ABSTRACT
Aryl hydrocarbon receptor (AHR) plays an important role in tumor development. However, its function in cervical cancer has not been fully elucidated. We evaluated the ten genes that are predicted to associate with AHR protein interaction. The comprehensive scores were: CYP1A1, ARNT2, HSP90AA1, ARNT, AIP, PTGES3, HSP90AB1, CYP1B1, ESR1, MAF, respectively. In addition, we showed that levels of AHR and its related genes were correlated with the immune infiltration and expression of immuno-regulators (immunoinhibitors, immunostimulators, MHC molecules) levels in cervical cancer. High expression of AHR, CYP1A1, HSP90AA1, and HSP90AB1 and low expression of ESR1 were negatively correlated with the prognoses of cervical cancer patients. The Cox multivariate regression showed that high expression of AHR (HR = 1.874, 95% CI = 1.069–3.285, P= 0.028) and CYP1A1 (HR = 1.822, 95%CI = 1.077–3.080, P= 0.025) were risk factors for prognosis in patients with cervical cancer. IHC results indicated that AHR and CYP1A1 were widely expressed in cervical cancer. These findings suggest that AHR and CYP1A1 may serve as prognostic biomarkers for determining prognosis and immune infiltration in cervical cancer.
Introduction
Cervical cancer is a common malignancy among women worldwide. According to the 2020 global cancer report, there were 570,000 new cases of cervical cancer, and 311,000 deaths in 2018 [Citation1]. Therefore, it is important to study the potential mechanisms and biomarkers involved in cervical cancer. Aryl hydrocarbon receptor (AHR) is a ligand-activated transcription factor that, can be activated by a variety of chemicals. AHR was originally investigated as a receptor of toxic substances in environmental pollution [Citation2]. Later studies showed that AHR plays an important role in the occurrence and development of various types of cancer [Citation3]. AHR is overexpressed in breast cancer, skin cancer, lung cancer, and other tumor tissues [Citation4,Citation5], suggesting that AHR is a potential target gene for the treatment of malignant tumors. However, there has not been much research have investigated the role of AHR and in cervical cancer.
Disorders of the immune response system in the tumor microenvironment are involved in the occurrence and development of cervical cancer [Citation6]. AHR is closely related to innate immunity and adaptive immunity [Citation7]. Inhibitory pathways related to AHR and its metabolites may be activated in the tumor microenvironment, thus promoting immune escape and tumor progression [Citation8]. In addition, AHR is involved in various immune cell regulation processes. The presence of endogenous AHR ligands may promote the development of T cells in the tumor microenvironment. AHR signal transduction in CD4 + T cells can promote the differentiation of CD4 + and CD8 + cell into adaptive T cells [Citation9]. AHR regulates the differentiation of B cells by inhibiting the transcription of EBF1 and PAX5 [Citation10]. AHR is also a key cofactor involved in the production of IL-10 by NK cells [Citation11]. Furthermore, the study has shown that AHR is a transcription factor that determines the differentiation of monocytes of mice, and that the activation of AHR promotes the differentiation of monocytes into dendritic cells and interferes with the differentiation of monocytes into macrophages [Citation12]. The level of immune cell infiltration in the tumor microenvironment is closely related to the survival and prognosis of patients [Citation13]. Wang et al. [Citation14] evaluated the landscape of tumor infiltrating immune cells as prognostic biomarkers in cervical cancer, which suggested that the level of tumor infiltrating immune cells is a decisive factor in the prognosis of cervical cancer. However, the mechanisms underlying the role of AHR in tumor progression and tumor immunology in cervical cancer remain unclear.
In the present study, we propose the hypothesis that AHR may play an important role in the occurrence and development of cervical cancer. To investigate the immune expression and clinical significance of AHR and related genes in cervical cancer. We analyzed the expression of AHR and related genes and its correlation with immune responses, and prognosis in cervical cancer using the Gene Expression Omnibus (GEO), and the Tumor Genome Atlas (TCGA) database. We used immunohistochemistry to verify the expression of AHR in cervical cancer. The results indicated that the AHR may be a potential immunotherapy target and biomarker for cervical cancer.
Materials and methods
Data collection
The transcriptome expression data sets of cervical cancer were obtained from the GEO databases (https://www.ncbi.nlm.nih.gov/geo/): GSE7410 (45 cases), GSE 67522 (42 cases), and GSE 6791 (28 cases), and the TCGA database (https://portal.gdc.cancer.gov/): 309 cases.
LinkedOmics database analysis
Co-expression genes of AHR were identified and gene set enrichment analysis in cervical cancer was performed using the LinkedOmics database (http://linkedomics.org/login.php) [Citation15]. The threshold was determined according to the following values: P < 0.05, FDR < 0.05.
STRING database analysis
The AHR protein-protein interaction network was searched using the STRING database (https://string-db.org/) [Citation16], the ten protein coding genes that are predicted to associate with AHR were extracted for subsequent analysis.
Differential expression analysis
GEO2R (https://www.ncbi.nlm.nih.gov/geo/geo2r/) tool was used to evaluate the differential expression of AHR and related genes in tumors and normal tissues (GSE7410, GSE67522, GSE6791).
TIMER database analysis
TIMER is a comprehensive resource for the systematic analysis of immune infiltrates across in diverse cancer types (https://cistrome.shinyapps.io/timer/) [Citation17]. The TIMER database includes 10,897 samples from 32 cancer types from TCGA to estimate the abundance of immune infiltrates. We analyzed the correlation between the expression of AHR and related genes and the abundance of immune infiltrates, including B cells, CD4 + T cells, CD8 + T cells, neutrophils, macrophages, and dendritic cells, via gene modules.
TISIDB database analysis
The correlations between the expression of AHR and related genes and immuno regulators (including immunoinhibitors, immunostimulators, and MHC molecules) were calculated using TISIDB database (http://cis.hku.hk/TISIDB/) [Citation18].
Survival analysis
The correlation between the expression of AHR and related genes and survival in cervical cancers was analyzed using Kaplan-Meier plotter (http://kmplot. com/analysis/) [Citation19]. The hazard ratio (HR) with 95% confidence interval and log-rank P-value were computed. The Cox regression model was used to explore the prognostic factors of patients with cervical cancer, using the forward step method (inclusion: P< 0.05, exclusion P> 0.1), P< 0.05 was considered statistically significant.
Immunohistochemistry analysis
Cervical paraffin tissue samples were obtained from the Sixth Division Hospital of Wujiaqu City, Xinjiang Uygur Autonomous Region. For immunohistochemical analysis, cervical sections were de-paraffinized with xylene, and dehydrated in a graded ethanol series. Endogenous peroxidase activity was eliminated with 3% H2O2-methanol-PBS (0.01 M, pH 7.2) for 20 min, and sections were then washed three times (every 5 min) in phosphate-buffered saline (PBS). The sections were boiled in 0.01 mol/L trisodium citrate buffer (pH 6.0) to retrieve the antigens, then allowed to cool. The sections were incubated overnight at 4°C with the relevant primary rabbit antirat polyclonal antibody. The primary antibodies included AHR and CYP1A1 (Beijing Bioss Biotechnology Co., Ltd, diluted 1:100). The sections were washed in PBS. The second antibody (Zhongshan Jinqiao, Beijing) was incubated at 37°C for 25 min, and overlaid with DAB Kit (Zhongshan Jinqiao, Beijing). Negative controls were obtained by omitting the primary antibodies. Sections were counterstained with hematoxylin. The images were acquired at 40× magnification using an optical microscope (Leica, Leica Corporation, German). The positive expression rate of AHR was calculated by Image J software. This study was approved by the ethics committee of the First Affiliated Hospital of Xinjiang Medical University (Number: K202103-19).
Statistical analysis
R 4.0.5 was used for statistical analysis. The ‘survival’ and ‘RMS’ package was used to draw the Nomogram. The difference in expression of AHR between cervical tissue and cervical cancer tissue was compared by the χ2 test.
Results
AHR and related genes may be closely associated with cervical cancer. Therefore, we investigated the immune response of these genes in cervical cancer and their relationship with prognosis. We found that AHR and its related genes associated significantly with immune response and prognosis.
AHR and co-expressed genes are involved in biological regulation in cervical cancer
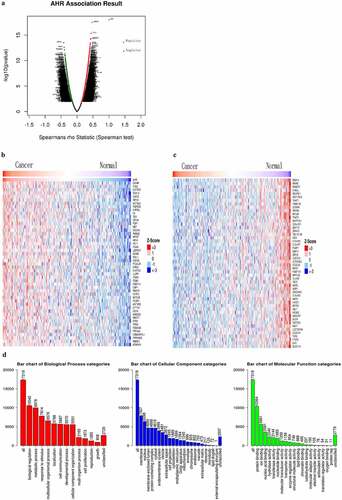
The relationship between the expression of AHR and related genes and biological regulation in cervical cancer was analyzed using the LinkedOmics database. As shown in ,209 genes were positively correlated with AHR and 2,651 genes were negatively correlated (P < 0.05, FDR < 0.05) (). The top 50 genes positively and negatively correlated with AHR are shown in a thermogram in . Gene set enrichment analysis showed that AHR-related genes were distributed in the cell membrane and nucleus (), and were involved in biological regulation, metabolic processes, stimulation response, and multicellular biological processes. The main molecular functions included binding protein, ions and nucleic acids.
Protein interaction comprehensive score of AHR with its related genes
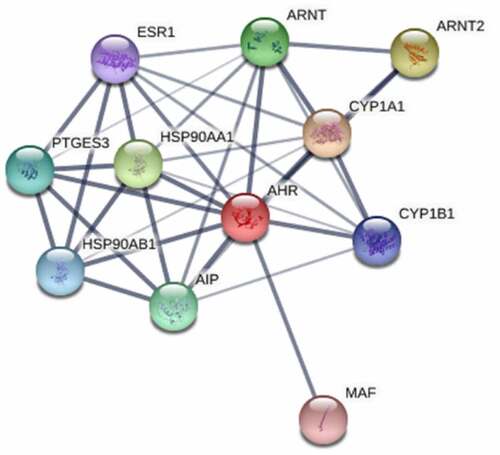
In order to further narrow the range of genes related to AHR. We extracted 10 genes that are predicted to associated with AHR from the STRING database. And we calculated the protein interaction score of AHR with its related genes, 10 genes: CYP1A1, ARNT2, HSP90AA1, AIP, PTGES3, HSP90AB1, CYP1B1, ESR1, and MAF (, ).
Table 1. The interaction scores of AHR with its related proteins
Expression of AHR and related genes in GEO datasets
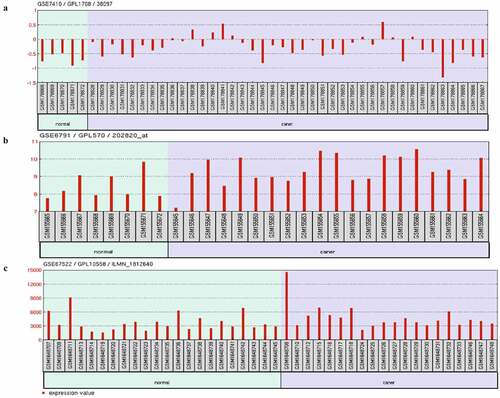
Three cervical cancer transcriptome datasets (GSE7410, GSE67522, and GSE6791) from the GEO database were selected, including 80 cases of cervical cancer and 35 cases of normal. The results showed that the expression of AHR in all data sets was statistically significantly different (P< 0.05), except AIP; the expression of nine other genes related to the AHR gene in at least one dataset was different in (P < 0.05). The mRNA expression of AHR was lower in the normal group than in the cervical cancer group ().
Table 2. The expression of AHR and related genes in GEO data sets
The expression of AHR and related genes is correlated with immune infiltration level in cervical cancers
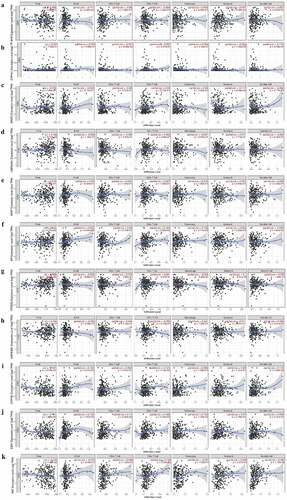
We investigated whether AHR and its related genes were correlated with immune infiltration levels in cervical cancer. According to the TIMER database (), CYP1A1 () was negatively correlated with CD8 + T cells (r= −0.201, P= 8.36e−04) and neutrophils (r= −0.253, P= 1.95e−05). HSP90AA1 () was positively associated with tumor purity (r= 0.158, P= 8.16e−03). AIP () was positively correlated with tumor purity (r = 0.164, P = 6.27e−03), B cells (r = 0.2633, P= 8.94e−06), CD4 + T cells (r = 0.184, P = 2.06e−03) and macrophages (r = 0.138, P= 0.05). PTGES3 () was positively correlated with tumor purity (r = 0.156, P = 8.99e−03). CYP1B1() was significantly associated with tumor purity (r= −0.197, P = 9.53e−04), B cells (r = 0.164, P= 3.81e−03), CD4 + T cells (r = 0.121, P = 4.43e−02), and dendritic cells (r= 0.119, P = 4.88e−02). ESR1() expression level was negatively associated with tumor purity (r= – 0.141, P= 1.88e−02), B cells (r = 0.141, P= 1.81e−02), CD4 + T cells (r = 0.185, P= 2.03e−03), and macrophages (r = 0.143, P = 1.76e−02). MAF () was significantly associated with CD4 + T cells (r= 0.129, P= 3.20e−02) and dendritic cells (r = 0.283, P= 1.75e−06), and negatively correlated with tumor purity (r = −0.146, P = 1.45e−02).
Figure 4. The expression of AHR and related genes was correlated with immune infiltration level in cervical cancer based on TIMER database. AHR(a), CYP1A1(b), ARNT2(c), HSP90AA1(d), ARNT(e), AIP(f), PTGES3(g), HSP90AB1(h), CYP1B1(i), ESR1(j)and MAF(k) with cervical cancer associated tumor sample purity, B cells, CD8 + T cells, CD4 + T cells, macrophages, neutrophils, and dendritic cells
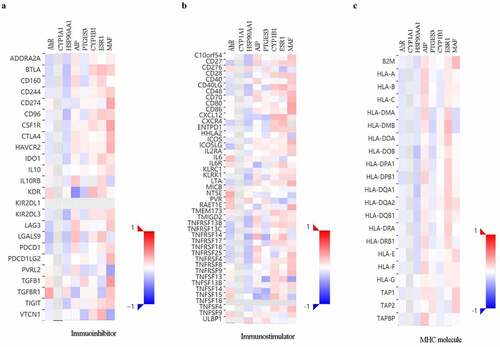
The expression of AHR-related genes is correlated with immuno regulators in cervical cancers
To further explore the effects of AHR and related genes on immune responses, the correlation between the expression of AHR and related genes and immune regulators was analyzed (). The results indicated that AHR, HSP90AA1, and PTGES3 were negatively correlated, whereas ESR1 and MAF were positively correlated with immunoinhibitors, immunostimulators, and MHC molecules.
Figure 5. Correlations between AHR and related genes and immuno regulators in cervical cancer based on TISIDB database. The correlations between the expression of AHR, CYP1A1, HSP90AA1, PTGES3, CYP1B1, ESR1, and MAF and immunoinhibitors (a), immunostimulator (b), and MHC molecules (c) were calculated based on TISIDB database
High AHR and CYP1A1 expression impacts the prognoses of cervical cancer patients
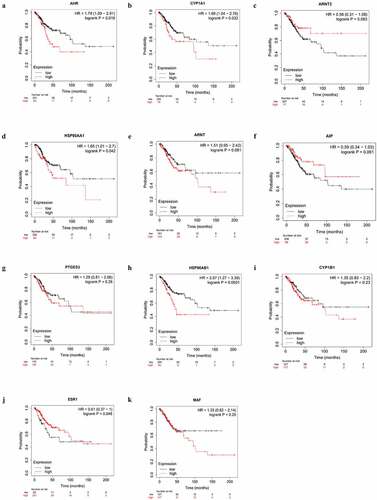
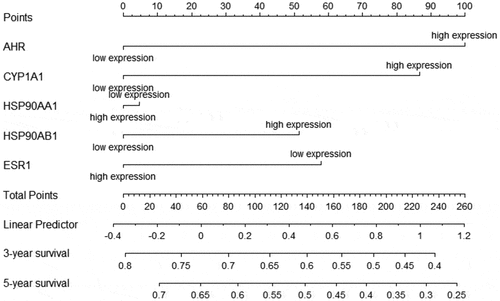
We investigated the relationship between the expression of AHR and related gene and the prognosis cervical cancer patients using the Kaplan-Meier plotter. Overexpression of AHR was associated with worse prognosis in cervical cancer patients (). Specifically, the median overall survival time was 48.43 months in the AHR high expression group and 136.2 months in the AHR low expression group (HR = 1.79, P= 0.019). The median overall survival time was 21.27 months in the CYP1A1 high expression group and 39.53 months in the low expression group (). The median overall survival time was 103.23 months in the ESR1 high expression group and 69.8 months in the ESR1 low expression group (). The median overall survival time was 31.83 months in HSP90AA1 high expression group and 37.27 months in the low expression group (), and 45.73 months in the HSP90AB1 high expression group and 136.2 months in the low expression group (). Generally, high expression of AHR, CYP1A1, HSP90AA1, and HSP90AB1 and low expression of ESR1 were not conducive to the prognosis of cervical cancer patients (). Analysis of AHR, CYP1A1, HSP90AA1, HSP90AB1 and ESR1 using the Cox multivariate regression model and the forward stepwise regression method (inclusion: P < 0.05, exclusion: P> 0.1) showed that high expression of AHR (HR = 1.874, 95% CI = 1.069–3.285, P= 0.028) and CYP1A1 (HR = 1.822, 95%CI = 1.077–3.080, P= 0.025) was a risk factor for the prognosis of patients with cervical cancer (, ).
Table 3. Multivariate Cox regression analysis
Immunohistochemistry
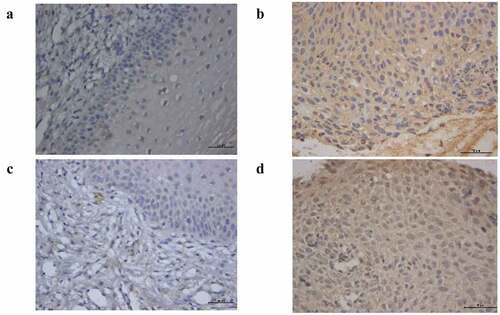
The expression of AHR and CYP1A1 were analyzed by immunohistochemistry in 30 normal cervical tissue samples and 30 cervical cancer tissue samples. As shown in , compared with the normal cervical group, AHR and CYP1A1 are widely expressed in cervical cancer tissues. We found that AHR and CYP1A1 were mainly expressed in cancer cells and immune cells, such as macrophages, and expressed in the nucleus and the cytoplasm. The positive expression of AHR and CYP1A1 differed significantly (P < 0.01) between the normal and cancer groups ().
Table 4. The positive expression of AHR and CYP1A1 in cervical tissue
Discussion
At present, cervical cancer ranks the fourth leading cause of cancer incidence and death among women in the world [Citation20]. At the time of initial diagnosis, 13% of women with cervical cancer were in advanced stage, which reduced the survival rate of patients. The 5-year disease-specific survival rates of cervical cancer patients was 12% [Citation21]. Many epidemiological evidences support that unmarried status, diseases stage, age at diagnosis, FIGO stage, metastasis, histology grade, treatment method, race, and lung, liver, brain, lymph node metastasis of cervical cancer patients were all associated with prognosis [Citation22,Citation23]. Therefore, it is still a necessary to study the potential mechanism and prognostic biomarkers of cervical cancer.
AHR, a member of the basic Helix-Loop-Helix/Per-Arnt-Sim protein family, is a ligand-activated transcription factor, that is involved in cell proliferation, differentiation, apoptosis, immune regulation and other life processes. AHR is an important regulator of environmental pollutants to modulate their toxic and carcinogenic effects [Citation15]. The roles of AHR in cancer were analyzed in several previous studies. AHR may act as a tumor suppressor in pituitary adenomas [Citation16]. AHR activation by tryptophan catabolites promotes tumor malignancy and suppresses anti-tumor immunity, thereby promoting tumor progression [Citation17]. AHR inhibits redox homeostasis and modulates the tumor promoting microenvironment in breast cancer [Citation18]. The activation of AHR and CYP1A1 and CYP1B1 related pathways promotes the proliferation and migration of gastric cancer cells. Epstein Barr virus can activate the phosphorylation of extracellular signal-regulated kinases in gastric cancer cells by encoding LMP2A. Thus, the functional pathway of AHR is regulated leading to the promotion of gastric cancer development [Citation19]. In female rats, dioxin containing AHR agonist induces, the occurrence of breast cancer and uterine tumors [Citation20], suggesting that AHR is related to the development of cervical cancer. However, the role of AHR in cervical cancer has not been fully elucidated to date.
In this study, we identified 5,860 genes associated with AHR and involved in the development of cervical cancer(P< 0.05, FDR < 0.05). Gene set enrichment analysis showed that AHR was involved in a variety of biological regulation processes related to the progression of cervical cancer. With the exception of AIP, AHR and related genes were differentially expressed between cervical cancer and normal tissues was observed in at least one GEO dataset. This indicated a potential interaction between AHR and related genes in cervical cancer. Recent studies indicate that AHR interacts with Estrogen Receptor alpha, thereby changing its conformation, which causes Estrogen Receptor alpha, transcription factors, and multiple protein complexes to stimulate the progress of cervical cancer [Citation21]. In addition, Estrogen Receptor alpha agonists can regulate the activation of AHR pathways in vitro and in vivo [Citation22]. For instance, Estrogen Receptor alpha agonists inhibit the activation of AHR in MCF-breast cancer cells [Citation23]. The AHR-ERS1 pathway promotes the development of cervical cancer. Heterodimer transcription factors comprising AHR and ARNT upregulate the expression of HSP90AA1, AHR, ARNT, SOD1, and SOD2 in correlation with the degree of oxidative stress, suggesting that AHR-ARNT are associated with cell damage [Citation24]. Beischlag [Citation25] et al. verified the protein interaction of ERα-AHR-ARNT in breast cancer cells, which mediated the estradiol-dependent repression of dioxin-induced gene transcription. Epidemiological studies suggest that activation of the AHR-CYP1A1 pathway is related to increased susceptibility in cervical cancer [Citation26]. The increasing expression of CYP1A1 and CYP1B1 is related to the AHR pathway and causes the production of electrophilic derivatives by cytochrome P450 metabolizing enzymes to form DNA adducts, resulting in the activation of oncogenes and inactivation of tumor suppressor genes [Citation27]. These studies together with the findings of the present study provide insight into the potential role of AHR and related genes in cervical cancer.
Increasing evidence supports the role of the tumor microenvironment in cancers, and tumor metastasis and prognosis depend on the level of tumor infiltrating immune cells to some extent [Citation28]. In this study, we examined the correlation between the expression of AHR and related genes and the immune response in cervical cancer using the TIMER and TISIDB databases. The results showed that the expression of CYP1A1, CYP1B1, AIP, ESR1, and MAF was significantly correlated with immune cells, immunoinhibitors, immunostimulators and MHC molecules. In addition, Kaplan-Meier plotter showed that high expression of AHR, CYP1A1, HSP90AA1, and HSP90AB1 and low expression of ESR1 were associated with a high hazard ratio for poor overall survival in cervical cancers. The Cox multivariate regression results further indicated that high expression of AHR and CYP1A1 was a risk factor for the prognosis of patients with cervical cancer. Increased activity of the AHR- IDO1-TDO2 pathway in the tumor microenvironment is related to poor prognosis in Merkel cell carcinoma [Citation29], which is consistent with the results of the present study. Moreover, AHR is a differentiation regulator of T cells. AHR combined with transcription factor c-Maf, promote the trans activation of the IL-10 and IL-21 promoters and the production of Tr1 [Citation30]. Zhang et al. [Citation31] demonstrated that AHR mediated the differentiation of infiltrating T cells in colon cancer, thereby increasing the risk of colon cancer.
In this study, we showed that the expression of CYP1A1, AIP, CYP1B1, ESR1, and MAF was significantly correlated with the level of T cell infiltration in cervical cancer. Moreover, the expression of CYP1A1 was negatively correlated with the level of T cells in cervical cancer, and high expression of CYP1A1 was not conducive to the prognosis of patients with cervical cancer. The expression of ESR1 was positively correlated with the level of T cells, and high expression of ESR1 associated with a favorable prognosis. These results indicated that CYP1A1 and ESR1 were not only potential biomarkers to predict the prognosis of cervical cancer patients, but may also serve as immune-related markers in cervical cancer. The potential of CYP1A1 and ESR1 for predicting prognosis and immune responses in other tumors was reported previously. Vasilis et al. [Citation32] demonstrated that overexpression of CYP1B1 and CYP1A1 in colon and bladder cancer adversely affected the prognosis of patients, which is consistent with the results of the present study. In addition, Zhou et al. [Citation33] suggested that gene polymorphisms of CYP1A1 improved the accuracy of prognosis in metastatic breast cancer, demonstrating the predictive potential of CYP1A1 for tumor therapy and prognosis. An increasing number of studies support the association between ESR1 and tumor immune defense [Citation34]. Increased expression of ESR1 is related to enhanced innate immunity, which inhibits the metastasis of lung cancer [Citation35]. Moreover, the activation of B cells is regulated by ESR1 signaling to some extent in different immune response periods [Citation36–45]. In this study, we found that the level of B cells infiltrated in cervical tumor was positively correlated with high expression of ESR1. This confirmed that the expression of ESR1 enhanced the immunity of the tumor microenvironment of cervical cancer, which may explain its role in the favorable prognosis of patients.
Conclusion
In conclusion, AHR and related genes were closely associated with cervical cancer. In particular, AHR, CYP1A1 and ESR1 were identified as potential prognostic biomarkers and shown to be correlated with immune responses in cervical cancer. This study provided useful information for the potential clinical diagnosis of cervical cancer, as well as potential molecular markers and immune targets in cervical cancer.
Appreciation
This work was supported by the National Natural Science Foundation of China (No.81760596) and Natural Science Foundation of Xinjiang Uygur Autonomous Region (No.2019D01C209)
Supplemental Material
Download MS Excel (331.8 KB)Disclosure statement
We declare that we do not have any commercial or associative interest that represents a conflict of interest in connection with the work submitted.
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Lu DJ, King B, Sandler HM, et al. Paid parental leave policies among U.S. news & world report 2020-2021 best hospitals and best hospitals for cancer[J]. JAMA Network Open. 2021;4(5):e218518.
- Suzuki T, Hidaka T, Kumagai Y, et al. Environmental pollutants and the immune response[J]. Nat Immunol. 2020;21(12):1486–1495.
- Paris A, Tardif N, Galibert MD, et al. AhR and cancer: from gene profiling to targeted therapy[J]. Int J Mol Sci. 2021;22(2):752.
- Takenaka MC, Gabriely G, Rothhammer V, et al. Control of tumor-associated macrophages and T cells in glioblastoma via AHR and CD39[J]. Nat Neurosci. 2019;22(5):729–740.
- Harms KL, Zhao L, Johnson B, et al. Virus-positive merkel cell carcinoma is an independent prognostic group with distinct predictive biomarkers[J]. Clin Cancer Res. 2021;27(9):2494–2504.
- Feng Q, Wei H, Morihara J, et al. Th2 type inflammation promotes the gradual progression of HPV-infected cervical cells to cervical carcinoma[J]. Gynecol Oncol. 2012;127(2):412–419.
- Trikha P, Lee DA. The role of AhR in transcriptional regulation of immune cell development and function[J]. Biochim Biophys Acta Rev Cancer. 2020;1873(1):188335.
- Murray IA, Patterson AD, Perdew GH. Aryl hydrocarbon receptor ligands in cancer: friend and foe[J]. Nat Rev Cancer. 2014;14(12):801–814.
- Funatake CJ, Marshall NB, Kerkvliet NI. 2,3,7,8-Tetrachlorodibenzo-p-dioxin alters the differentiation of alloreactive CD8+ T cells toward a regulatory T cell phenotype by a mechanism that is dependent on aryl hydrocarbon receptor in CD4+ T cells[J]. J Immunotoxicol. 2008;5(1):81–91.
- Li J, Bhattacharya S, Zhou J, et al. Aryl hydrocarbon receptor activation suppresses EBF1 and PAX5 and impairs human B lymphopoiesis[J]. J Immunol. 2017;199(10):3504–3515.
- Wagage S, John B, Krock BL, et al. The aryl hydrocarbon receptor promotes IL-10 production by NK cells[J]. J Immunol. 2014;192(4):1661–1670.
- Goudot C, Coillard A, Villani AC, et al. Aryl hydrocarbon receptor controls monocyte differentiation into dendritic cells versus macrophages[J]. Immunity. 2017;47(3):582–596.
- Mei J, Xing Y, Lv J, et al. Construction of an immune-related gene signature for prediction of prognosis in patients with cervical cancer[J]. Int Immunopharmacol. 2020;88:106882.
- Wang J, Li Z, Gao A, et al. The prognostic landscape of tumor-infiltrating immune cells in cervical cancer[J]. Biomed Pharmacother. 2019;120:109444.
- Vasaikar SV, Straub P, Wang J, et al. LinkedOmics: analyzing multi-omics data within and across 32 cancer types[J]. Nucleic Acids Res. 2018;46(D1):D956–D963.
- Szklarczyk D, Franceschini A, Wyder S, et al. STRING v10: protein-protein interaction networks, integrated over the tree of life[J]. Nucleic Acids Res. 2015 Database issue;43:D447–D452.
- Li T, Fan J, Wang B, et al. TIMER: a web server for comprehensive analysis of tumor-infiltrating immune cells[J]. Cancer Res. 2017;77(21):e108–e110.
- Ru B, Wong CN, Tong Y, et al. TISIDB: an integrated repository portal for tumor-immune system interactions[J]. Bioinformatics. 2019;35(20):4200–4202.
- Lanczky A, Nagy A, Bottai G, et al. miRpower: a web-tool to validate survival-associated miRNAs utilizing expression data from 2178 breast cancer patients[J]. Breast Cancer Res Treat. 2016;160(3):439–446.
- Lu DJ, King B, Sandler HM, et al. Paid parental leave policies among U.S. news & world report 2020-2021 best hospitals and best hospitals for cancer[J]. JAMA Network Open. 2021;4(5):e218518.
- Gardner AB, Charo LM, Mann AK, et al. Ovarian, uterine, and cervical cancer patients with distant metastases at diagnosis: most common locations and outcomes[J]. Clin Exp Metastasis. 2020;37(1):107–113.
- Zhang Y, Guo X, Wang G, et al. Real-World study of the incidence, risk factors, and prognostic factors associated with bone metastases in women with uterine cervical cancer using Surveillance, Epidemiology, and End Results (SEER) data analysis[J]. Med Sci Monit. 2018;24:6387–6397.
- Hou Y, Guo S, Lyu J, et al. Prognostic factors in Asian and white American patients with cervical cancer, considering competing risks[J]. Curr Oncol. 2019;26(3):e277–e285.
- Bekki K, Vogel H, Li W, et al. The aryl hydrocarbon receptor (AhR) mediates resistance to apoptosis induced in breast cancer cells[J]. Pestic Biochem Physiol. 2015;120:5–13.
- Formosa R, Borg J, Vassallo J. Aryl hydrocarbon receptor (AHR) is a potential tumour suppressor in pituitary adenomas[J]. Endocr Relat Cancer. 2017;24(8):445–457.
- Sadik A, Somarribas PL, Ozturk S, et al. IL4I1 Is a metabolic immune checkpoint that activates the AHR and promotes tumor progression[J]. Cell. 2020;182(5):1252–1270.
- Kubli SP, Bassi C, Roux C, et al. AhR controls redox homeostasis and shapes the tumor microenvironment in BRCA1-associated breast cancer[J]. Proc Natl Acad Sci U S A. 2019;116(9):3604–3613.
- Jiang Y, Xiao H, Sun L, et al. LMP2A suppresses the role of AHR pathway through ERK signal pathway in EBV-associated gastric cancer[J]. Virus Res. 2021;297:198399.
- Kociba RJ, Keyes DG, Beyer JE, et al. Results of a two-year chronic toxicity and oncogenicity study of 2,3,7,8-tetrachlorodibenzo-p-dioxin in rats[J]. Toxicol Appl Pharmacol. 1978;46(2):279–303.
- Mei J, Xing Y, Lv J, et al. Construction of an immune-related gene signature for prediction of prognosis in patients with cervical cancer[J]. Int Immunopharmacol. 2020;88:106882.
- Kluxen FM, Hofer N, Kretzschmar G, et al. Cadmium modulates expression of aryl hydrocarbon receptor-associated genes in rat uterus by interaction with the estrogen receptor[J]. Arch Toxicol. 2012;86(4):591–601.
- Dunlap TL, Howell CE, Mukand N, et al. Red clover Aryl Hydrocarbon Receptor (AhR) and Estrogen Receptor (ER) agonists enhance genotoxic estrogen metabolism[J]. Chem Res Toxicol. 2017;30(11):2084–2092.
- Esakky P, Hansen DA, Drury AM, et al. Cigarette smoke condensate induces aryl hydrocarbon receptor-dependent changes in gene expression in spermatocytes[J]. Reprod Toxicol. 2012;34(4):665–676.
- Beischlag TV, Perdew GH. ER alpha-AHR-ARNT protein-protein interactions mediate estradiol-dependent transrepression of dioxin-inducible gene transcription[J]. J Biol Chem. 2005;280(22):21607–21611.
- Wongpratate M, Ishida W, Phuthong S, et al. Genetic polymorphisms of the human cytochrome P450 1A1 (CYP1A1) and cervical cancer susceptibility among northeast thai women[J]. Asian Pac J Cancer Prev. 2020;21(1):243–248.
- Stejskalova L, Pavek P. The function of cytochrome P450 1A1 enzyme (CYP1A1) and aryl hydrocarbon receptor (AhR) in the placenta[J]. Curr Pharm Biotechnol. 2011;12(5):715–730.
- Liu Y, Wu L, Tong R, et al. PD-1/PD-L1 inhibitors in cervical cancer[J]. Front Pharmacol. 2019;10:65.
- Wardhani LO, Matsushita M, Iwasaki T, et al. Expression of the IDO1/TDO2-AhR pathway in tumor cells or the tumor microenvironment is associated with Merkel cell polyomavirus status and prognosis in merkel cell carcinoma[J]. Hum Pathol. 2019;84:52–61.
- Ehrlich AK, Pennington JM, Tilton S, et al. AhR activation increases IL-2 production by alloreactive CD4(+) T cells initiating the differentiation of mucosal-homing Tim3(+) Lag3(+) Tr1 cells[J]. Eur J Immunol. 2017;47(11):1989–2001.
- Zhang X, Liu X, Zhou W, et al. Blockade of IDO-kynurenine-AhR axis ameliorated colitis-associated colon cancer via inhibiting immune tolerance[J]. Cell Mol Gastroenterol Hepatol. 2021;12(4):1179–1199.
- Androutsopoulos VP, Spyrou I, Ploumidis A, et al. Expression profile of CYP1A1 and CYP1B1 enzymes in colon and bladder tumors[J]. PLoS One. 2013;8(12):e82487.
- Zhou X, Qiao G, Wang X, et al. CYP1A1 genetic polymorphism is a promising predictor to improve chemotherapy effects in patients with metastatic breast cancer treated with docetaxel plus thiotepa vs. docetaxel plus capecitabine[J]. Cancer Chemother Pharmacol. 2018;81(2):365–372.
- Chen D, Li Q, Chen H, et al. Estrogen receptor regulates immune defense by suppressing NF-kappaB signaling in the crassostrea hongkongensis[J]. Fish Shellfish Immunol. 2020;106:796–803.
- Zhao L, Huang S, Mei S, et al. Pharmacological activation of estrogen receptor beta augments innate immunity to suppress cancer metastasis[J]. Proc Natl Acad Sci U S A. 2018;115(16):E3673–E3681.
- Asaba J, Bandyopadhyay M, Kindy M, et al. Estrogen receptor signal in regulation of B cell activation during diverse immune responses[J]. Int J Biochem Cell Biol. 2015;68:42–47.