.
- Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, et al. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–5465.
- Halterman JA, Kwon HM, Wamhoff BR. Tonicity-independent regulation of the osmosensitive transcription factor TonEBP (NFAT5). Am J Physiol Cell Physiol. 2012;302:C1–8.
- Kim DH, Kim KS, Ramakrishna S. NFAT5 promotes in vivo development of murine melanoma metastasis. Biochem Biophys Res Commun. 2018;505:748–754.
- Kuper C. NFAT5-mediated expression of S100A4 contributes to proliferation and migration of renal carcinoma cells. Front Physiol. 2014;5:293.
- Duan YR, Chen BP, Chen F, et al. LncRNA lnc-ISG20 promotes renal fibrosis in diabetic nephropathy by inducing AKT phosphorylation through miR-486-5p/NFAT5. J Cell Mol Med. 2021;25:4922–4937.
