ABSTRACT
Osteosarcoma is one of the most common primary malignant tumors of bone in adolescents. Human umbilical vein endothelial cells (HUVECs) derived exosomes are associated with osteosarcoma cell stemness. Little is known about the function of HUVECs-exosomes in osteosarcoma cell stemness. This work aimed to investigate the mechanism of action of HUVECs-exosomes in regulating stem cell-like phenotypes of osteosarcoma cells. HUVECs were treated with GW4869 (exosome inhibitor). Human osteosarcoma cells (U2OS and 143B) were treated with HUVECs supernatant, HUVECs-exosomes with or without RO4929097 (γ secretase inhibitor, used to block Notch signaling pathway). We found that HUVECs supernatant and HUVECs-exosomes enhanced the proportions of STRO-1+CD117+ cells and the expression of stem cell-related proteins Oct4 and Sox2. Both HUVECs supernatant and HUVECs-exosomes promoted the sarcosphere formation efficiency of U2OS and 143B cells. These stem-like phenotypes of U2OS and 143B cells conferred by HUVECs-exosomes were repressed by GW4869. Moreover, HUVECs-exosomes promoted the expression of Notch1, Hes1 and Hey1 in the U2OS and 143B cells. RO4929097 treatment reversed the impact of HUVECs-exosomes on Notch1, Hes1, and Hey1 expression by inhibiting Notch1 signaling pathway. In conclusion, this work demonstrated that HUVECs-exosomes promoted cell stemness in osteosarcoma through activating Notch signaling pathway. Thus, our data reveal the mechanism of HUVECs-exosomes in regulating cell stemness of osteosarcoma, and provide a theoretical basis for osteosarcoma treatment by exosomes.
Introduction
Osteosarcoma is a kind of mesenchymal tissue tumor, and one of the common primary malignant tumors of bone in adolescents [Citation1]. Osteosarcoma has obvious heterogeneity, high invasive ability, and the tendency for early metastasis [Citation2]. Cancer stem cells (CSCs) are cells that have the potential to regenerate tumors. Osteosarcoma cells with stem cell-like characteristics are closely associated with the malignant phenotype of osteosarcoma [Citation3]. Clarifying the regulation mechanism of stem cell-like phenotype in osteosarcoma cells may be of great significance for seeking new therapeutic strategies for osteosarcoma.
CSCs rely on the microenvironment to maintain their biological characteristics [Citation4]. It also continuously affects and modifies the microenvironment. The vascular microenvironment of CSCs includes some cells located next to vein endothelial cells, such as CSCs, vein endothelial cells, immune cells, and fibroblasts. Among them, endothelial cells have a crucial role in the tumor microenvironment, and have the ability to promote tumor metastasis [Citation5]. Osteosarcoma is a malignant vascular tumor, and osteosarcoma cells can undergo hematogenous metastasis through the bloodstream. Thus, it is very important to study the interaction between vein endothelial cells and stem cell-like phenotypes in osteosarcoma cells. Previous studies have reported that the vein endothelial cell supernatant induces tumor cells to acquire a CSC-like characteristics [Citation6,Citation7]. Moreover, Lu et al. have found that endothelial cells activate neurogenic locus notch homolog protein 1 (Notch1) signaling pathway by secreting soluble form of Jagged-1, which contributes to promote the stem cell-like characteristics of colorectal cancer cells [Citation8]. In glioma, the vein endothelial cells in the tumor microenvironment promote stemness of glioma cells and accelerate the progression of glioma by activating the Hedgehog signaling pathway [Citation9]. Vein endothelial cells also induce glioma cells to differentiate into stem cell-like glioma cells through secreting bFGF [Citation10]. In liver cancer, lymphatic endothelial cells promote the self-renewal and immune escape of liver cancer stem cells through secreting IL-17A [Citation11]. Whether endothelial cells can affect osteosarcoma cells is still unknown.
Additionally, endothelial cells also affect the function of target cells in a paracrine or endocrine manner by producing exosomes [Citation12,Citation13]. Exosomes are one of the main components of the tumor microenvironment, which carries biomolecules such as proteins, RNA, lipids. Exosomes interact with target cells, participate in cell communication, and guide various physiological and pathological processes. Previously, we have found that the activated Notch signaling pathway enhances the expression of cancer stem cell surface markers (cluster of differentiation 9, CD117; stromal cell antigen 1, Stro-1) and stem cell-related genes (octamer-binding transcription factor 4, Oct4; SRY-box transcription factor 2, Sox2), and promotes stem cell-like characteristics in osteosarcoma cells [Citation14]. Whether exosomes can affect stem cell-like characteristics of osteosarcoma cells by regulating Notch signaling pathway has not been reported.
We hypothesized that human umbilical vein endothelial cell (HUVEC)-derived exosomes may regulate cell stemness of osteosarcoma by regulating Notch signaling pathway. Thus, the aim of this work was to investigate the functional role of HUVECs-exosomes in regulating cell stemness of osteosarcoma. This work confirmed that HUVECs can activate the Notch signaling pathway through exosome manner, thereby regulating the stem cell-like characteristics in osteosarcoma.
Materials and methods
Cell culture
HUVECs and human osteosarcoma cell lines (U2OS and 143B) were purchased from China Centre for Type Culture Collection (CCTCC; Wuhan, China). These cells were cultured in Dulbecco’s Modified Eagle’s medium (DMEM; Gibco, Camarillo, CA, USA) containing 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/streptomycin (Solarbio, Beijing, China). Cells were cultured in a constant temperature incubator at 37°C and 5% CO2.
HUVECs were incubated with 10 μM GW4869 (exosome inhibitor; Sigma-Aldrich, St. Louis, MO, USA) or 0.1% DMSO for 24 h. GW4869 is a cell-permeable, noncompetitive N-Smase (neutral sphingomyelinase) inhibitor. GW4869 blocks the ceramide-mediated sprouting of multivesicular bodies, thereby inhibiting the release of exosomes from multivesicular bodies. Thus, GW4869 is a commonly used as an exosome inhibitor [Citation15]. For co-culture, U2OS and 143B cells were incubated with the HUVEC supernatant, HUVEC-derived exosomes (VECs-Exo) or PBS. U2OS and 143B cells were treated with 1 mM RO4929097 (γ secretase inhibitor; Sigma-Aldrich) for 24 h. The Notch receptor is cleaved by furin convertase, ADAM and γ-secretase to produce Notch intracellular domain. The Notch intracellular domain is released to the nucleus and then activates the transcription of related genes [Citation16]. Thus, RO4929097 blocks the Notch signaling pathway by inhibiting the activity of γ secretase.
Isolation and identification of exosomes
Exosomes were isolated from HUVEC supernatant by ultracentrifugation as previous reported [Citation17]. HUVECs were seed into six-welll plates and cultured in exosome-free DMEM at 37°C and 5% CO2 for 24 h. HUVEC supernatant was collected by centrifugation, and then filtrated through a 0.22 µm filter to remove the debris. To collect the pellets, the HUVEC supernatant were centrifuged at 200,000 g, 4°C for 2 h. The collected pellets were washed with PBS for several times, and then dissolved in PBS. The ultrastructure and particle size of BMSCs-exosomes was analyzed using transmission electron microscopy (Thermo Fisher Scientific, Waltham, MA, USA) or NanoSight nanoparticle tracking analysis (NanoSight, Salisbury, UK). The expression of exosome positive proteins, cluster of differentiation 9 (CD9) and apoptosis-linked gene-2 interacting protein X (ALIX), and exosome negative protein, cis-Golgi matrix protein (GM130), was assessed to identify exosomes by performing Western blot (WB). Exosomes are positive for CD9 and ALIX, and negative for GM130.
Flow cytometry
Flow cytometry was performed to examine the proportions of STRO-1+CD117+ cells in U2OS and 143B cells as previous described [Citation18]. U2OS and 143B cells were cultured in DMEM medium, and the cell suspension (106 cells/100 μL) was incubated with 10 μL STRO-1-PE or CD117-APC (Abcam) at 4°C for 30 min in darkness. After that, the proportions of STRO-1+CD117+ cells in U2OS and 143B cells were detected using a BD FACScaliber flow cytometer (BD Biosciences, San Jose, CA, USA). The fluorescent intensity was analyzed using Cell Quest Software (BD Biosciences).
Quantitative real-time PCR (qRT-PCR)
The qRT-PCR was performed to assess gene expression following the description of previous study [Citation19]. Total RNA was extracted from cells using RNeasy Plus Mini Kit (Qiagen, Hilden, Germany). ND-1000 spectrophotometer (Thermo Fisher Scientific) was used to examine the concentration of RNA. RNA integrity was examined by 1.5% agarose gel electrophoresis. Complementary DNA was generated applying PrimeScript™ RT reagent Kit (Takara, Tokyo, Japan) as the protocol described. The relative expression of genes was estimated by qRT-PCR using TB Green® Premix Ex Taq II (Tli RNaseH Plus) (Takara). The PCR system (50 μL) contained 4 μL cDNA, 25 μL Ex Taq II, 2 μL of 10 μM forward and reverse primers, 1 μL ROX Reference Dye and 16 μL sterile water. The following thermocycling conditions were used for qPCR: Initial denaturation at 95°C for 5 min; followed by 40 cycles of 15 s at °C, 1 min at 60°C and 30 s at 72°C; and a final extension for 10 min at 72°C. The primer sequences were shown in . The qRT-PCR was carried out on a 7900 Real-Time PCR machine (Applied Biosystems, Foster City, CA, USA). The results were analyzed using 2−∆∆CT method for quantification.
Table 1. The primers used in Qrt-PCR
WB
WB was performed to assess protein expression according to the description of previous study [Citation19]. Total protein was extracted from exosomes or cells using Exosomal RNA and Protein Extraction Kit (Nwbiotec, Beijing, China) or Total Protein Extraction Kit (Solarbio). The concentration of proteins was examined using BCA Protein Assay Kit (Solarbio). Protein samples (25 μg) were separated by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis. The separated proteins were transferred onto the polyvinylidene fluoride membranes (Merck Millipore, Billerica, MA, USA). The membranes were blocked with 5% skimmed milk at room temperature for 1 h. Then, the membranes were incubated with the primary antibodies, CD9 (1:1000; Proteintech, Wuhan, China), ALIX (1:1000; Proteintech), GM130 (1:2000; Proteintech), Oct4 (1:1000; Proteintech), Sox2 (1:1000; Proteintech), Notch1 (1:1000; Proteintech), Hes family bHLH transcription factor 1 (Hes1; 1:1000; Abcam, Cambridge, MA, USA) or Hes-related family BHLH transcription factor with YRPW motif 1 (Hey1; 1:1000; Abcam) at 4°C for 12 h. After washed with Tris Buffered saline Tween, horseradish peroxidase-conjugated second antibody (1:5000; Proteintech) was incubated with the membranes. β-actin antibody (1:5000; Proteintech) was used as a reference protein for normalization. The WB bands were visualized by electrochemiluminescence detection reagent (Beyotime, Shanghai, China). The data were analyzed by Image J software.
Tumor spheroid assay
Tumor spheroid assay was carried out to detect the sarcosphere formation efficiency of U2OS and 143B cells as previous described [Citation20]. U2OS and 143B cells were seeded into an ultra-low attachment plate with 5000 cells each well. Each well contained DMEM, B27 supplement (1: 50; Invitrogen, Carlsbad, CA, USA), 20 ng/mL human basic fibroblast growth factor (bFGF; Sigma-Aldrich) and 20 ng/mL human epidermal growth factor (EGF; Invitrogen). bFGF is widely distributed in various tissues and is an important cell proliferation and differentiation regulator, which can stimulate proliferation of various cells. Similarly, EGF is an active substance in the human organism that promotes cell proliferation and differentiation. Thus, the role of bFGF and EGF is to promote proliferation of U2OS and 143B cells. U2OS and 143B cells were cultured at 37°C and 5% CO2 for 2 weeks, and 100 µL DMEM was added into each well every 2–3 days. After culture, colonies with a diameter ≥ 50 μm were regarded as sarcospheres. The spheres were quantified and photographed under an inverted-phase contrast microscopy (Leica, Wetzlar, Germany).
Statistical analysis
Each assay was performed for three times. All data reported as mean ± standard deviation. SPSS 22.0 statistical software (IBM, Armonk, NY, USA) was used for statistical analysis. Two-tailed Student’s t test and one-way ANOVA was used to analyze the statistical difference. P < 0.05 was considered as a significant difference.
Results
Osteosarcoma cells with stem cell-like characteristics are closely associated with the malignant phenotype of osteosarcoma. This work aimed to investigate whether HUVECs-exosomes can affect cell stemness of osteosarcoma and osteosarcoma progression. We hypothesized that HUVECs-exosomes may regulate cell stemness of osteosarcoma through Notch signaling pathway. We treated osteosarcoma cells with HUVECs-exosomes, and examined the impact of HUVECs-exosomes on the stem cell-like characteristics of osteosarcoma cells. The data revealed that HUVECs-exosomes activated Notch signaling pathway, which contributed to promote cell stemness in osteosarcoma.
HUVECs promoted osteosarcoma cell stemness through the secretion of exosomes
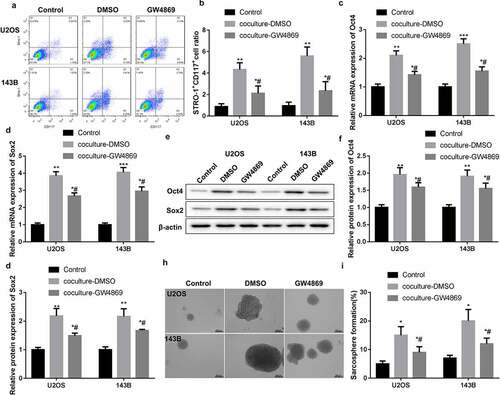
To explore whether HUVEC-exosomes can regulate osteosarcoma cell stemness, GW4869 was used to inhibit the secretion of exosomes from HUVECs. Then, flow cytometry was performed to examine the proportions of STRO-1+CD117+ cells in the U2OS and 143B cells. Compared with normal U2OS and 143B cells, coculture-DMSO and coculture-GW4869 significantly enhanced the proportions of STRO-1+CD117+ cells in U2OS and 143B cells. GW4869 treatment caused a decrease of STRO-1+CD117+ cell proportions in U2OS and 143B cells (). Moreover, the expression of stem cell-related proteins Oct4 and Sox2 in the U2OS and 143B cells was examined by qRT-PCR. The mRNA expression of Oct4 and Sox2 was notably enhanced in both coculture-DMSO and coculture-GW4869 groups. Coculture-DMSO-mediated up-regulation of Oct4 and Sox2 was reversed by coculture-GW4869 treatment (). Consistently, coculture-DMSO and coculture-GW4869 groups displayed an up-regulation of Oct4 and Sox2 proteins. The expression of Oct4 and Sox2 proteins in the U2OS and 143B cells was decreased by coculture-GW4869 (). Subsequently, we assessed the sarcosphere formation efficiency of U2OS and 143B cells by tumor spheroid assay. reveals that both coculture-DMSO and coculture-GW4869 promoted the sarcosphere formation efficiency of U2OS and 143B cells. However, coculture-GW4869 impaired the impact of coculture-DMSO on sarcosphere formation efficiency of U2OS and 143B cells (). Taken together, HUVECs promoted osteosarcoma cell stemness by secreting exosomes.
Figure 1. HUVECs promoted stem-like phenotype of U2OS and 143B cells by secreting exosomes. HUVECs were treated with GW4869 or DMSO. U2OS and 143B cells were co-cultured with HUVECs supernatant. Normal U2OS and 143B cells were served as control. (a-b) Flow cytometry was performed to assess the proportions of STRO-1+CD117+ cells in the U2OS and 143B cells. (c-g) The gene and protein expression of Oct4 and Sox2 in the U2OS and 143B cells was examined by qRT-PCR and WB. (h-i) Tumor spheroid assay was performed to estimate the sarcosphere formation efficiency of U2OS and 143B cells. *P < 0.05, **P < 0.01, ***P < 0.001 vs. Control; #P < 0.05 vs. coculture-DMSO
Identification of exosomes derived from HUVECs
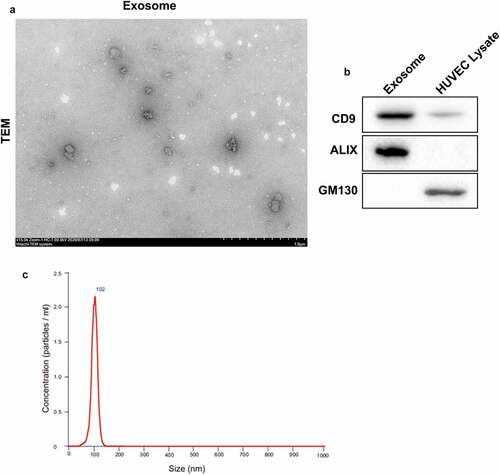
To further explore the biological role of HUVECs-exosomes in osteosarcoma cell stemness, we separated exosomes from HUVECs, and identified the ultrastructure and particle size of exosomes. showed that exosomes exhibited a typical cup-shaped morphological feature formed by a lipid bilayer membrane, and the diameter of exosomes was approximately 102 nm. Then, WB was performed to examine the expression of exosome markers. The expression of exosome-positive proteins CD9 and ALIX were increased in exosomes. The exosome-negative protein GM130 was highly expressed in HUVEC lysate, while GM130 was not expressed in exosomes (). These data showed that exosomes were successfully separated from HUVECs and can be used for further analysis.
Figure 2. Identification of exosomes. Exosomes were separated from HUVECs. (a) The ultrastructure of exosomes was observed under transmission electron microscopy. (b) The expression of CD9, ALIX and GM130 in exosomes or HUVEC lysate was assessed by WB. (c) The particle size of exosomes was analyzed by NanoSight nanoparticle tracking analysis
HUVECs-exosomes promoted osteosarcoma cell stemness by activating Notch signaling pathway
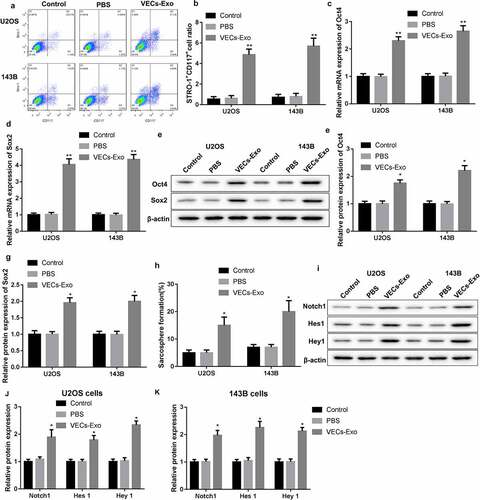
U2OS and 143B cells were treated with HUVEC-derived exosomes to explore the molecular mechanism of HUVEC-exosomes in regulating osteosarcoma cell stemness. The results obtained from flow cytomerty revealed that the proportions of STRO-1+CD117+ cells were notably enhanced in the U2OS and 143B cells in the presence of VECs-Exo (). Then, we performed qRT-PCR and WB to estimate the gene and protein expression of Oct4 and Sox2 in the U2OS and 143B cells. VECs-Exo treatment led to an increase in the mRNA and protein expression of Oct4 and Sox2 in the U2OS and 143B cells (). The results of tumor spheroid assay showed that VECs-Exo treatment significantly enhanced the sarcosphere formation efficiency of U2OS and 143B cells (). In addition, we determined the expression of Notch signaling pathway-related proteins by WB analysis. VECs-Exo-treated U2OS and 143B cells displayed an up-regulation of Notch1, Hes1, and Hey1 ().
Figure 3. HUVECs-exosomes promoted stem-like phenotype of U2OS and 143B cells through Notch signaling pathway. U2OS and 143B cells were treated with the HUVEC-derived exosomes or PBS. Normal U2OS and 143B cells served as control. (a-b) Flow cytometry was performed to assess the proportions of STRO-1+CD117+ cells in the U2OS and 143B cells. (c-g) The gene and protein expression of Oct4 and Sox2 in the U2OS and 143B cells was examined by qRT-PCR and WB. (h) Tumor spheroid assay was performed to estimate the sarcosphere formation efficiency of U2OS and 143B cells. (i-k) The expression of Notch1, Hes1 and Hey1 in the U2OS and 143B cells was examined by WB. *P < 0.05, **P < 0.01 vs. PBS
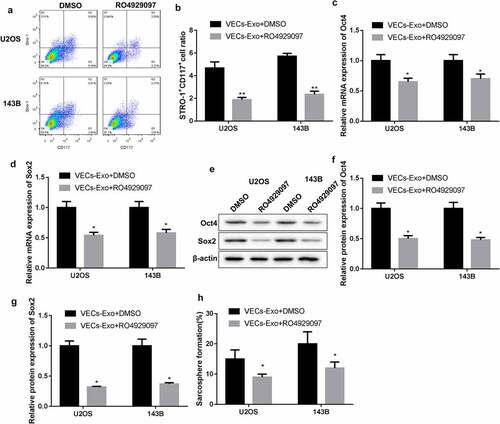
Next, U2OS and 143B cells were treated with RO4929097 to inhibit Notch signaling pathway in the presence of VECs-Exo. The results of flow cytomerty showed that RO4929097 treatment significantly reduced the proportions of STRO-1+CD117+ cells in the U2OS and 143B cells (). Compared with VECs-Exo treatment, RO4929097 treatment caused a decrease in the mRNA expression of Oct4 and Sox2 in the VECs-Exo-treated U2OS and 143B cells (). Oct4 and Sox2 proteins were also decreased in VECs-Exo-treated U2OS and 143B cells following RO4929097 treatment (). In addition, compared with VECs-Exo treatment, the sarcosphere formation efficiency of U2OS and 143B cells was notably repressed by RO4929097 treatment ().
Figure 4. RO4929097 treatment inhibited stem-like phenotype of U2OS and 143B cells. U2OS and 143B cells were treated with the HUVEC-derived exosomes combined with RO4929097. U2OS and 143B cells treated with the HUVEC-derived exosomes combined with DMSO served as control. (a-b) Flow cytometry was performed to assess the proportions of STRO-1+CD117+ cells in the U2OS and 143B cells. (c-g) The gene and protein expression of Oct4 and Sox2 in the U2OS and 143B cells was examined by qRT-PCR and WB. (h) Tumor spheroid assay was performed to estimate the sarcosphere formation efficiency of U2OS and 143B cells. *P < 0.05, **P < 0.01 vs. VECs-Exo + DMSO
These data taken together revealed that HUVECs-exosomes promoted osteosarcoma cell stemness by activating Notch signaling pathway.
Discussion
Osteosarcoma cells with stem cell-like properties have been reported to play a vital role in the occurrence, development, recurrence, and drug resistance of osteosarcoma [Citation21]. Accumulating researches have confirmed the biological role of exosomes in cancer stem cells. Cancer stem cell-derived exosomes function as a carcinogenic factor and promote the stem-like phenotype in papillary thyroid carcinoma through delivering long non-coding RNA DOCK9-AS2 [Citation22]. MiR-500a-3p is highly expressed in the plasma exosomes of gastric cancer patients. Exosomal miR-500a-3p promotes stem-like phenotype and cisplatin resistance of gastric cancer, which attributes to repress FBXW7 expression [Citation23]. For example, Li et al. have demonstrated that exosomal PSGR enhances the expression of Sox2 and Oct4a, and promotes epithelial-mesenchymal transition in low aggressive prostate cancer cells [Citation24]. Consistently, we found that HUVECs-exosomes enhanced the proportions of STRO-1+CD117+ cells and the expression of Oct4 and Sox2 in the U2OS and 143B cells. HUVECs-exosomes also enhanced the sarcosphere formation efficiency of U2OS and 143B cells. STRO-1 and CD117 are important markers for identification of stem cell-like osteosarcoma cells [Citation25]. Oct4 and Sox2 are two main transcription factors that associated with self-renewal and differentiation of stem cells. Oct4 is expressed in the undifferentiated embryonic stem cells, embryonic cells and embryonic germ cells [Citation26]. As a common stem gene, Sox2 regulates the self-renewal of different types of stem cells and tissues [Citation27]. Thus, our data suggested that HUVECs supernatant promoted stem-like phenotype of U2OS and 143B cells. However, GW4869 treatment impaired the impact of HUVECs-exosomes on the stem-like phenotype of U2OS and 143B cells. GW4869 inhibits the release of exosomes from multivesicular bodies by blocking the sprouting of multivesicular bodies [Citation15]. All these findings demonstrated that HUVECs promoted stem-like phenotype of U2OS and 143B cells by secreting exosomes.
Notch signaling pathway is highly conserved in evolution, and participates in regulating cell proliferation, survival, apoptosis, and differentiation [Citation28]. The dysfunction of Notch signaling pathway usually hinders cell differentiation and leads to malignant transformation [Citation29,Citation30]. A previous report has found that Notch signaling pathway is abnormally expressed in osteosarcoma [Citation31]. Inhibition of Notch signaling pathway accelerates the tumor growth of osteosarcoma by regulating the balance between Th1 and Th2 cells and promoting M2 polarization of tumor-associated macrophages [Citation32]. Jin et al. have found that activation of Notch signaling pathway is contribute to cell stemness of osteosarcoma, and CSC-induced recurrence in osteosarcoma by activating Notch signaling pathway [Citation33]. Consistently, we also confirmed the involvement of Notch signaling pathway in cell stemness of osteosarcoma. HUVECs-exosomes activated Notch signaling pathway and enhanced the stem-like phenotype of U2OS and 143B cells. However, RO4929097 treatment reversed the impact of HUVECs-exosomes on stem-like phenotypes by inhibiting Notch signaling pathway. Therefore, these results supported that HUVECs-exosomes promoted stem-like phenotype of osteosarcoma cells through activating Notch signaling pathway.
Stem cell-derived extracellular vesicles have a crucial role in maintaining stemness, differentiation and repairing tissue injuries [Citation34,Citation35]. Additionally, previous study has found that glioblastoma stem cell-derived exosomes promote stemness and tumorigenicity of non-glioblastoma stem cell-like glioma cells by activating Notch1 signaling pathway [Citation36]. Our data revealed that HUVECs-exosomes promoted osteosarcoma cell stemness by activating Notch signaling pathway. Mu et al. have demonstrated that murine osteosarcoma cell-derived exosomes are rich in Notch-activating factors, and activate Notch signaling pathway in muscle-derived stem cells [Citation37]. Thus, we speculated that exosomes derived from osteosarcoma cells may promote stemness of itself through activating Notch signaling pathway. We will further explore whether osteosarcoma cell-exosomes can affect cell stemness of itself in our further work.
Conclusions
In conclusion, this work demonstrated that HUVECs-exosomes promoted cell stemness in osteosarcoma through activating Notch signaling pathway. Thus, our data reveal the molecular mechanism of HUVECs-exosomes in regulating cell stemness of osteosarcoma, and provides a theoretical basis for exosomes as a treatment for osteosarcoma.
Data Availability
The data used to support the findings of this study are available from the corresponding author on reasonable request.
Authors’ contributions
JY designed the study; JY, YH, LW, XS, LY, WG performed the experiments; JY, YH analyzed the data; JY drafted the paper. All authors read and approved the paper.
Supplemental Material
Download Zip (2.6 MB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Xu N, Kang Y, Wang W, et al. The prognostic role of CD133 expression in patients with osteosarcoma. Clin Exp Med. 2020;20(2):261–267.
- Sadykova LR, Ntekim AI, Muyangwa-Semenova M, et al. Epidemiology and risk factors of osteosarcoma. Cancer Invest. 2020;38(5):259–269.
- Brown HK, Tellez-Gabriel M, Heymann D. Cancer stem cells in osteosarcoma. Cancer Lett. 2017;386:189–195.
- Das M, Law S. Role of tumor microenvironment in cancer stem cell chemoresistance and recurrence. Int J Biochem Cell Biol. 2018;103:115–124.
- Chouaib S, Kieda C, Benlalam H, et al. Endothelial cells as key determinants of the tumor microenvironment: interaction with tumor cells, extracellular matrix and immune killer cells. Crit Rev Immunol. 2010;30(6):529–545.
- Wang R, Bhattacharya R, Ye X, et al. Endothelial cells activate the cancer stem cell-associated NANOGP8 pathway in colorectal cancer cells in a paracrine fashion. Mol Oncol. 2017;11(8):1023–1034.
- Wang R, Bhattacharya R, Ye X, et al. Endothelial cells promote colorectal cancer cell survival by activating the HER3-AKT pathway in a paracrine fashion. Mol Cancer Res. 2019;17(1):20–29.
- Lu J, Ye X, Fan F, et al. Endothelial cells promote the colorectal cancer stem cell phenotype through a soluble form of Jagged-1. Cancer Cell. 2013;23(2):171–185.
- Yan GN, Yang L, Lv YF, et al. Endothelial cells promote stem-like phenotype of glioma cells through activating the hedgehog pathway. J Pathol. 2014;234(1):11–22.
- Fessler E, Borovski T, Medema JP. Endothelial cells induce cancer stem cell features in differentiated glioblastoma cells via bFGF. Mol Cancer. 2015;14(1):157.
- Wei Y, Shi D, Liang Z, et al. IL-17A secreted from lymphatic endothelial cells promotes tumorigenesis by upregulation of PD-L1 in hepatoma stem cells. J Hepatol. 2019;71(6):1206–1215.
- Xu ZH, Miao ZW, Jiang QZ, et al. Brain microvascular endothelial cell exosome-mediated S100A16 up-regulation confers small-cell lung cancer cell survival in brain. FASEB J. 2019;33(2):1742–1757.
- Zeng Y, Yao X, Liu X, et al. Anti-angiogenesis triggers exosomes release from endothelial cells to promote tumor vasculogenesis. J Extracell Vesicles. 2019;8(1):1629865.
- Yu L, Fan Z, Fang S, et al. Cisplatin selects for stem-like cells in osteosarcoma by activating Notch signaling. Oncotarget. 2016;7(22):33055–33068.
- Trajkovic K, Hsu C, Chiantia S, et al. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science (New York, NY). 2008;319(5867):1244–1247.
- Wang H, Zang C, Liu X, et al. The role of Notch receptors in transcriptional regulation. J Cell Physiol. 2015;230(5):982–988.
- Yan Z, Dutta S, Liu Z, et al. A label-free platform for identification of exosomes from different Sources. ACS Sens. 2019;4(2):488–497.
- Yu L, Fan Z, Fang S, et al. Cisplatin selects for stem-like cells in osteosarcoma by activating Notch signaling. Oncotarget. 2016;7(22):33055–33068.
- Cai J, Qiao B, Gao N, et al. Oral squamous cell carcinoma-derived exosomes promote M2 subtype macrophage polarization mediated by exosome-enclosed miR-29a-3p. Am J Physiol Cell Physiol. 2019;316(5):C731–C740.
- Sambi M, Samuel V, Qorri B, et al. A triple combination of metformin, acetylsalicylic acid, and oseltamivir phosphate impacts tumour spheroid viability and upends chemoresistance in triple-negative breast cancer. Drug Des Devel Ther. 2020;14:1995–2019.
- Lin YH, Jewell BE, Gingold J, et al. Osteosarcoma: molecular pathogenesis and iPSC modeling. Trends Mol Med. 2017;23(8):737–755.
- Dai W, Jin X, Han L, et al. Exosomal lncRNA DOCK9-AS2 derived from cancer stem cell-like cells activated Wnt/β-catenin pathway to aggravate stemness, proliferation, migration, and invasion in papillary thyroid carcinoma. Cell Death Dis. 2020;11(9):743.
- Lin H, Zhang L, Zhang C, et al. Exosomal MiR-500a-3p promotes cisplatin resistance and stemness via negatively regulating FBXW7 in gastric cancer. J Cell Mol Med. 2020;24(16):8930–8941.
- Li Y, Li Q, Li D, et al. Exosome carrying PSGR promotes stemness and epithelial-mesenchymal transition of low aggressive prostate cancer cells. Life Sci. 2020;264:118638.
- Adhikari AS, Agarwal N, Wood BM, et al. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010;70(11):4602–4612.
- Hr S, Ak H, Balling R, et al. A family of octamer-specific proteins present during mouse embryogenesis: evidence for germline-specific expression of an Oct factor. EMBO J. 1989;8(9):2543–2550.
- Driessens G, Blanpain C. Long live sox2: sox2 lasts a lifetime. Cell Stem Cell. 2011;9(4):283–284.
- Previs RA, Coleman RL, Harris AL, et al. Molecular pathways: translational and therapeutic implications of the Notch signaling pathway in cancer. Clin Cancer Res off J Am Assoc Cancer Res. 2015;21(5):955–961.
- Kranenburg O. Prometastatic NOTCH signaling in colon cancer. Cancer Discov. 2015;5(2):115–117.
- Hu Y, Su H, Li X, et al. The NOTCH ligand JAGGED2 promotes pancreatic cancer metastasis independent of NOTCH signaling activation. Mol Cancer Ther. 2015;14(1):289–297.
- Tanaka M, Setoguchi T, Hirotsu M, et al. Inhibition of Notch pathway prevents osteosarcoma growth by cell cycle regulation. Br J Cancer. 2009;100(12):1957–1965.
- Ren S, Zhang X, Hu Y, et al. Blocking the Notch signal transduction pathway promotes tumor growth in osteosarcoma by affecting polarization of TAM to M2 phenotype. Ann Transl Med. 2020;8(17):1057.
- Jin H, Luo S, Wang Y, et al. miR-135b stimulates osteosarcoma recurrence and lung metastasis via Notch and Wnt/β-Catenin signaling. Mol Ther Nucleic Acids. 2017;8:111–122.
- Nawaz M. Extracellular vesicle-mediated transport of non-coding RNAs between stem cells and cancer cells: implications in tumor progression and therapeutic resistance. Stem Cell Investig. 2017;4(10):83.
- Hur Y, Feng S, Wilson K, et al. Embryonic stem cell-derived extracellular vesicles maintain ESC Stemness by activating FAK. Dev Cell. 2021;56(3):277–291.e6.
- Sun Z, Wang L, Zhou Y, et al. Glioblastoma stem cell-derived exosomes enhance stemness and tumorigenicity of glioma cells by transferring Notch1 protein. Cell Mol Neurobiol. 2020;40(5):767–784.
- Mu X, Agarwal R, March D, et al. Notch signaling mediates skeletal muscle atrophy in cancer cachexia caused by osteosarcoma. Sarcoma. 2016;2016:3758162.
