ABSTRACT
Postmenopausal osteoporosis (PMOP) is known as one of the prevalent diseases among middle-aged and elderly women. This paper revolves around the alteration of miR-211-5p in PMOP patients and its function in osteogenic differentiation. Quantitative real-time polymerase chain reaction (qRT-PCR) was implemented to check the miR-211-5p level in the plasma of PMOP patients. Knockdown and overexpression experiments were done to verify the influence of miR-211-5p on human-derived mesenchymal stem cell (hMSC) osteogenic differentiation and osteogenesis. The alkaline phosphatase (ALP) assay kit was taken to test ALP activity. Alizarin red staining monitored osteogenic differentiation, while oil red O staining examined adipogenesis. Western blot confirmed the profiles of osteoclastogenesis-concerned factors (TRAP, NFAT2, c-FOS, Runx2, OCN, CTSK), dual specific phosphatase 6 (DUSP6), ERK, SMAD, and β-catenin. Dual-luciferase reporter and RNA immunoprecipitation assays were implemented to identify the association between miR-211-5p and DUSP6. Our data displayed that miR-211-5p was down-regulated in the PMOP patients’ plasma (in contrast with the healthy controls), and it was positively correlated with Vit-D and BMD levels. miR-211-5p overexpression vigorously facilitated hMSC osteogenic differentiation, while miR-211-5p inhibition contributed to the opposite situation. miR-211-5p initiated the ERK/SMAD/β-catenin pathway and repressed DUSP6’s expression. Overexpression of DUSP6 counteracted the miR-211-5p-mediated function to a great extent and inactivated ERK/SMAD/β-catenin, whereas enhancing ERK phosphorylation weakened the DUSP6 overexpression-induced function. Consequently, this research unveiled that miR-211-5p promotes osteogenic differentiation by interfering with the DUSP6-mediated ERK/SMAD/β-catenin pathway.
1. Introduction
Postmenopausal osteoporosis (PMOP) is a metabolic disease stemming from a decline in the ovarian endocrine function following menopause, which contributes to a reduction in the estrogen level and thus makes osteoclast bone resorption bigger than osteoblast bone formation. The patients are prone to fracture due to a decrease in bone mass and an increase in osteopsathyrosis, and fracture may be followed by bone deformation, pain, other complications, and even death [Citation1]. With no specific clinical manifestations, osteoporosis (OP) will not be taken into account until a minor trauma incurs a fracture [Citation2]. Therefore, OP prevention is the top priority in clinical practice.
MicroRNAs (miRNAs), small non-coding RNAs, suppress messenger RNA (mRNA) translation or expedite mRNA degradation to modulate gene expression at the post-transcriptional level, hence regulating umpteen physiological and pathological processes like cancers, cardiovascular diseases, and metabolic diseases [Citation3–5]. What’s more, the functions of miRNAs in OP have been substantiated by several studies [Citation6]. For instance, by engineering a PMOP model in rats through bilateral oophorectomy (OVX), some researchers have discovered that miR-133a knockdown alters the serum levels of factors pertaining to osteoclast formation, augments the lumbar bone mineral density (BMD), and changes bone morphology [Citation7]. miR-19a-3p cramps HDAC4’s expression to step up human-derived mesenchymal stem cell (hMSC) osteogenic differentiation, thereby mitigating OP [Citation8]. All these findings have unraveled that different miRNAs exert positive or negative regulatory functions in OP.
Dual specific phosphatase 6 (DUSP6), a pivotal gene of OP [Citation9], is a member of the bi-specific protein phosphatase subfamily [Citation10]. So far, researches on DUSP6 have primarily concentrated on tumors and inflammatory diseases. For instance, inhibition of DUSP6 modulates ERK signal response genes to boost ovarian cancer cells’ sensitivity to chemotherapy drugs [Citation11]. (E/Z)-BCI hydrochloride (BCI), a small molecular inhibitor of DUSP6, attenuates lipopolysaccharide (LPS)-elicited inflammatory mediators and ROS generation in macrophages by initiating Nrf2 and inactivating NF-κB. These anti-inflammatory functions suggest that BCI can be adopted as a therapeutic agent for blocking inflammatory diseases [Citation12]. On the other hand, DUSP6 mediates glycolysis involving T cell receptors and dampens TFH cell differentiation through IL-21 production inhibition [Citation13]. Notwithstanding, we are still in the dark about the certain function and corresponding molecular mechanism of DUSP6 in the context of OP.
As evidenced by a multitude of studies, ERK [Citation14], SMAD [Citation15], and β-catenin [Citation16,Citation17] all participate in OP, and their activation enhances osteogenic differentiation. Of note, uncarboxylated osteocalcin, a hormone derived from osteoblasts, has been uncovered to boost the osteoblastic differentiation of mouse bone marrow-derived mesenchymal stem cells (MSCs) via ERK/SMAD/β-catenin pathway activation [Citation18]. Nevertheless, the function of the ERK/SMAD/β-catenin pathway in hMSCs remains elusive, and whether it is regulated by miR-211-5p or DUSP6 has not been disclosed.
Predicated on the above studies, we posit that miR-211-5p makes a potential role in OP progression. We found that miR-211-5p was down-regulated in the PMOP plasma compared with healthy controls, and miR-211-5p overexpression vigorously facilitated hMSC osteogenic differentiation. In addition, miR-211-5p promoted ERK/SMAD/β-catenin pathway. DUSP6, as revealed by bioinformatic analysis, was a potential target of miR-211-5p. Therefore, we guessed that miR-211-5p gets involved in OP by targeting UDSP6. In conclusion, this paper aims to reveal a novel miR-211-5p-DUSP6-ERK/SMAD/β-catenin axis in OP development, which provides a new reference for OP diagnosis and treatment.
2. Materials and Methods
2.1 Clinical specimen collection
Here, 95 postmenopausal women at the age of 58 to 68 were selected from the Second Hospital of Shanxi Medical University. Among them, 75 were patients suffering from OP, and 20 were adopted as healthy controls. OP was diagnosed if the femoral neck and/or lumbar T-score was less than or equal to −2.5, with a T-score of −1 or above considered normal. Postmenopausal women were enrolled if they had not had a period for at least 12 months. Patients with metabolic diseases, cardiovascular diseases, malignancies, or other severe diseases over the past five years were ruled out of the study. Also, subjects were excluded if they had been treated with glucocorticoids, estrogens, thyroid hormones, parathyroid hormones, fluoride, bisphosphonates, calcitonin, thiazide diuretics, barbiturates, or antiepileptic drugs [Citation19]. This study had received the imprimatur from the Medical Ethics Committee of Second Hospital of Shanxi Medical University. All participants had signed the informed consent. The morning fasting peripheral venous blood (4–8 mL) of all subjects was harvested, put into a K2-EDTA-coated tube (BD, NJ, USA), and stored at 4°C. Within 24 hours, the blood samples were centrifuged at 1500 RPM for 15 min at 4°C and then at 14,000 RPM for another 15 min at 4°C. Subsequent to centrifugation, the supernatant (plasma) was administered equally to RNase/DNase-free tubes and stored at −80°C in preparation for the following procedures.
2.2 Cell culture
hMSCs (HUXMA-01001) were supplied by Cyagen Biosciences (Suzhou, China). This cell line was characterized to be CD73-, CD90-, and CD105-positive (≥ 95%), and CD11b-, CD19-, CD45-, CD34-, and CD HLA-DR-negative (≤ 5%) [Citation20]. The cells were cultured with an oriCellTM human Mesenchymal Stem Cell growth medium (HUXMA-90011, Cyagen Biosciences) incorporating 10% fetal bovine serum (FBS), 1% glutamine, and 1% penicillin, and kept in an incubator at 37°C with 5% CO2. Next, the cells were trypsinized employing 0.25% trypsin-EDTA (Gibco; Thermo Fisher Scientific, Inc.). For inducing osteogenic differentiation, the hMSCs were seeded in 60-mm dishes, dealt with 50 mM ascorbic acid, 10 mM β-glycerophosphate, and 100 nM dexamethasone (Sigma-Aldrich, St. Louis, MO) for 20 days (the medium was refreshed every 3 days). For inducing adipogenic differentiation, the hMSCs were seeded in 60-mm dishes, and the culture medium encompassed human recombinant-insulin (SingleQuots™), L-glutamine, MCGS, dexamethasone, indomethacin, IBMX (3-isobutyl-l-methyl-xanthine), and GA-1000 (Lonza, Walkersville, MD, USA).
2.3 Cell transfection
hMSCs were transfected along with miR-211-5p mimics to get a miR-211-5p overexpression model established. On the contrary, the miR-211-5p inhibitor was transfected into hMSCs to engineer a miR-211-5p knockdown model. Meanwhile, hMSCs were transfected along with pcDNA-DUSP6 and pcDNA-vector for the construction of a DUSP6 overexpression model. The cell transfection procedure was performed according to a previous study [Citation21]. The ERK Agonist PAF-C16 (Santa Cruz Biotechnology Inc., 4 μM) was utilized to activate the ERK pathway. Under the same conditions of cell culture, the blank group received no treatment.
2.4 Quantitative real-time polymerase chain reaction (qRT-PCR)
As per miR-211-5p and DUSP6 mRNA sequences in Genebank, primers were designed with the assistance of the Primer3.0 software and synthesized by Sangon Biotechnology Co., Ltd (Shanghai). TRIzol was taken to separate total cellular RNA, with the RNA purity and concentration determined using an ultraviolet spectrophotometer. Reverse transcription was implemented to synthesize the first strand of cDNA. With the strand as a template, the mRNA fragments were amplified employing the ABI7300 fluorescent quantitative PCR instrument. GAPDH was regarded as the internal parameter of DUSP6, while U6 was adopted as that of miR-211-5p. The relative profile of the target gene was calculated using the 2−∆∆CT method [Citation22]. The primer sequences of each gene are detailed in .
Table 1. The primer sequence of each gene
2.5 Plasma 25 (OH) D concentration determination
The blood samples harvested from 75 PMOP patients and 20 healthy controls were examined. On the basis of prior studies [Citation23], the plasma concentration of 25 (OH) D [coefficient of variation (CV) was 3.2%] was determined through enzyme-linked immunosorbent assay (ELISA) (Immunodiagnostic Systems Inc., Fountain Hills, AZ). This assay was sensitive to the concentration of 25 (OH) D at 2.0 ng/mL.
2.6 BMD determination
A Hologic 4500A dual-energy X-ray absorptiometry (DXA) scanner (Hologic Inc., Bedford, MA) was adopted to check the BMD (g/cm2) level of the lumbar spine (L1-4). The instrument disclosed that the coefficient of variation (CV) of the spine was 0.9%.
2.7 Alkaline phosphatase (ALP) activity
After being inoculated into 6-well plates (2 × 106 cells/well), hMSCs steadily transfected were flushed with PBS, dealt with 0.25% trypsin (Thermo Fisher HyClone, Utah, USA) for 15–30 min, and centrifuged at 14,000 RPM for 15 min. The ALP assay kit (Merck, Shanghai, China) was utilized to determine ALP activity in the cell supernatant as instructed by the manufacturer [Citation24].
2.8 Western blot (WB)
hMSCs were dealt with different factors, and then the culture medium was discarded. The protein lysate (Roche) was administered to isolate the total protein. Afterward, 50 g of the total protein was loaded onto 12% polyacrylamide gel for 2 hours’ electrophoresis at 100 V and then electrically moved onto polyvinylidene fluoride membranes. After being sealed with 5% skimmed milk at room temperature for an hour and flushed with TBST three times (10 min each time), the membranes were incubated overnight at 4°C along with the following primary antibodies: Anti-TRAP antibody (1:5000, ab52750), Anti-NFAT2 antibody (1:1000, ab2796), Anti-c-FOS antibody (1:1000, ab222699), Anti-Runx2 antibody (1:1000, ab23981), Anti-OCN antibody (1:1000, ab93876), Anti-CTSK antibody (1:1000, ab19027), Anti-p-Erk1 antibody (1:1000, ab131438), Anti-Erk1 antibody (1:1000, ab32537), Anti-p-Smad3 antibody (1:1000, ab74062), Anti-Smad3 antibody (1:1000, ab40854), Anti-β-catenin antibody (1:5000, ab32572), Anti-DUSP6 antibody (1:1000, ab76310), and Anti-GAPDH antibody (1:2500, ab9485). TBST was taken to rinse the membranes, which were next incubated along with the horseradish peroxidase (HRP)-labeled anti-rabbit secondary antibody (1:2500, ab6721) at RT for an hour. TBST was applied to rinse the membranes three times, 10 min each. At last, WB reagent (Invitrogen) was utilized for color development, and Image J was adopted to evaluate the gray value of each protein. All antibodies employed in this study were ordered from Abcam (MA, USA) [Citation25].
2.9 Alizarin red staining
Following osteogenic differentiation induction, hMSCs were flushed in PBS 3 times and immobilized in 10% formalin for 30 min. Then, the cells were rewashed with PBS and maintained for 3 hours with alizarin red solution at RT. After being washed twice with distilled water, the cells were photographed [Citation26].
2.10 Oil red O staining
Subsequent to adipogenic differentiation induction, hMSCs (1 × 105 cells/mL) were dyed employing oil red O and flushed twice in PBS. Then, the cells were immobilized with 10% formalin at 37°C for 10 min, stained adopting filtered oil red O solution at 37°C for an hour, and monitored with a Leica Microsystem fluorescence microscope (DFC300 FX; Leica Microsystems GmbH, Wetzlar, Germany) [Citation27].
2.11 Statistical analysis
The SPSS16.0 software (SPSS Inc., Chicago, IL, USA) was introduced for statistical analysis, with the outcomes presented as mean ± SD (x ± s). The t-test was taken to compare two groups, while one-way ANOVA with Tukey post hoc test was applied for comparison among multiple groups. Pearson correlation analysis was utilized to assess the correlation among miR-211-5p, vitamin D (Vit-D), and BMD. P < 0.05 was regarded as statistically meaningful.
3. Results
Aiming at exploring the role of miR-211-5p in OP development, we first detected miR-211-5p level in the plasma samples of PMOP patients and heathy donors. miR-211-5p was downregulated in PMOP patients, we guessed that miR-211-5p has effects on osteogenic differentiation. Thus, we performed gain-and loss-of functional assays of miR-211-5p and DUSP6 on hMSCs for verifying the effects of the miR-211-5p-DUSP6 axis in osteogenic differentiation.
3.1 miR-211-5p was knocked down in the plasma of PMOP patients and positively correlated with the plasma levels of Vit-D and BMD
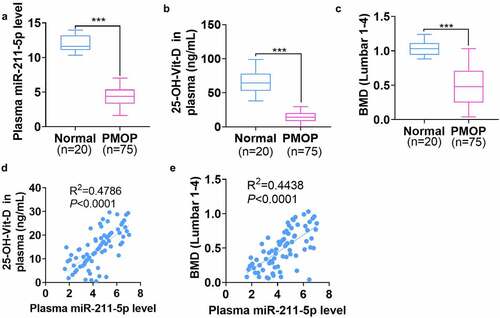
The plasma was harvested from 20 healthy controls and 75 PMOP patients. qRT-PCR exhibited that miR-211-5p was knocked down in the plasma of PMOP patients (P < 0.001, ). The enzyme immunoassay displayed that 25-OH-D concentration was notably attenuated in the plasma of PMOP patients as opposed to that of the healthy controls (P < 0.001, ). Besides, the lumbar BMD of PMOP patients was evidently lower than that of the healthy patients (P < 0.001, ). Next, Pearson analysis revealed that miR-211-5p’s level was positively associated with both the 25-OH-D level (R2 = 0.4786, P < 0.0001, ) and the lumbar BMD level (R2 = 0.4438, P < 0.0001, ) in the patients’ plasma. All these discoveries suggested that miR-211-5p, down-regulated in PMOP patients’ plasma, was positively correlated with Vit-D and BMD levels in the plasma.
Figure 1. miR-211-5p was knocked down in the plasma of PMOP patients and positively correlated with plasma Vit-D and BMD levels.
3.2 miR-211-5p overexpression enhanced hMSCs’ osteogenic differentiation and abated their adipogenic differentiation
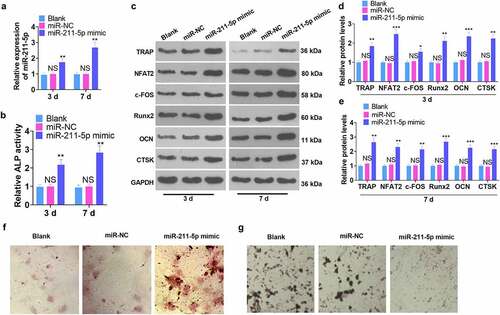
A miR-211-5p overexpression model was engineered in hMSCs. qRT-PCR demonstrated that miR-211-5p’s profile was dramatically higher than that of the miR-NC group on the 3rd and 7th day following the miR-211-5p mimic transfection (P < 0.05, ). The ALP assay illustrated that miR-211-5p overexpression obviously uplifted ALP activity (vs. the miR-NC group) (P < 0.05, ). WB confirmed protein expression in hMSCs, suggesting that osteogenic specific proteins (TRAP, NFAT2, c-FOS, Runx2, OCN, and CTSK) were all up-regulated on the 3rd and 7th days subsequent to miR-211-5p overexpression (P < 0.05, ). Alizarin red and oil red O staining further examined the function of miR-211-5p in hMSC osteogenic and adipogenic differentiation. The findings reflected that overexpression of miR-211-5p boosted mineralized matrix formation and lessened lipid droplets in hMSCs (). These discoveries signified that miR-211-5p overexpression bolstered the osteogenic differentiation and attenuated the adipogenic differentiation of hMSCs.
Figure 2. miR-211-5p overexpression enhanced hMSC osteogenic differentiation.
3.3 miR-211-5p down-regulation impeded the osteogenic differentiation and stepped up the adipogenic differentiation of hMSCs
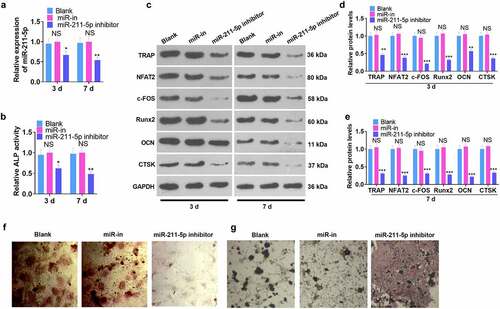
hMSCs were transfected along with the miR-211-5p inhibitor and miR-NC, and all the relevant experimental indicators were detected on the 3rd and 7th day after miR-211-5p knockdown. qRT-PCR examined the validity of the transfection. In contrast with the miR-NC group, the miR-211-5p inhibitor evidently suppressed miR-211-5p expression (P < 0.05, ). ELISA unveiled that miR-211-5p inhibition weakened ALP activity in hMSCs (P < 0.05, ). WB confirmed that miR-211-5p knockdown hampered the protein profiles of TRAP, NFAT2, c-FOS, Runx2, OCN, and CTSK (P < 0.05, ). Additionally, alizarin red and oil red O staining confirmed that miR-211-5p knockdown abated mineralized matrix formation and augmented lipid droplets in hMSCs (). In conclusion, miR-211-5p knockdown exerted an inhibitory impact on hMSC osteogenic differentiation.
Figure 3. miR-211-5p knockdown hampered hMSC osteogenic differentiation.
3.4 DUSP6 was targeted by miR-211-5p
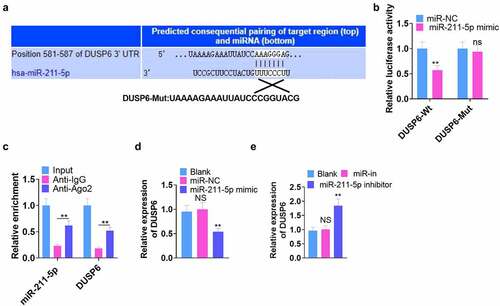
We browsed Targetscan (http://www.targetscan.org/) to further investigate the mechanism by which miR-211-5p took effect in hMSCs and discovered that DUSP6 was the binding site of miR-211-5p (). Next, dual-luciferase reporter gene assay verified the binding correlation between the two. As displayed in , miR-211-5p overexpression abated the luciferase activity of the DUSP6-Mt group (P < 0.05) but exerted little impact on that of DUSP6-Mut (). RIP analysis disclosed that the Ago2 antibody strengthened miR-211-5p and DUSP6 enrichment (vs. the IgG group) (P < 0.05, ). qRT-PCR determined DUSP6’s expression, reflecting that miR-211-5p overexpression repressed its expression, whereas miR-211-5p knockdown culminated in the opposite phenomenon (P < 0.05, ). These findings unraveled that DUSP6 was targeted by miR-211-5p.
Figure 4. DUSP6 was targeted by miR-211-5p.
3.5 Overexpression of DUSP6 attenuated the osteogenic differentiation of hMSCs
We engineered a DUSP6 overexpression model to probe the function of DUSP6 in hMSCs’ osteogenic differentiation. All the relevant experimental indicators were examined on the 7th day following cell transfection. qRT-PCR checked the cell transfection efficiency. DUSP6 was conspicuously up-regulated in the pcDNA-DUSP6 group as opposed to the pcDNA-vector group (P < 0.05, ). As exhibited in , DUSP6 overexpression dampened ALP activity (P < 0.05). WB disclosed that overexpression of DUSP6 lowered the profiles of osteogenic specific proteins (TRAP, NFAT2, c-FOS, Runx2, OCN, and CTSK) (vs. the pcDNA-vector group) (P < 0.05, ). Alizarin red and oil red O staining tracked matrix mineralization and lipid droplet formation in hMSCs, signaling that overexpression of DUSP6 frustrated matrix mineralization generation () and augmented the number of lipid droplets in hMSCs (). Thus, it could be concluded that overexpression of DUSP6 impaired hMSCs’ osteogenic differentiation.
Figure 5. DUSP6 overexpression dampened the osteogenic differentiation of hMSCs.
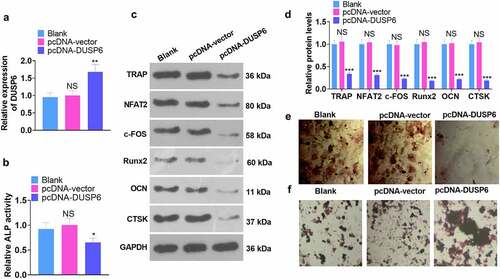
3.6 miR-211-5p overexpression attenuated DUSP6-mediated hMSC osteogenic differentiation inhibition
To confirm the interplay between miR-211-5p and DUSP6, we adopted miR-211-5p mimics to transfect hMSCs already transfected along with pcDNA-DUSP6. All the relevant experimental indicators were tested on the 7th day following the cell transfection. qRT-PCR displayed that DUSP6’s expression was restrained in the pcDNA-DUSP6+ miR-211-5p group (vs. the pcDNA-DUSP6 group) (P < 0.05, ). As exhibited in , in contrast with the pcDNA-DUSP6 group, overexpression of miR-211-5p vigorously boosted ALP activity (P < 0.05). WB unveiled that miR-211-5p overexpression based on DUSP6 overexpression heightened the profiles of TRAP, NFAT2, c-FOS, Runx2, OCN, and CTSK (P < 0.05, ). The images of alizarin red and oil red O staining signaled that as compared with the pcDNA-DUSP6 group, the PCDNA-DUSP6+ miR-211-5p group witnessed a notable rise in matrix mineralization formation () and a reduction in lipid droplets (). In brief, miR-211-5p lowered DUSP6’s expression, hence weakening the DUSP6-mediated inhibition of hMSC osteogenic differentiation.
Figure 6. miR-211-5p overexpression abated the DUSP6-mediated inhibition of hMSC osteogenic differentiation.
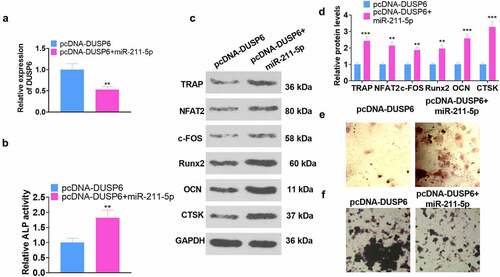
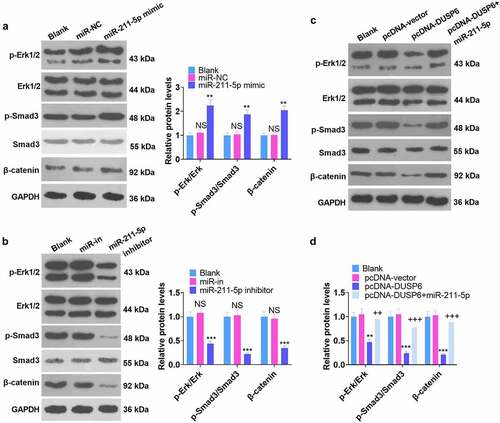
3.7 Modulation of the ERK-SMAD/β-catenin pathway by miR-211-5p and DUSP6
Corresponding experimental indicators were examined on Day 7 subsequent to cell transfection. The influence of miR-211-5p and DUSP6 on the ERK-Smad/β-catenin pathway was investigated by WB. As displayed in -B, by contrast to the miR-NC group, miR-211-5p overexpression elevated the levels of p-ERK1/ERK1, p-Smad3/Smad3, and β-catenin, whereas miR-211-5p inhibition completely inverted this situation (P < 0.05). disclosed that the levels of p-ERK1/ERK1, p-Smad/Smad, and β-catenin were hampered in the pcDNA-DUSP6 group as compared with the pcDNA-vector group, but overexpression of miR-211-5p on this basis impaired such a function of DUSP6 (P < 0.05). These phenomena demonstrated that DUSP6 curbed the ERK-SMAD/β-catenin pathway, whereas miR-211-5p boosted the pathway.
Figure 7. Regulation of the ERK-Smad/β-catenin pathway by miR-211-5p and DUSP6.
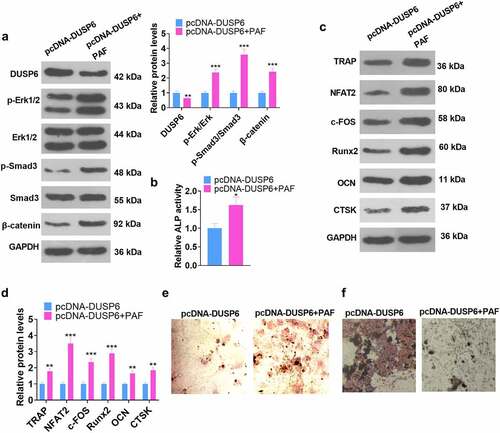
3.8 Activating the ERK pathway attenuated the DUSP6-mediated inhibition of hMSC osteogenic differentiation
To better understand the functions of ERK on DUSP6 in hMSCs, we adopted the ERK agonist PAF-C16 to treat the cells following pcDNA-DUSP6 transfection. Relevant experimental indicators were determined on Day 7 subsequent to the cell transfection. WB unraveled that by contrast to the pcDNA-DUSP6 group, the profile of DUSP6 was lowered, whereas the levels of p-ERK1/ ERK1, p-Smad/Smad, and β-catenin were uplifted (P < 0.05, ), and those of TRAP, NFAT2, c-FOS, Runx2, OCN, and CTSK were heightened in the pcDNA-DUSP6+ PAF group (P < 0.05, ). ELISA checked ALP activity. As displayed in , as compared with the pcDNA-DUSP6 group, PAF boosted ALP activity (P < 0.05). Alizarin red and oil red O staining images exhibited that ERK pathway activation by PAF culminated in increased matrix mineralization formation and lessened lipid droplets generation in hMSCs (). These findings unveiled that ERK pathway activation weakened the inhibitory impact of DUSP6 on the osteoblastic differentiation of hMSCs.
Figure 8. ERK pathway activation impeded the DUSP6-mediated inhibition of hMSC osteogenic differentiation.
4. Discussion
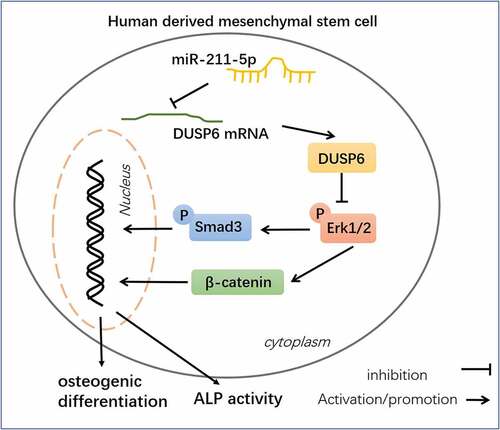
OP is characterized as the most prevalent chronic metabolic bone disease, and reduced osteogenic differentiation is a significant feature of PMOP [Citation28–30]. Over the years, several studies have verified the correlation between miRNAs and PMOP [Citation31–33]. Here, we discovered that miR-211-5p, down-regulated in the plasma of PMOP patients, targeted DUSP6 to modulate ERK-Smad/β-catenin, hence influencing hMSC osteogenic differentiation ().
Figure 9. Schematic diagram of miR-211-5p in hMSC osteogenic differentiation.
As a member of miRNAs, miR-211-5p has been confirmed with multiple biological functions [Citation34]. During the progression of tumors, miR-211-5p is usually downregulated and exerts a tumor-suppressive role by mediating migration, proliferation and apoptosis [Citation35]. MiR-211-5p mitigates inflammation, apoptosis and cell viability inhibition after cerebral ischemia-reperfusion injury [Citation36]. In terms of bone-related diseases, miR-211-5p also makes a role. For instance, miR-211-5p has a lower level in articular cartilage tissues of osteoarthritis (OA) rat model and enhances chondrocyte differentiation by downregulation Fibulin-4 of ATDC5 cells [Citation37]. In another study, lncRNA PRNCR1 up-regulates CXCR4 by hampering miR-211-5p, thereby repressing osteoblast differentiation and leading to osteolysis following hip replacement [Citation38]. Supported by those studies, we supposed that miR-211-5p can alos affect OP. We examined miR-211-5p’s expression in the plasma samples of 75 PMOP patients and 20 healthy controls. As a result, miR-211-5p was declined in the PMOP group as opposed to the control group. Vit-D is a pivotal factor in OP prevention, and menopause expedites bone loss [Citation39]. Pearson analysis outcomes, aligned with prior works, unveiled that miR-211-5p was positively associated with Vit-D and lumbar BMD levels in the plasma of PMOP patients. Highly efficient osteogenic differentiation of MSCs is critical in treating bone defects and bone diseases [Citation40]. Furthermore, we explored the effects of miR-211-5p on hMSCs differentiation. Our data disclosed that miR-211-5p overexpression expanded the osteogenic differentiation of hMSCs. Nonetheless, knockdown of miR-211-5p gave rise to the opposite effect. Hence, we posit that miR-211-5p up-regulation is conducive to enhance osteogenic differentiation.
DUSP family members, which contain a class of non-receptor-type protein-tyrosine phosphatases, have been found with potent role in mediating osteogenic differentiation [Citation41,Citation42]. For example, METTL3 significantly facilitated osteogenesis ability of BMSCs by mediating the m6A modification of DUSP14 [Citation43]. In addition to the marked effects on mediating inflammation and oxidative stress [Citation44,Citation45], DUSP6 has also gained a function on regulation cell differentiation [Citation46–48]. Interestingly, a recent study revealed that miR-181a promotes differentiation and inhibits apoptosis of osteoclasts by directly targeting DUSP6 [Citation49]. Here, TargetScan, dual-luciferase reporter assay, and RIP assay confirmed that miR-211-5p targeted DUSP6 and negatively modulated its profile in hMSCs. Here, we uncovered that overexpression of DUSP6 attenuated hMSCs’ osteogenic differentiation. Moreover, overexpression of miR-211-5p weakened the inhibitory impact of DUSP6 on osteogenic differentiation, which further substantiated the negative regulatory influence of miR-211-5p on DUSP6.
A review of preceding works has enabled us to realize that ERK [Citation50], Smad [Citation51], and β-catenin [Citation52] are crucial in osteogenic differentiation and that activating the ERK, Smad, and β-catenin pathways favors osteogenic differentiation. Here, WB demonstrated that overexpression of miR-211-5p initiated the ERK-Smad/β-catenin pathway. Besides, overexpression of miR-211-5p impaired the inhibitory impact of DUSP6 on ERK-Smad/β-catenin. Further works displayed that ERK pathway activation hindered the DUSP6-mediated inhibition of hMSC osteogenic differentiation. This is consistent with prior studies.
5. Conclusion
To summarize, this research unveils miR-211-5p can forecast PMOP, and further mechanism studies display that miR-211-5p curbs DUSP6 to initiate the ERK-Smad/β-catenin pathway, hence boosting hMSC osteogenic differentiation (). Nevertheless, other molecular mechanisms that modulate osteogenic differentiation need to be further investigated.
Abbreviations
| ALP: | = | alkaline phosphatase |
| BMD: | = | bone mineral density |
| DUSP6: | = | dual specific phosphatase 6 |
| hMSCs: | = | human-derived mesenchymal stem cells |
| LPS: | = | lipopolysaccharide; miRNA: microRNA |
| miR-in: | = | microRNA inhibitor negative control |
| MPNST: | = | malignant peripheral nerve sheath tumor |
| NC: | = | negative control |
| NS: | = | no significance |
| OA: | = | osteoarthritis |
| OP: | = | Osteoporosis |
| PMOD: | = | Postmenopausal osteoporosis |
| qRT-PCR: | = | quantitative real-time polymerase chain reaction |
| SMAD3: | = | SMAD family member 3 |
| Vit-D: | = | vitamin D |
Authors’ contribution
Conceived and designed the experiments: Zhi Lv;
Performed the experiments: Huan Wang, Xiaoyan Shi, Zhenye Guo;
Statistical analysis: Weifu He, Mingming Kang, Feng Zhao;
Wrote the paper: Huan Wang, Xiaoyan Shi.
All authors read and approved the final manuscript.
Ethical Approval and Consent for Publication
All procedures in this study were conducted in accordance with the Medical Ethics Committee of Second Hospital of Shanxi Medical University approved protocols.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
The data sets used and analyzed during the current study are available from the corresponding author on reasonable request.
Additional information
Funding
References
- Li J, Chen X, Lu L, et al. The relationship between bone marrow adipose tissue and bone metabolism in postmenopausal osteoporosis. Cytokine Growth Factor Rev. 2020Apr; 52: 88–98 Epub 2020 Feb 10. PMID: 32081538
- Sözen T, Özışık L, Başaran NÇ. An overview and management of osteoporosis. Eur J Rheumatol. 2017Mar;4(1):46–56. Epub 2016 Dec 30. PMID: 28293453; PMCID: PMC5335887.
- Correia de Sousa M, Gjorgjieva M, Dolicka D, et al. Deciphering miRNAs’ action through miRNA editing. Int J Mol Sci. 2019Dec11;20(24):6249. PMID: 31835747; PMCID: PMC6941098.
- Ji Y, Ji J, Yin H, et al. Exosomes derived from microRNA-129-5p-modified tumor cells selectively enhanced suppressive effect in malignant behaviors of homologous colon cancer cells. Bioengineered. 2021Nov15;12(2):12148–12156. Epub ahead of print. PMID: 34775889.
- Lou X, Wang D, Gu Z, et al. Mechanism of microRNA regulating the progress of atherosclerosis in apoE-deficient mice. Bioengineered. 2021Dec;12(2):10994–11006. PMID: 34775883.
- Mei L, Li M, Zhang T. MicroRNA miR-874-3p inhibits osteoporosis by targeting leptin (LEP). Bioengineered. 2021 Nov 24. Epub ahead of print. PMID: 34818977. 10.1080/21655979.2021.2009618
- Li Z, Zhang W, Huang Y. MiRNA-133a is involved in the regulation of postmenopausal osteoporosis through promoting osteoclast differentiation. Acta Biochim Biophys Sin (Shanghai). 2018Mar1;50(3):273–280. PMID: 29425279.
- Chen R, Qiu H, Tong Y, et al. MiRNA-19a-3p alleviates the progression of osteoporosis by targeting HDAC4 to promote the osteogenic differentiation of hMSCs. Biochem Biophys Res Commun. 2019Aug27;516(3):666–672. Epub 2019 Jun 24. PMID: 31248594.
- Xu F, Gao F. Liuwei Dihuang pill cures postmenopausal osteoporosis with kidney-Yin deficiency: potential therapeutic targets identified based on gene expression profiling. Medicine (Baltimore). 2018Aug;97(31):e11659. PMID: 30075554; PMCID: PMC6081159.
- Higa T, Takahashi H, Higa-Nakamine S, et al. Up-regulation of DUSP5 and DUSP6 by gonadotropin-releasing hormone in cultured hypothalamic neurons, GT1-7 cells. Biomed Res. 2018;39(3):149–158. PMID: 29899190.
- James NE, Beffa L, Oliver MT, et al.Inhibition of DUSP6 sensitizes ovarian cancer cells to chemotherapeutic agents via regulation of ERK signaling response genes.Oncotarget.2019May21;10:3315–3327. PMID: 31164954; PMCID: PMC653436136
- Zhang F, Tang B, Zhang Z, et al. DUSP6 Inhibitor (E/Z)-BCI hydrochloride attenuates lipopolysaccharide-induced inflammatory responses in murine macrophage cells via activating the Nrf2 signaling axis and inhibiting the NF-κB Pathway. Inflammation. 2019Apr;42(2):672–681. PMID: 30506106.
- Hsu WC, Chen MY, Hsu SC, et al. DUSP6 mediates T cell receptor-engaged glycolysis and restrains TFH cell differentiation. Proc Natl Acad Sci U S A. 2018Aug21;115(34):E8027–E8036. Epub 2018 Aug 7. PMID: 30087184; PMCID: PMC6112722.
- Yin Z, Zhu W, Wu Q, et al. Glycyrrhizic acid suppresses osteoclast differentiation and postmenopausal osteoporosis by modulating the NF-κB, ERK, and JNK signaling pathways. Eur J Pharmacol. 2019Sep15;859:172550. Epub 2019 Jul 16. PMID: 31323222.
- Wang Y, Han X, Zang T, et al. miR-29b enhances the proliferation and migration of bone marrow mesenchymal stem cells in rats with castration-induced osteoporosis through the PI3K/AKT and TGF-β/Smad signaling pathways. Exp Ther Med. 2020Oct;20(4):3185–3195. Epub 2020 Jul 24. PMID: 32855687; PMCID: PMC7444379.
- Zhou P, Li Y, Di R, et al. H19 and Foxc2 synergistically promotes osteogenic differentiation of BMSCs via Wnt-β-catenin pathway. J Cell Physiol. 2019Aug;234(8):13799–13806. Epub 2019 Jan 11. PMID: 30633332.
- Peng Z, Lu S, Lou Z, et al. Exosomes from bone marrow mesenchymal stem cells promoted osteogenic differentiation by delivering miR-196a that targeted Dickkopf-1 to activate Wnt/β-catenin pathway. Bioengineered. 2021 Nov 1. Epub ahead of print. PMID: 34720039. 10.1080/21655979.2021.1996015
- Liu Z, Yang J. Uncarboxylated osteocalcin promotes osteogenic differentiation of mouse bone marrow-derived mesenchymal stem cells by activating the ERK-Smad/β-catenin signalling pathways. Cell Biochem Funct. 2020Jan;38(1):87–96. Epub 2019 Oct 31. PMID: 31674048.
- Zhu J, Wang H, Liu H. Osteoclastic miR-301-b knockout reduces ovariectomy (OVX)-induced bone loss by regulating CYDR/NF-κB signaling pathway. Biochem Biophys Res Commun. 2020Aug13;529(1):35–42. Epub 2020 Jun 5. PMID: 32560816.
- Wang T, Zhang C, Wu C, et al. miR-765 inhibits the osteogenic differentiation of human bone marrow mesenchymal stem cells by targeting BMP6 via regulating the BMP6/Smad1/5/9 signaling pathway. Stem Cell Res Ther. 2020;11(1):62.
- Wang B, Wu W, Xu K, et al. MicroRNA-223-3p is involved in fracture healing by regulating fibroblast growth factor receptor 2. Bioengineered. 2021Nov9;12(2):12040–12048. Epub ahead of print. PMID: 34753389.
- Yin CM, Suen WC, Lin S, et al. Dysregulation of both miR-140-3p and miR-140-5p in synovial fluid correlate with osteoarthritis severity. Bone Joint Res. 2017Nov;6(11):612–618. PMID: 29092816; PMCID: PMC5717073.
- Forman JP, Curhan GC, Taylor EN. Plasma 25-hydroxyvitamin D levels and risk of incident hypertension among young women. Hypertension. 2008Nov;52(5):828–832. Epub 2008 Oct 6. PMID: 18838623; PMCID: PMC2747298.
- Sun Y, Chen R, Zhu D, et al. Osteoking improves OP rat by enhancing HSP90-β expression. Int J Mol Med. 2020May;45(5):1543–1553. Epub 2020 Mar 6. PMID: 32323753; PMCID: PMC7138285.
- Rocha-Martins M, Njaine B, Silveira MS. Avoiding pitfalls of internal controls: validation of reference genes for analysis by qRT-PCR and Western blot throughout rat retinal development. PLoS One. 2012;7(8):e43028. Epub 2012 Aug 20. PMID: 22916200; PMCID: PMC3423434.
- Li ZH, Hu H, Zhang XY, et al. MiR-291a-3p regulates the BMSCs differentiation via targeting DKK1 in dexamethasone-induced osteoporosis. Kaohsiung J Med Sci. 2020Jan;36(1):35–42. Epub 2019 Nov 15. PMID: 31729834.
- Qiu J, Huang G, Na N, et al. MicroRNA-214-5p/TGF-β/Smad2 signaling alters adipogenic differentiation of bone marrow stem cells in postmenopausal osteoporosis. Mol Med Rep. 2018May;17(5):6301–6310. Epub 2018 Mar 9. PMID: 29532880; PMCID: PMC5928609.
- Noh JY, Yang Y, Jung H. Molecular mechanisms and emerging therapeutics for osteoporosis. Int J Mol Sci. 2020Oct15;21(20):E7623. PMID: 33076329.
- Anupama DS, Norohna JA, Acharya KK, et al. Effect of exercise on bone mineral density and quality of life among postmenopausal women with osteoporosis without fracture: a systematic review. Int J Orthop Trauma Nurs. 2020Jul; 39: 100796 Epub ahead of print. PMID: 33041224
- Ma W, Dou Q, Ha X. Let-7a-5p inhibits BMSCs osteogenesis in postmenopausal osteoporosis mice. Biochem Biophys Res Commun. 2019Feb26;510(1):53–58. Epub 2019 Jan 16. PMID: 30660362.
- Fu Y, Xu Y, Chen S, et al. MiR-151a-3p promotes postmenopausal osteoporosis by targeting SOCS5 and activating JAK2/STAT3 signaling. Rejuvenation Res. 2020Aug;23(4):313–323. Epub 2019 Sep 23. PMID: 31411118.
- Wang WW, Yang L, Wu J, et al. The function of miR-218 and miR-618 in postmenopausal osteoporosis. Eur Rev Med Pharmacol Sci. 2017Dec;21(24):5534–5541. PMID: 29271983.
- Li LY, Wang XL, Wang GS, et al. MiR-373 promotes the osteogenic differentiation of BMSCs from the estrogen deficiency induced osteoporosis. Eur Rev Med Pharmacol Sci. 2019Sep;23(17):7247–7255. PMID: 31539111.
- Wang L, Shen YF, Shi ZM, et al. Overexpression miR-211-5p hinders the proliferation, migration, and invasion of thyroid tumor cells by downregulating SOX11. J Clin Lab Anal. 2018Mar;32(3):e22293. Epub 2017 Jul 13. PMID: 28703321; PMCID: PMC6817049.
- Quan J, Pan X, He T, et al. Tumor suppressor miR-211-5p is associated with cellular migration, proliferation and apoptosis in renal cell carcinoma. Exp Ther Med. 2018Apr;15(4):4019–4028. Epub 2018 Feb 28. PMID: 29581751; PMCID: PMC5863605.
- Peng Z, Li M, Tan X, et al. miR-211-5p alleviates focal cerebral ischemia-reperfusion injury in rats by down-regulating the expression of COX2. Biochem Pharmacol. 2020Jul; 177: 113983 Epub 2020 Apr 18. PMID: 32311346
- Liu H, Luo J. miR-211-5p contributes to chondrocyte differentiation by suppressing Fibulin-4 expression to play a role in osteoarthritis. J Biochem. 2019Dec1;166(6):495–502. PMID: 31396630.
- Gong ZM, Tang ZY, Sun XL. LncRNA PRNCR1 regulates osteogenic differentiation in osteolysis after hip replacement by targeting miR-211-5p. Biosci Rep. 2018May11;BSR20180042. 10.1042/BSR20180042 Epub ahead of print. PMID: 29752342
- Lane JM, Russell L, Khan SN. Osteoporosis. Clin Orthop Relat Res. 2000Mar; 372: 139–150 PMID: 10738423
- Ghorbaninejad M, Khademi-Shirvan M, Hosseini S, et al. Epidrugs: novel epigenetic regulators that open a new window for targeting osteoblast differentiation. Stem Cell Res Ther. 2020Oct28;11(1):456. PMID: 33115508.
- Yu L, Hu M, Cui X, et al. M1 macrophage-derived exosomes aggravate bone loss in postmenopausal osteoporosis via a microRNA-98/DUSP1/JNK axis. Cell Biol Int. 2021Dec;45(12):2452–2463. Epub 2021 Aug 30. PMID: 34431160.
- Liu X, Liu X, Du Y, et al. DUSP5 promotes osteogenic differentiation through SCP1/2-dependent phosphorylation of SMAD1. Stem Cells. 2021Oct;39(10):1395–1409. Epub 2021 Jul 10. PMID: 34169608; PMCID: PMC8518947.
- Lei H, He M, He X, et al. METTL3 induces bone marrow mesenchymal stem cells osteogenic differentiation and migration through facilitating M1 macrophage differentiation. Am J Transl Res. 2021 May 15;13(5):4376–4388. PMID: 34150020; PMCID: PMC8205672.
- Ma R, Ma L, Weng W, et al. DUSP6 SUMOylation protects cells from oxidative damage via direct regulation of Drp1 dephosphorylation. Sci Adv. 2020Mar25;6(13):eaaz0361. PMID: 32232156; PMCID: PMC7096176.
- Mendell AL, MacLusky NJ. The testosterone metabolite 3α-androstanediol inhibits oxidative stress-induced ERK phosphorylation and neurotoxicity in SH-SY5Y cells through an MKP3/DUSP6-dependent mechanism. Neurosci Lett. 2019Mar23;696:60–66. Epub 2018 Dec 12. PMID: 30552945.
- Ma J, Yu X, Guo L, et al. DUSP6, a tumor suppressor, is involved in differentiation and apoptosis in esophageal squamous cell carcinoma. Oncol Lett. 2013Dec;6(6):1624–1630. Epub 2013 Oct 7. PMID: 24260056; PMCID: PMC3834198.
- Wang HT, Song YQ, Ren YW, et al. Dual specificity phosphatase 6 (DUSP6) polymorphism predicts prognosis of inoperable non-small cell lung cancer after chemoradiotherapy. Clin Lab. 2016;62(3):301–310. PMID: 27156317.
- Ramkissoon A, Chaney KE, Milewski D, et al. Targeted inhibition of the dual specificity phosphatases DUSP1 and DUSP6 suppress MPNST growth via JNK. Clin Cancer Res. 2019Jul1;25(13):4117–4127. Epub 2019 Apr 1. PMID: 30936125; PMCID: PMC6606396.
- Zhang B, Yuan P, Xu G, et al. DUSP6 expression is associated with osteoporosis through the regulation of osteoclast differentiation via ERK2/Smad2 signaling. Cell Death Dis. 2021Sep2;12(9):825. PMID: 34475393; PMCID: PMC8413376.
- Wang H, Li C, Li J, et al. Naringin enhances osteogenic differentiation through the activation of ERK signaling in human bone marrow mesenchymal stem cells. Iran J Basic Med Sci. 2017Apr;20(4):408–414. PMID: 28804610; PMCID: PMC5425923.
- Cao J, Wei Y, Lian J, et al. Notch signaling pathway promotes osteogenic differentiation of mesenchymal stem cells by enhancing BMP9/Smad signaling. Int J Mol Med. 2017Aug;40(2):378–388. Epub 2017 Jun 22. PMID: 28656211; PMCID: PMC5504972.
- Gong YY, Peng MY, Yin DQ, et al. Long non-coding RNA H19 promotes the osteogenic differentiation of rat ectomesenchymal stem cells via Wnt/β-catenin signaling pathway. Eur Rev Med Pharmacol Sci. 2018Dec;22(24):8805–8813. PMID: 30575922.
