ABSTRACT
Gastric cancer (GC) is among the most frequent malignancies originating from the digestive system worldwide, while the role and specific mechanism of integrin-subunit alpha 11 (ITGA11) in GC remain unclear. This study probes the expression characteristics and function of ITGA11 in GC. Firstly, the ITGA11 profile in GC tissues and paracancerous non-tumor tissues was assessed by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot (WB), and the association between ITGA11 and GC patients’ clinicopathological indicators was evaluated. ITGA11 knockdown models were set up in GC cell lines MKN45 and AGS. Cell proliferation was determined by the cell counting kit-8 (CCK-8) assay and colony formation assay. WB was utilized to gauge the expression of apoptosis-related proteins (Bax, Bcl2, Bad, and C-Caspase3) and the PI3K/AKT pathway. We discovered that the ITGA11 expression was boosted in GC tissues and was related to the unfavorable prognosis of GC patients. Additionally, ITGA11 knockdown abated GC cell proliferation, invasion and migration, and enhanced cell apoptosis. In animal experiments, the tumorigenesis of GC cells knocking down ITGA11 was reduced. Mechanically, knocking down ITGA11 notably inactivated the PI3K/AKT axis. The tumor-suppressive effect mediated by ITGA11 knockdown was attenuated after activating the PI3K/AKT pathway with insulin-like growth factor 1 (IGF-1). Overall, this study substantiated that the ITGA11 expression was heightened in GC tissues, which affected GC progression by modulating the PI3K/AKT pathway.
1. Introduction
Gastric cancer (GC), among the common malignancies originating from the digestive system, is the second risk factor of cancer-associated mortality and the fourth most common cancer globally [Citation1,Citation2]. With the development of clinical technology, great progress has been made in the diagnosis and staging, genome classification, surgical resection and treatment, systemic radiotherapy and chemotherapy, targeted therapy, and immunotherapy of peritoneal diseases [Citation3]. However, due to the insidious onset and highly postoperative recurrence and metastasis rate, GC treatment is still challenging [Citation4]. Therefore, it is crucial to find strategies for GC development and treatment.
Integrin acts as a bridge between the extracellular matrix (ECM) and the cytoskeleton, controlling cell proliferation, differentiation, tumor invasion, and metastasis [Citation5]. Recently, the role of integrin in tumors has attracted extensive attention. The integrin family consists of 18 distinct α subunits and eight distinct β subunits, including integrin Subunit Alpha 11 (ITGA11), also known as recombinant integrin Alpha 11. It is reported that overexpressed ITGA11 is strongly linked to breast cancer suffers’ unfavorable prognosis [Citation6]. In addition, Wu P and Ando T et al. claimed that ITGA11 is up-regulated in non-small cell lung cancer (NSCLC), and overexpressing ITGA11 facilitates tumor progression and postoperative recurrence [Citation7,Citation8]. These reports illustrate that ITGA11 is an underlying therapeutic molecule for various tumors. Nevertheless, ITGA11’s mechanism of action in GC has seldom been reported.
According to reports, the PI3K/AKT axis exerts a vital role in regulating cell proliferation and differentiation as a classical signaling pathway. Abnormal activation of PI3K/AKT substantially affects multiple tumors [Citation9], and PI3K/AKT is considered a key pathway in the development of prostate cancer [Citation10], endometrial cancer [Citation11], and laryngeal cancer [Citation12]. Additionally, PI3K/AKT contributes to GC. For example, casein kinase 2a1 (CSNK2A1) facilitates GC evolvement via PI3K/AKT/mTOR [Citation13]. Similarly, Zhao H et al. indicated that knocking down CEACAM19 represses GC cell proliferation, migration, and invasion by blocking PI3K/AKT and NF-κB [Citation14]. However, whether ITGA11 affects GC through PI3K/AKT remains to be further explored.
Previous studies suggest that ITGA11 functions as an oncogene in tumors. Therefore, we suppose that ITGA11 is also involved in GC progression. Here, we detected the ITGA11 expression in GC tissues and probed the relationship between ITGA11 and the PI3K/AKT pathway. The novel ITGA11/PI3K/AKT axis may exert a dominant role in GC evolvement and metastasis. These outcomes reveal a new molecular mechanism in GC evolvement and provide a reference value for the therapy and prognosis of GC.
2 Materials and Methods
2.1 Specimen collection and manipulations
The cancerous tissues of 49 GC patients who underwent gastrectomy in our hospital were obtained from March 2018 to March 2020. Before the operation, patients did not receive chemotherapy, radiotherapy, or other adjuvant treatment. The control samples were collected from the same patient’s adjacent non-tumor tissues (3 cm from the surgical margin at any rate), and no tumor cells were observed on postoperative pathological examination. The diagnosis of GC was confirmed following World Health Organization (WHO) criteria. All specimens were removed and preserved in liquid nitrogen at −196°C until adopted for RNA separation. Our research was granted by the research ethics committee of our hospital and received informed consent from all participating patients.
2.2 Cell culture and manipulation
GC cell lines MKN45 and AGS were ordered from the Cell Center of the Chinese Academy of Sciences (Shanghai, China). They were cultured with RPMI1640 comprising 10% fetal bovine serum (FBS) and 1% penicillin/streptomycin (Invitrogen, CA, USA) in an incubator at 37°C with 5% CO2. RPMI1640 and FBS were afforded by Thermo Fisher Scientific (MA, USA). Cells were trypsinized and sub-cultured with 0.25% trypsin (Thermo Fisher HyClone, Utah, USA) during the logarithmic growth phase.
2.3 Cell transfection and treatment
siRNA against ITGA11 (Si-ITGA11#1, Si-ITGA11#2, Si-ITGA11 #3) and the corresponding negative control (Si-NC) were afforded by Geneharma (Shanghai, China). The PI3K/AKT agonist IGF-1 was obtained from MedChemExpress (Cat.No. HY-P70788, Shanghai, China) and adopted for activating the PI3K/AKT pathway at a dose of 50 ng/mL [Citation15]. MKN45 and AGS cells were seeded on 24-well plates (2 × 104 cells/well), kept at 37°C with 5% CO2 for 24 hours, and transfected with si-ITGA11 or si-NC along with Lipofectamine® 3000 (Invitrogen; ThermoFisherScientific, Inc.). Following 24 hours, the culture medium was discarded and refreshed with a complete one. After another 24-hour culture, the transfection validity was measured by quantitative reverse transcription-polymerase chain reaction (qRT-PCR) and Western blot (WB) [Citation16].
2.4 Cell counting kit-8 (CCK-8) assay
MKN45 and AGS cells were seeded on 24-well plates (2 × 104 cells/well), incubated at 37°C with 5% CO2 for 24 hours, and transfected with si-ITGA11 or si-NC along with Lipofectamine® 3000 (Invitrogen; ThermoFisher Scientific, Inc.). Following 24 hours, the culture medium was refreshed with a complete one. After another 24-hour culture, the ITGA11 mRNA and protein level was gauged by qRT-PCR or WB for confirming stable transfection. For evaluating cell viability, MKN45 and AGS cells were inoculated into 96-well plates (1 × 103 cells/well) and maintained for 24 hours. Then, 10 μL CCK-8 reagent (Dojindo Molecular Technologies, Kumamoto, Japan) was added to each well, following the manufacturer’s guidelines. After one hour of incubation at 37°C, the optical density (OD) value at 450 nm was reviewed on a spectrophotometer (Bio-Rad, CA, USA). Each test was implemented in triplicate.
2.5 Colony formation experiment
The transfected MKN45 and AGS cells were inoculated into 6-well plates (1000 cells/well). They were then kept in the RPMI-1640 medium (contained 10% FBS and 1% penicillin/streptomycin) for 2 weeks. Afterward, the cells underwent immobilization with formaldehyde (10 min) and staining with 0.5% crystal violet (10 min). After being rinsed with PBS buffer, the stained colonies were imaged and calculated using a light microscope (Bx23, Olympus) [Citation17]. Each test was conducted three times.
2.6 Transwell assay
After dispersion with 0.25% trypsin, MKN45 and AGS cells were centrifuged, resuspended, and seeded in individual wells of 24-well plates. The 8 µM pore size Matrigel Chambers (Corning, Beijing, China) were employed for the invasion assay but not for the migration test. The upper chamber was supplemented with 5 × 104 transfected cells, and Matrigel matrix gel was added. In parallel, the medium comprising 10% FBS was put in the lower chamber, which was filled with 400 μL of RPMI-1640. Following incubation at 37°C for 24 hours, the cells that failed to migrate were cleared. Transwell membranes were secured with 4% paraformaldehyde for 10 min and then stained with 0.5% crystalline violet. After being flushed with tap water, the cells were calculated under an inverted microscope. All tests were conducted three times.
2.7 Tissues immunofluorescence
Paraffin sections underwent dewaxing and hydration, antigen repair, blocking and incubation with the primary Anti-p-PI3K antibody (ab182651) and Anti-p-AKT antibody (ab18785). Next, they were maintained with secondary antibodies, maintained with 4,6 diamidino-2-phenylindole (DAPI), nucleated and photographed on a fluorescence microscope (BX53, Olympus, China) [Citation18]. Three sections of each specimen were taken, and five non-overlapping high-powered fields of view were chosen at random for each section.
2.8 Terminal deoxynucleotidyl transferase-mediated dUTP-biotin nick end labeling assay (TUNEL) staining
MKN45 and AGS cells were centrifuged, resuspended, and seeded in individual wells of 24-well plates (each well contained 2 × 105 cells). 24 hours later, the culture medium was discarded, and the cells were fixed in 4% paraformaldehyde at room temperature at room temperature for 30 min. Next, the cells were staining using TUNEL staining solution (Beyotime, Shanghai, China) for 1 hour in 37°C. The nucleus was staining DAPI (Beyotime, Shanghai, China) as per the manufacturer’s directions.
To evaluate apoptosis in vivo, all samples were fastened in 4% paraformaldehyde overnight at 4°C, permeabilized with 0.1% Triton X-100 and then maintained with the TUNEL staining solution (Beyotime, Shanghai, China) for 1 hour as per the manufacturer’s directions [Citation19]. TUNEL-positive cells were counted in five randomly chosen areas of the slide under a microscope (Olympus).
2.9 qRT-PCR
Total cellular RNA was separated by adopting the Trizol reagent and then transcribed into cDNA with the PrimeScript™ RT Reagent kit (Invitrogen, Shanghai, China). The qPCR was implemented using the Bio-Rad CFX96 quantitative PCR system and SYBR, with initial denaturation (95°C, 5 min), denaturation (95°C, 15 s) and annealing (60°C, 30 s). β-actin was the housekeeping gene of ITGA11, with the 2 (-ΔΔCt) method for statistics. Each test was performed in triplicate. All primers were devised and synthesized by Guangzhou Ribo Biotechnology Co. Ltd. Primer sequences are shown in .
2.10 Western blot (WB)
After cells handling, the culture medium was removed, and protein lysate (Roche) was added to isolate the total protein. Following gel electrophoresis, 50 µg total protein was transferred to polyvinylidene fluoride (PVDF) membranes. The membranes were then sealed with 5% skimmed milk at room temperature (RT) for one hour, cleared with TBST three times (10 min each), and incubated with the primary antibodies (1:1000; Abcam, MA, USA) of Bcl-2 (ab32124), Bax (ab32503), p-PI3K (ab182651), PI3K (ab154598), p-AKT (ab18785), AKT (Ab131168), ITGA11 (ab198826), and β-actin (ab115777) overnight at 4°C. After being flushed with TBST, the membranes were kept at RT with horseradish peroxidase (HRP)-tagged anti-rabbit secondary antibody (1:300) for 1 hour. Subsequently, they were rinsed with TBST three times (10 min each). Finally, the Western blot reagent (Invitrogen) was adopted for color imaging, and the gray intensity was analyzed by Image J.
2.11 Xenograft tumor experiment
Sixty BALB/c-nu nude mice (4 to 6 weeks old, ordered from the Animal Center of Harbin Medical University) were selected to establish an in-vivo xenograft tumor model. MKN45 and AGS cells knocking down ITGA11 were adjusted to reach a density of 2 × 107 mL−1. Then, 0.1 mL cell suspension was administered subcutaneously into the left forelimb armpit of each mouse. Within 30 days following the administration, the mice’s livability, body weight and survival conditions were recorded. Tumor volume (V) = 0.5 × a (the longest diameter of the tumor) ×b (the shortest diameter perpendicular to the long diameter)2 [Citation20]. After continuous drug administration, mice were dislocated and killed on the 30th day. Finally, the tumor was stripped off and weighed. All animal tests were authorized by the Ethics Committee of the First Hospital of Qiqihar.
2.12 Data Process
The SPSS22.0 statistical software (SPSS Inc., Chicago, IL, USA) was adopted for data analysis. Data conforming to normal distribution were presented as mean ± standard deviation (x ± s). Two groups of data were compared by t test. The comparison between multiple groups was made by one-way ANOVA, and the LSD-t method was employed for multiple comparisons between groups. When P < 0.05, the statistics were significant.
3. Results
3.1 Expression of ITGA11 in gastric cancer tissues
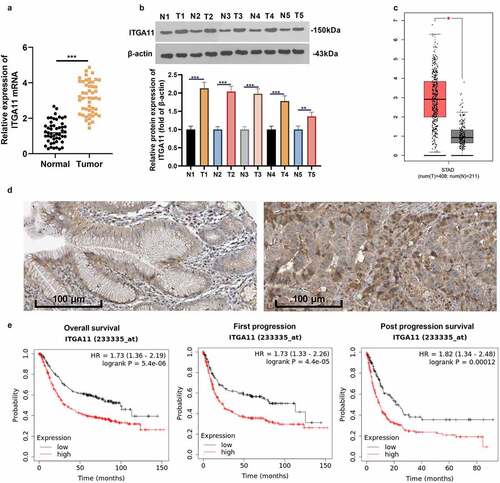
We analyzed the ITGA11 expression in 49 clinical samples to probe the expression characteristics of ITGA11 in GC tissues. As a result, ITGA11 was highly expressed in GC tissues versus non-tumor tissues (P < 0.05, )). As substantiated by WB outcomes, the ITGA11 profile was obviously uplifted in GC tissues versus normal tissues (P < 0.05, )). By querying the GEPIA database (http://gepia.cancer-pku.cn/) and The Human Protein Atlas (https://www.proteinatlas.org/), we discovered that the ITGA11 profile in GC tissues was overtly higher than that in paracancerous non-tumor tissues (). More importantly, patients with high ITGA11 expression had larger tumor volume, later clinical staging, and more distant metastasis and vascular invasion (vs. patients with lower ITGA11 expression, ). Meanwhile, the data from the Kaplan–Meier Plotter database (http://kmplot.com/analysis/) corroborated that the higher ITGA11 expression was linked to poorer overall survival, first progression and post-progression survival of GC sufferers ()). Thus, ITGA11 was a potential carcinogenic gene in GC and was an unfavorable predictor of GC patients.
Table 1. Relationship between the ITGA11 expression in GC tissue samples and clinical characteristics
Table 2. Primer sequences
Figure 1. Expression of ITGA11 in GC
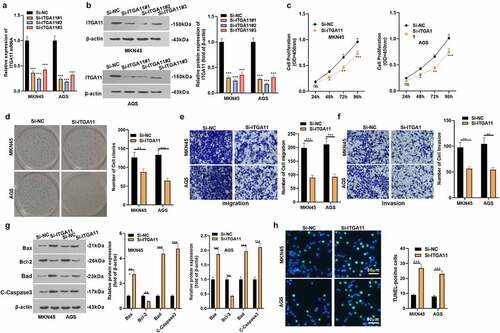
3.2 Inhibiting ITGA11 attenuated gastric cancer cell proliferation and invasion and facilitated apoptosis
To explore the effect of ITGA11 on the evolvement of GC, we constructed an ITGA11 knockdown model in MKN45 and AGS and confirmed the transfection validity by qRT-PCR and WB to figure out the influence of ITGA11 on GC progression (P < 0.05, ). The CCK-8 assay and colony formation test disclosed MKN45 and AGS cell viability declined after ITGA11 knockdown (P < 0.05, ). Transwell assay manifested that attenuating ITGA11 weakened the migrative and invasive abilities of MKN45 and AGS (P < 0.05, ). We applied WB to examine the impact of ITGA11 knockdown on cell apoptosis and discovered that ITGA11 knockdown impeded the anti-apoptotic protein Bcl-2 level but up-regulated the pro-apoptotic protein Bax, Bad and C-Caspase3 (P < 0.05, )). At the same time, TUNEL outcomes displayed that by contrast with the control group, the positive cell number of ITGA11 was distinctly augmented by knocking down ITGA11(P < 0.05, )). In conclusion, hindering ITGA11 attenuated the malignant behaviors of MKN45 and AGS.
Figure 2. Inhibiting ITGA11 attenuated GC cell proliferation and invasion and strengthened apoptosis
3.3 The regulatory effect of ITGA11 on the ITGA11/ PI3K/AKT pathway
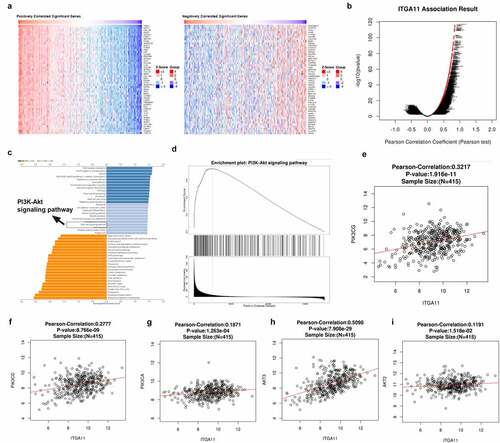
In order to investigate the downstream mechanism of ITGA11 in GC, we analyzed the co-expressed genes of ITGA11 in GC with the LinkedOmics database (http://linkedomics.org/login.php). The Top-50 positively co-expressed genes and the Top-50 negatively co-expressed genes of ITGA11 in GC were shown ()). The overall closely co-expressed genes of ITGA11 in GC were exhibited as a volcano plot ()). Through LinkedOmics database, we performed Gene Set Enrichment Analysis (GSEA) using those co-expressed genes in LUAD. We observed that the PI3K/AKT pathway was associated with ITGA11 (p < 0.05, ). Moreover, we found that ITGA11 has positive relationships with PIK3CG, PIK3CD, PIK3CA, AKT2, and AKT3 (). These data hinted that ITGA11 is significantly associated with the PI3K/AKT pathway in GC.
Figure 3. The regulatory effect of ITGA11 on the ITGA11/ PI3K/AKT pathway
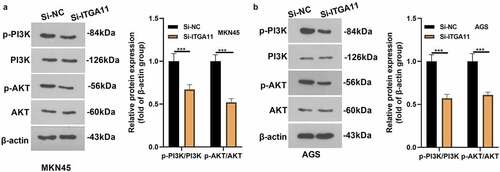
3.4 Inhibiting ITGA11 blocked the PI3K/AKT pathway
To confirm whether PI3K/AKT pathway involves in ITGA11-mediated GC progression, the PI3K/AKT level in MKN45 and AGS was tested by WB. As a result, the PI3K/AKT profile was hindered after the ITGA11 knockdown (P < 0.05, ), manifesting that inhibiting ITGA11 choked the PI3K/AKT pathway.
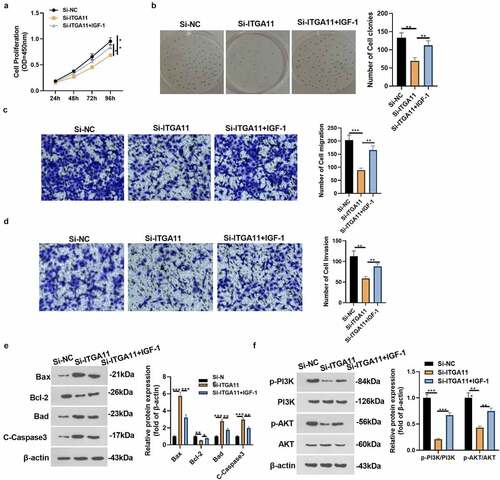
3.5 Activating PI3K abated the effect of ITGA11 knockdown in gastric cancer progression
For the aim of confirming PI3K pathway activation can reversed the anti-tumor role-mediated by si-ITGA11, we added the ITGA11 knockdown plasmid and/or PI3K/AKT agonist IGF-1 into MKN45 to explore the specific mechanism of ITGA11 in GC progression. The CCK-8 assay and colony formation test were adopted to verify cell proliferation. It was discovered that cell proliferation was reduced after knocking down ITGA11 (vs. the Si-NC group). However, IGF-1 intervention facilitated cell proliferation versus the Si-ITGA11 group (P < 0.05, ). Transwell assay manifested that MKN45 cell migration and invasion were dampened after ITGA11 knockdown (vs. the Si-NC group). Nonetheless, IGF-1 intervention strengthened cell migration and invasion versus the Si-ITGA11 group (P < 0.05, ). Moreover, WB substantiated that knocking down ITGA11 suppressed the Bcl-2 profile and up-regulated Bax, Bad and C-Caspase3. In contrast, the results were completely opposite after IGF-1 interventions (P < 0.05, )). Meanwhile, knockdown of ITGA11 caused inhibition of PI3K/AKT phosphorylation levels (vs. the Si-NC group), whereas PI3K/AKT phosphorylation levels were augmented after IGF-1 application (vs. the Si-ITGA11 group) (P < 0.05, )). These conclusions corroborated that attenuating ITGA11 exerted anti-tumor effects by restraining PI3K/AKT in GC cells.
Figure 5. Activating PI3K repressed the effect of ITGA11 knockdown on GC progression
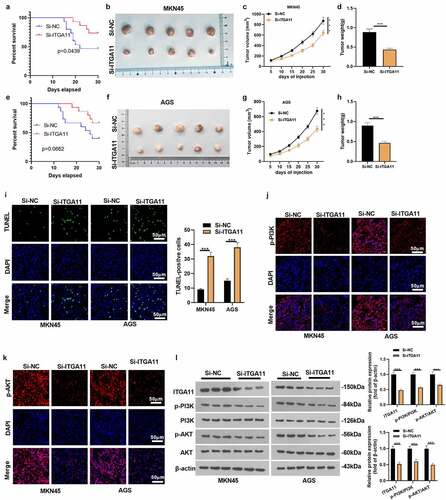
3.6 Knocking down ITGA11 repressed gastric cancer cell growth in vivo
We constructed ITGA11 knockdown tumor models in MKN45 and AGS to clarify the influence of ITGA11 on tumor growth and metastasis in vivo. An analysis of the survival rate of mice displayed that knockdown of ITGA11 improved the mice’s survival rate (), e). Knocking down ITGA11 choked tumor volume and weight (P < 0.05, ). As manifested by TUNEL data, knockdown of ITGA11 resulted in distinct facilitation in TUNEL-positive cell number versus the control group (P < 0.05, )). In parallel, tissue immunofluorescence outcomes uncovered that knockdown of ITGA11 brought about notably declined fluorescence intensity of PI3K/AKT versus the control group (P < 0.05, ). Besides, WB established that si-ITGA11 down-regulated ITGA11 and repressed PI3K/AKT pathway activation (P < 0.05, )), further confirming that abating ITGA11 repressed GC growth in vivo.
Figure 6. Knocking down ITGA11 dampened GC cell growth cells in vivo.
4. Discussion
GC is the second risk factor of tumor-associated mortality in China, second only to lung cancer. It is a multi-factorial and highly invasive disease with stages [Citation21]. However, its molecular mechanisms are not fully understood. Therefore, the exploration of GC pathogenesis and the search for treatment are helpful in improving GC prognosis. Here, we found that ITGA11 facilitated GC cell proliferation and metastasis and inhibited apoptosis. Further experiments demonstrated that inhibiting ITGA11 weakened the PI3K/AKT activation, contributing to developing new therapeutic targets for GC.
ITGA11 is a novel collagen-binding integrin, which acts as a mesenchymal collagen receptor and mediates the migration and collagen recombination of mesenchymal non-muscle cells [Citation22]. Studies have shown that ITGA11 contributes to regulating fibroblast differentiation in diabetic cardiomyopathy [Citation23], mouse embryo implantation [Citation24], atherosclerosis [Citation25], and osteoarthritis [Citation26]. More importantly, ITGA11 has been widely reported to have significant values in glioblastoma [Citation27], head and neck squamous cell carcinoma [Citation28], and breast cancer [Citation29]. However, the biological role of ITGA11 in GC remains unclear. Here, we probed the impact of ITGA11 on GC. The results revealed that human GC cell lines and tissues exhibited enhanced expression levels of ITGA11. The database showed that ITGA11 was a poor prognostic factor in GC patients. Meanwhile, ITGA11 knockdown bridled GC cell proliferation, migration and invasion, and enhanced the apoptosis. Mechanism research also manifested that ITGA11 exerted its biological function through PI3K/Akt, suggesting that ITGA11 was a positive regulator of GC, which was confirmed in vivo.
The PI3K/AKT axis is a common pathway contributing to various cancers. Multiple reports have shown that PI3K/AKT exerts crucial functions in GC. For instance, Zhao et al. found that the deletion of PDZK1 expression in GC activated PI3K/AKT, leading to poor prognosis [Citation30]. XLOC_006753 enhances the resistance of GC cells through PI3K/AKT [Citation31]. At the same time, PI3K/AKT induces GC cells’ stemness [Citation32]. Therefore, the PI3K/AKT pathway has attracted extensive attention as a potential therapeutic target. Although PI3K/AKT’s biological function in GC progression has been well-established, the function of ITGA11 in modulating PI3K/AKT remains unclear. This study manifested that ITGA11 regulated PI3K/AKT. Inhibiting ITGA11 inactivated the PI3K/AKT pathway. Further, we used IGF-1 to activate the PI3K/AKT pathway and discovered that IGF-1 elevated tumor growth and weakened the tumor-suppressive effect mediated by ITGA11 knockdown in GC. Hence, the PI3K/AKT activator can promote GC progression, which is consistent with previous reports [Citation33,Citation34].
Conclusion
Overall, ITGA11 is involved in GC progression and plays a carcinogenic role in GC by activating PI3K/AKT. Inhibiting ITGA11 reduces GC cell proliferation, migration, and invasion and facilitates apoptosis by suppressing PI3K/AKT. These results provide valuable insights for GC treatment and prognosis. However, we have not confirmed the reliability of this mechanism in vivo, which will be further explored in subsequent experiments.
List of abbreviations
| Abbreviations | = | Full names |
| CCK-8 | = | Cell counting kit-8 |
| CSNK2A1 | = | Casein kinase 2a1 |
| ECM | = | Extracellular matrix |
| FBS | = | Fetal bovine serum |
| GC | = | Gastric cancer |
| GEPIA | = | Gene Expression Profiling and Interactive Analyses |
| GSEA | = | Gene Set Enrichment Analysis |
| HRP | = | Horseradish peroxidase |
| IGF-1 | = | insulin like growth factor 1 |
| ITGA11 | = | Integrin subunit alpha 11 |
| KEGG | = | Kyoto Encyclopedia of Genes and Genomes |
| NSCLC | = | Non-small cell lung cancer |
| OD | = | Optical density |
| PI3K | = | phosphatidylinositol 3-kinase |
| PVDF | = | Polyvinylidene fluoride |
| qRT-PCR | = | quantitative reverse transcription-polymerase chain reaction |
| RT | = | Room temperature |
| WB | = | Western blot |
| WHO | = | World Health Organization |
Author’s contributions
Conceived and designed the experiments: Haijun Zhang; Performed the experiments: Haijun Zhang, Lin Zhang; Statistical analysis: Ming Lu; Wrote the paper: Haijun Zhang.
All authors read and approved the final manuscript.
Ethics statement
Our study was approved by the Ethics Committee of the First Hospital of Qiqihar.
Availability of data
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Song Z, Wu Y, Yang J, et al. Progress in the treatment of advanced gastric cancer. Tumour Biol. 2017 Jul;39(7):1010428317714626.
- Ang TL, Fock KM. Clinical epidemiology of gastric cancer. Singapore Med J. 2014 Dec;55(12):621–628.
- Johnston FM, Beckman M. Updates on Management of Gastric Cancer. Curr Oncol Rep. 2019 Jun 24;21(8):67.
- Petryszyn P, Chapelle N, Matysiak-Budnik T. Gastric Cancer: where Are We Heading? Dig Dis. 2020;38(4):280–285. Epub 2020 Feb 17.
- Bianconi D, Unseld M, Prager GW. Integrins in the Spotlight of Cancer. Int J Mol Sci. 2016 Dec 6;17(12):2037.
- Pan Y, Liu G, Yuan Y, et al. Analysis of differential gene expression profile identifies novel biomarkers for breast cancer. Oncotarget. 2017 Dec 8;8(70):114613–114625.
- Wu P, Wang Y, Wu Y, et al. Expression and prognostic analyses of ITGA11, ITGB4 and ITGB8 in human non-small cell lung cancer. PeerJ. 2019;7:e8299.
- Ando T, Kage H, Matsumoto Y, et al. Integrin α11 in non-small cell lung cancer is associated with tumor progression and postoperative recurrence. Cancer Sci. 2020Jan;1112:200–208. Epub 2019 Dec 18
- Pompura SL, Dominguez-Villar M. The PI3K/AKT signaling pathway in regulatory T-cell development, stability, and function. J Leukoc Biol. 2018 Jan 22. 10.1002/JLB.2MIR0817-349R. Epub ahead of print.
- Shorning BY, Dass MS, Smalley MJ, et al. The PI3K-AKT-mTOR Pathway and Prostate Cancer: at the Crossroads of AR, MAPK, and WNT Signaling. Int J Mol Sci. 2020 Jun 25;21(12):4507.
- Slomovitz BM, Coleman RL. The PI3K/AKT/mTOR pathway as a therapeutic target in endometrial cancer. Clin Cancer Res. 2012Nov1;18(21):5856–5864. Epub 2012 Oct 18.
- Jiang T, Zhou ML, Fan J. Inhibition of GLUT-1 expression and the PI3K/Akt pathway to enhance the chemosensitivity of laryngeal carcinoma cells in vitro. Onco Targets Ther. 2018 Nov 6;11: 7865–7872.
- Jiang C, Ma Z, Zhang G, et al. CSNK2A1 Promotes Gastric Cancer Invasion Through the PI3K-Akt-mTOR Signaling Pathway. Cancer Manag Res. 2019;11:10135–10143.
- Zhao H, Xu J, Wang Y, et al. Knockdown of CEACAM19 suppresses human gastric cancer through inhibition of PI3K/Akt and NF-κB. Surg Oncol. 2018Sep;273:495–502. Epub 2018 May 3
- Binsila BK, Selvaraju S, Ghosh SK, et al. EGF, GDNF, and IGF-1 influence the proliferation and stemness of ovine spermatogonial stem cells in vitro. J Assist Reprod Genet. 2020;37(10):2615–2630.
- Zhu Q, Li Y, Li L, et al. MicroRNA-889-3p restrains the proliferation and epithelial-mesenchymal transformation of lung cancer cells via down-regulation of Homeodomain-interacting protein kinase 1 [published online ahead of print, 2021 Nov 1]. Bioengineered 2021. 10.1080/21655979.2021.2000283.
- Zhang N, Liu JF. MicroRNA (MiR)-301a-3p regulates the proliferation of esophageal squamous cells via targeting PTEN. Bioengineered. 2020;11(1):972–983.
- Tang Y, Li Y, Xin D, et al. Icariin alleviates osteoarthritis by regulating autophagy of chondrocytes by mediating PI3K/AKT/mTOR signaling. Bioengineered. 2021;12(1):2984–2999.
- Zhao J, Zhou K, Ma L, et al. MicroRNA-145 overexpression inhibits neuroblastoma tumorigenesis in vitro and in vivo. Bioengineered. 2020;11(1):219–228.
- Ning L, Zhang M, Zhu Q, et al. miR-25-3p inhibition impairs tumorigenesis and invasion in gastric cancer cells in vitro and in vivo. Bioengineered. 2020;11(1):81–90.
- Smyth EC, Nilsson M, Grabsch HI, et al. Gastric cancer. Lancet. 2020 Aug 29;396(10251):635–648.
- Tiger CF, Fougerousse F, Grundström G, et al. alpha11beta1 integrin is a receptor for interstitial collagens involved in cell migration and collagen reorganization on mesenchymal nonmuscle cells. Dev Biol. 2001 Sep 1;237(1):116–129.
- Talior-Volodarsky I, Connelly KA, Arora PD, et al. α11 integrin stimulates myofibroblast differentiation in diabetic cardiomyopathy. Cardiovasc Res. 2012Nov1;96(2):265–275. Epub 2012 Aug 6.
- Li Z, Jia J, Gou J, et al. Mmu-miR-126a-3p plays a role in murine embryo implantation by regulating Itga11. Reprod Biomed Online. 2015Sep;313:384–393. Epub 2015 Jun 4
- Mao Z, Wu F, Shan Y. Identification of key genes and miRNAs associated with carotid atherosclerosis based on mRNA-seq data. Medicine (Baltimore). 2018 Mar;97(13):e9832.
- Guo SM, Wang JX, Li J, et al. Identification of gene expression profiles and key genes in subchondral bone of osteoarthritis using weighted gene expression network analysis. J Cell Biochem. 2018Sep;1199:7687–7695. Epub 2018 Jun 15
- Shin J, Shim HG, Hwang T, et al. Restoration of miR-29b exerts anti-cancer effects on glioblastoma. Cancer Cell Int. 2017;17:104.
- Ju JA, Godet I, Ye IC, et al. Hypoxia Selectively Enhances Integrin α5β1 Receptor Expression in Breast Cancer to Promote Metastasis. Mol Cancer Res. 2017Jun;156:723–734. Epub 2017 Feb 17
- Parajuli H, Teh MT, Abrahamsen S, et al. Integrin α11 is overexpressed by tumour stroma of head and neck squamous cell carcinoma and correlates positively with alpha smooth muscle actin expression. J Oral Pathol Med. 2017Apr;464:267–275. Epub 2016 Oct 4
- Zhao C, Tao T, Yang L, et al. Loss of PDZK1 expression activates PI3K/AKT signaling via PTEN phosphorylation in gastric cancer. Cancer Lett. 2019 Jul 1;453:107–121. Epub 2019 Mar 29.
- Zeng L, Liao Q, Zou Z, et al. Long Non-Coding RNA XLOC_006753 Promotes the Development of Multidrug Resistance in Gastric Cancer Cells Through the PI3K/AKT/mTOR Signaling Pathway. Cell Physiol Biochem. 2018;51(3):1221–1236. Epub 2018 Nov 27.
- Ni SJ, Zhao LQ, Wang XF, et al. CBX7 regulates stem cell-like properties of gastric cancer cells via p16 and AKT-NF-κB-miR-21 pathways. J Hematol Oncol. 2018 Feb 8;11(1):17.
- Wang G, Lu M, Yao Y, et al. Esculetin exerts antitumor effect on human gastric cancer cells through IGF-1/PI3K/Akt signaling pathway. Eur J Pharmacol. 2017 Nov 5;814:207–215. Epub 2017 Aug 25.
- Wang J, Zhang Y, Dou Z, et al. Knockdown of STIL suppresses the progression of gastric cancer by down-regulating the IGF-1/PI3K/AKT pathway. J Cell Mol Med. 2019Aug;238:5566–5575. Epub 2019 Jun 11