ABSTRACT
Elevated evidences manifest that circular RNAs (circRNAs) are vital in human tumor advancement and chemotherapy resistance. The study was to explore the character of Circ-CUL2 in non-small cell lung cancer (NSCLC). Firstly, the expression of circ-CUL2, microRNA (miR)-888-5p and RB1CC1 was detected in human NSCLC tissues and cell lines by reverse transcription quantitative polymerase chain reaction or Western blot. Then, cell counting kit (CCK)-8, plate clone, Transwell assays, and flow cytometry were applied to separately detect the impacts of circ-CUL2 on proliferation, migration, invasion, apoptosis and cisplatin (DDP) resistance of A549/DDP cells. In this study, exploration of the biological function of Circ-CUL2 was via the Circ-CUL2/miR-888-5p/RB1CC1 axis. The results manifested circ-CUL2 and RB1CC1 were down-regulated in NSCLC tissues and cell lines, while miR-888-5p was up-regulated. Elevated Circ-CUL2 or refrained miR-888-5p repressed A549/DDP cell progression with depressive DDP resistance. Circ-CUL2 curbed miR-888-5p, which targeted RB1CC1. Restrained RB1CC1 turned around the impacts of Circ-CUL2 on the cells. All in all, Circ-CUL2 is anti-NSCLC via miR-888-5p/RB1CC1 axis, enhancing the sensitivity of A549/DDP cells to DDP. Hence, Circ-CUL2 is supposed to be a novel biomarker offering a brand-new strategy for NSCLC therapy.
Graphical abstract
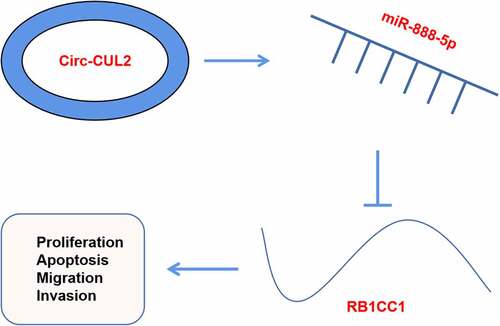
1 Introduction
Lung cancer (LC) is a momentous cancer with surprising morbidity and mortality, and listed as a main reason of cancer-linked deaths worldwide [Citation1,Citation2]. It is reported the link of LC’s incidence with ongoing lung transfection, smoking, exposure to carcinogens, and susceptibility induced via gene mutations [Citation3–5]. Nevertheless, the molecular mechanism of LC is still far from completely explicit. Non-small cell LC (NSCLC)’s histological and cytological features are apparently disparate from small cell LC, and epidemiological reports clarify that non-small cell LC takes up over 80% of LC cases worldwide [Citation6]. A great deal of chemotherapy drugs, like cisplatin (DDP) and Vinorelbine, are extensively applicated in NSCLC therapy, but frequent drug resistance usually results in adverse reactions and elevated recurrence rates, seriously influencing their efficacy [Citation7,Citation8]. Uncovering the molecular mechanism of chemotherapeutic drug resistance is vital for overcoming chemotherapeutic drug resistance and new drugs exploitation.
Circular RNAs (circRNAs), a group of non-coding RNA molecules, modulate functional genes via sponge-binding miRNA and interacting with RNA-binding proteins [Citation9,Citation10]. Recent reports have established the crucial modulators of circRNAs in all kinds of biological processes and pathogenic conditions, like the presence of NSCLC [Citation11,Citation12]. For instance, CircRNA_101237 is memorably elevated in both tissues and cell lines of NSCLC, and supposed to function as a brand-new oncogene in NSCLC advancement via targeting miR-490-3p [Citation13]. What’s more, circRNA is also apparently implicated with the multi-resistance of LC cells to chemotherapy drugs [Citation14,Citation15]. The character of circRNA CUL2 as a brand-new RNA for DDP chemotherapy resistance in NSCLC cells has not been clarified.
MicroRNAs (miRNAs), diminutive non-coding RNAs, modulate their expression via targeting specific 3ʹuntranslated region of mRNA. MiRNAs are linked with all kinds of physiological and pathological processes of cancer [Citation16], and available to control disparateiation, infiltration, radiation resistance and chemotherapy resistance of cancer cells [Citation17]. A study has proposed elevation of miR-888-5p in hepatocellular carcinoma and facilitation of metastasis of cancer cells [Citation18].
RB1CC1, also named as FIP200 (200kda FAK family interacting protein), is a tumor suppressant [Citation19,Citation20]. A recent study has manifested the repression of RB1CC1 in renal cell carcinoma (RCC) cells advancement [Citation21]. The study was to clarify the relative Circ-CUL2 in NSCLC tissues and cells, its impacts on the biological behavior of A549/DDP cells, and the potential character in DDP resistance. The results affirmed that Circ-CUL2 repressed A549/DDP cell advancement, and enhanced DDP sensitivity of A549/DDP cells via miR-888-5p/RB1CC1 axis. Hence, Circ-CUL2 is supposed to be a biomarker and therapeutic target for NSCLC.
2 Materials and methods
2.1 Patients and specimens
Gain of 46 pairs NSCLC and the paired adjacent anticancer tissues was from NSCLC patients undergoing surgery in Cancer Hospital Chinese Academy of Medical Sciences. No reception of chemotherapy or radiotherapy before surgery was in all patients with confirming pathological examination as NSCLC. Store of all tissue samples was in liquid nitrogen for RNA extraction. All patients were followed up for 60 months.
2.2 Cell Culture
Culture of human NSCLC cell lines (A549, NCI-H1299, PC-9 and LLC) and normal human liver cell line (BEAS-2B) (China Type Culture Collection Center, Wuhan, China) was in Roswell Park Memorial Institute-1640 medium (Gibco) consisting of 10% fetal bovine serum (FBS) (PAN, Bavaria, Germany).
2.3 Drug-resistant cell model
Seeding of the logarithmic A549 cells was in petri dishes at a density of 5 × 105 cells/mL. Introduction of the cells was with 1 mg/L DDP (Jiangsu Hengrui Pharmaceutical Co., Ltd.). After 3 times of passages, stable growth of the cells was in the medium consisting of drugs. During this time, elevation of DDP concentration was to 2 mg/L and gradual augment was done until the cells clarified steady growth at 10 mg/L. Name of the established cell line was A549/DDP cells.
2.4 Cell transfection
Synthesis of oe/sh-CUL2, miR-888-5p inhibitor/mimic, si-RB1CC1 and their negative control (NC) lentivirus vectors was via GenePharma (Shanghai, China). In line with the manufacturer’s instructions, introduction of lentiviruses was into cells via polypropylene (Shanghai Hanssen Biotechnology Co., Ltd., China). Removing the un-transfected cells was with puromycin (2 µg/mL).
2.5 Cell counting kit (CCK)-8
Seeding of A549/DDP cells was into 96-well plates. Culture of the cells was with disparate doses of DDP and then addition was with 10 mg/mL CCK-8 reagent (Beyotime, Shanghai). Determination of cell viability was in line with manufacturer’s instructions. Measurement of the absorbance at 450 nm was with a microplate reader, and determination of the half maximal inhibitory concentration (IC50) of DDP in A549/DDP cells was conducted.
2.6 Plate cloning
Seeding of A549/DDP cells in the logarithmic phase was into 6-well plates at a density of 600 cells per well. Culture of the 6-well plates was clarified until a cell clone was seen to the unaided eye in a petri dish. Fixation was with 10% formaldehyde and stain was with crystal violet (Beyotime, Shanghai), and count of clones with over 50 cells was under a low power microscope.
2.7 Transwell
Determination of cell migration and invasion capacities was via Transwell chambers (pore size 8.0 mTM; EMD Millipol, Billerica, Ma, USA) and Matrigel (1: 9) (Corning, USA). Suspension of the transfected A549/DDP cells (4 × 105 cells) was in 200 µL serum-free Dulbecco’s Modified Eagle Medium, and addition of the upper chamber was with or without 10 μL matterige, and the lower chamber was full of 500 μL complete medium consisting of 10% FBS. After incubation, removing of the cells remaining on the upper surface of the membrane was done. Fixation of the basal cells was with 4% paraformaldehyde, and stain was with 0.1% crystal violet (Beyotime, Shanghai), and count via photographing was under an upright microscope (Nikon, Japan).
2.8 Flow cytometry
Harvest of A549/DDP cells in logarithmic growth period was to prepare single-cell suspension and seeding was in 25 cm2 culture flask. The old culture medium was abandoned behind overnight adherence. Culture of the experimental group was with 0.3% FBS culture medium, and that of the control was with equal volume phosphate buffer saline culture medium. Behind harvest of the cell supernatant, detachment of the cells was with trypsin without ethylene diamine tetraacetic acid with collection. The rest steps were followed in line with the instructions of AnnexinV-propidium iodide apoptosis kit. Detection of the cell apoptosis was via flow cytometry.
2.9 Quantitative Real-Time PCR (RT-qPCR)
Extraction of total RNA was from tissues or cells via TRIzol reagent (Invitgen, USA) with synthesis of complementary DNA via CircRNA reverse transcription kit (Guangzhou GYSai Biotechnology Co., Ltd.). Conduction of RT-qPCR was via CircRNA real-time PCR detection kit on ABI7500 real-time PCR system (Applied Biosystems, Foster City, California, USA). Detection of Circ-CUL2 was with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) as a loading control and calculation of the relative circRNA was via 2ΔΔCt method. Design and synthesis of all the primers were via Guangzhou RiboBio, as clarified in .
Table 1. RT-qPCR primers
2.10 Western blot Analysis
Lysis of the cells was with a lysis buffer consisting of 1 nM phenylmethylsulfonyl fluoride (Beyotime, Shanghai, China) and determination of the protein concentration was via bicinchoninic acid protein analysis kit (Beyotime). Determination of protein separation was via sulfate polyacrylamide gel electrophoresis (Beyotime) and electroblot was onto polyvinylidene fluoride microporous membrane. Incubation was with anti-β-catenin (1: 800), anti-c-Myc (1: 500) and anti-GAPDH (1: 500), and horseradish peroxidase (1: 5000) (all Ruiying, China) coupling with the mouse anti-rabbit Immunoglobulin G secondary antibody. Manifestation of protein bands was with the WestrenBright electrogenerated chemiluminescence kit (Advansta, USA). Finally, application of Image Lab software (Bio-Rad, Hercules, California, USA) was for analysis of the signal strength of the bands.
2.11 The luciferase activity assay
Prediction of the binding sites of Circ-CUL2 to miR-888-5p and miR-888-5p to RB1CC1 was via the bioinformatics website (starBase). Verification of the binding of the factors was affirmed. Inserting of corresponding sequences was into pmirGLO (Promega, Madison, Wisconsin, USA) to construct CUL2/RB1CC1-wild and mut types (WT/MUT), which were then co-transfected with miR-888-5p mimic or its NC. Finally, assessment of the luciferase activity was via a dual luciferase reporter gene assay kit (Promega).
2.12 Statistical analysis
Results analysis was via GraphPad Software 6.0 (GraphPad Inc., San Diego, CA, USA) and SPSS software (version 24.0, Chicago, IL, USA) (N = 3). Presentation of the data was as mean ± standard deviation (SD). Application of student t test and one-way analysis of variance (ANOVA) was for comparison of the statistical differences of two or multiple groups, with χ2 test for evaluation of the association of CirC-CUL2 with clinicopathological features of NSCLC patients. Kaplan–Meier survival analysis and logarithmic rank test were applied to analyze the survival curve. P < 0.05 was considered statistically significant.
3 Results
3.1 Repressive Circ-CUL2 is in NSCLC; the circRNA is implicated with unpleasing prognosis in NSCLC patients
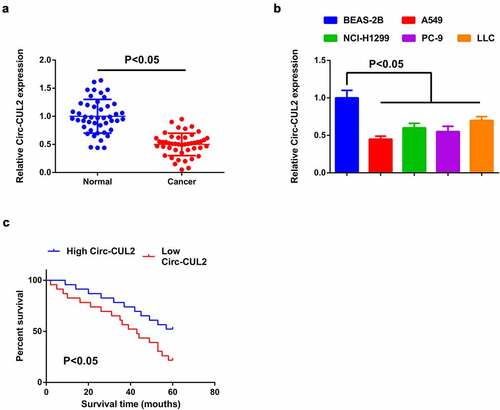
For exploration of the character of Circ-CUL2 in NSCLC, detection of Circ-CUL2 in NSCLC and tissues and cells was done, manifesting the decline ()). Meanwhile, A549 cells clarified the most apparent difference, hence, choice of the cells was for subsequent experiments. In view of the dysregulation of Circ-CUL2 in NSCLC tissues and cells, analysis of the association of Circ-CUL2 with the clinicopathological features of NSCLC patients was done. Assignation of the patients was into Circ-CUL2 elevation and repression groups (n = 23 separately). Comparison of the clinicopathological parameters of the two was via Pearson Chi-square test. The association of plasma Circ-CUL2 with clinicopathological features was clarified, like age, gender, smoking status, histological classification, tumor size, lymph node metastasis and clinical stage (). The results manifested that plasma Circ-CUL2 was extremely linked with clinical stage and lymph node metastasis, assuring that CirC-CUL2 was supposed to be a biomarker for NSCLC diagnosis and prognosis. Meanwhile, via Kaplan–Meier survival analysis, it was clarified the elevated overall survival rate of patients with augmented Circ-CUL2 versus that with reduced one ()), manifesting patients with elevated CirC-CUL2 were more sensitive to drugs, while that with reduced one were more likely to develop drug resistance.
Table 2. Association of Circ-CUL2 with clinical features in NSCLC patients
Figure 1. Reduction of Circ-CUL2 is in NSCLC tissues and cells.
3.2 Elevated Circ-CUL2 represses A549/DDP cell advancement with depressive DDP resistance
For exploration of the modulation of Circ-CUL2 on DDP resistance in NSCLC, construction of a DDP resistance cell model was via A549 cells with name as A549/DDP cells. Analysis of IC50 value of DDP in A549 cells and A549/DDP cells was clarified, manifesting the apparent elevation in A549/DDP cells ()). Then separate transfection of oe/sh-CUL2 and their NC was into A549/DDP cells and the efficiency of lentivirus transfection was confirmed ()). The results manifested that elevated Circ-CUL2 was available to repress cell advancement with depressive DDP IC50 value, while reduced one was opposite with strengthening DDP resistance ().
Figure 2. Circ-CUL2 curbs A549/DDP cell advancement.
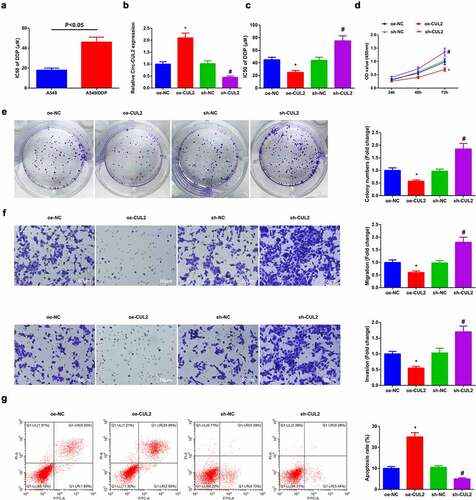
Briefly, Circ-CUL2 restrained A549/DDP cell advancement, with enhancing DDP sensitivity.
3.3 Circ-CUL2 refrains miR-888-5p
Detection of miR-888-5p in NSCLC tissues and cells affirmed the augment ().
Figure 3. Circ-CUL2 represses miR-888-5p.
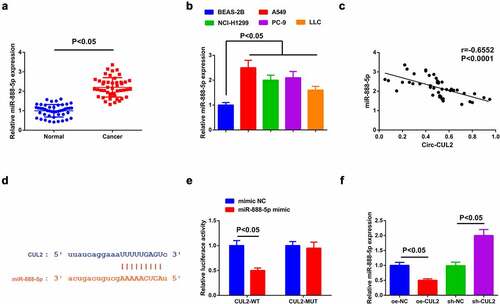
A speculation was clarified the certain association of Circ-CUL2 and miR-888-5p in NSCLC. To explore whether Circ-CUL2 directly curbed miR-888-5p, application of Pearson test was for analysis of the link of the two in clinical samples, clarifying the negative link of them ()). Then, via the bioinformatics website was discovered the binding sites of the two ()). Additionally, reduced luciferase activity was manifested after co-transfection of CUL2-WT and miR-888-5p mimic ()). Further verification clarified descending miR-888-5p behind augmented Circ-CUL2, which was opposite behind depressing one ()). Shortly, Circ-CUL2 refrained miR-888-5p. In addition, it was also found that circ-CUL2 promoted the expression of RB1CC1 by repressing the expression of miR-888-5p, and down-regulation of RB1CC1 reversed the acceleration of elevated circ-CUL2 on the expression of RB1CC1 (attached ).
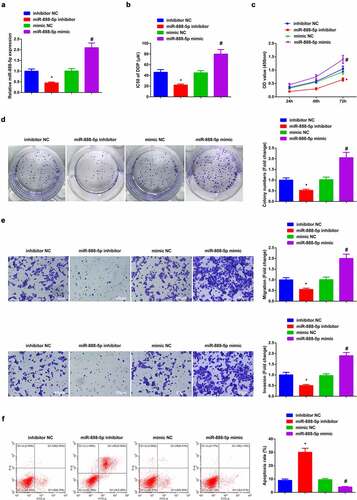
3.4 Depressive miR-888-5p refrains A549/DDP cell advancement with constrained DDP resistance
Verification of the successful transfection of miR-888-5p was done ()). The results clarified that restrained miR-888-5p constrainedA549/DDP cell advancement with depressive IC50 value of DDP, while ascending one was available to enhance DDP resistance with expediting cell progression (). Briefly, refrained miR-888-5p curbed A549/DDP cell advancement with constrained DDP resistance.
Figure 4. Repressive miR-888-5p refrains A549/DDP cell advancement with descending DDP resistance.
3.5 MiR-888-5p targets RB1CC1
Detection of RB1CC1 in NSCLC tissues and cells clarified the decline (). Meanwhile, a negative correlation of miR-888-5p with RB1CC1 was observed in clinical samples ()), while positive link was clarified in Circ-CUL2 and RB1CC1 ()).
Figure 5. MiR-888-5p targets RB1CC1.
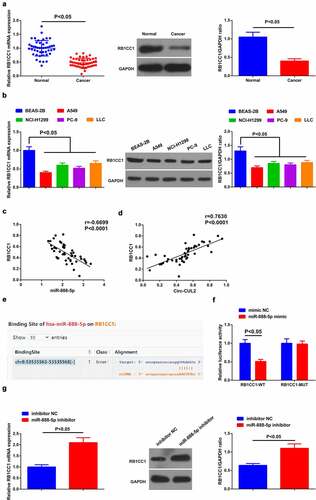
As predicted via starBase, targeted binding sites was manifested in miR-888-5p and RB1CC1 ()). Subsequently, descending luciferase activity of A549/DDP cells was clarified behind co-transfection of RB1CC1-WT and miR-888-5p mimic ()). Moreover, repressive miR-888-5p elevated RB1CC1 ()). Shortly, miR-888-5p negatively controlled RB1CC1.
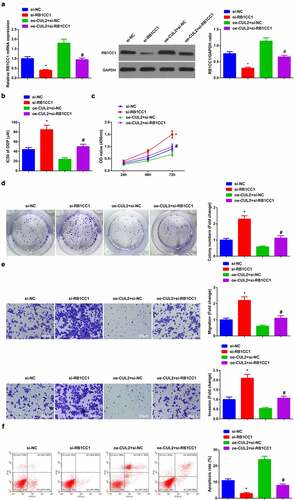
3.6 Depressive RB1CC1 turns around the impacts of Circ-CUL2 elevation on A549/DDP cells
Next, exploration of the modulatory character of Circ-CUL2/miR-888-5p/RB1CC1 axis was clarified. Transfection of si-RB1CC1, oe-CUL2 + si-RB1CC1, si-NC and oe-CUL2 + si-NC was into A549/DDP cells, and the successful transfection was verified ()). Meanwhile, depressive RB1CC1 turned around the repression of elevated Circ-CUL2 on IC50 value of DDP ()). Moreover, behind descending RB1CC1, the refraining of Circ-CUL2 on A549/DDP cell advancement was of reversion (). Briefly, reduced RB1CC1 turned around the impacts of Circ-CUL2 enhancing on A549/DDP cells. In the meantime, it was also investigated the effect of circ-CUL2/miR-888-5p/RB1CC1 axis on DDP nonresistant cell lines, and the experimental results were similar to those of A549/DDP cells (attached ). This further supported the conclusion.
Figure 6. Refraining RB1CC1 turns around the impacts of elevated Circ-CUL2 on A549/DDP cells.
4 Discussion
LC has been diagnosed as an extremely familiar malignant tumor worldwide, with surprising morbidity and mortality. In terms of LC, nearly 1.8 million newly diagnosed cases and 1.6 million deaths each year are present, and the deaths induced via LC takes up 19% of all cancer-linked deaths [Citation22]. LC is regarded as the major reason of cancer-linked death, with a steadily enhancing incidence and has become a principal obstacle to human health [Citation23]. Surprising recurrence and metastasis rates are considered main elements resulting in unpleasing prognosis of LC patients [Citation24]. Despite the persistent perfection of diagnosis and treatment, the 5-year overall survival rate of LC patients is still less than 20% [Citation25]. More importantly, drug resistance in LC therapy is extremely linked with abnormal oncogenes or anti-tumor genes, consisting of changes in biological features of malignant tumors, cell proliferation, metastasis etc [Citation26]. Though DDP has been applied as an anticancer chemotherapy drug, all kinds of cancers (covering LC) may produce acquired resistance to DDP, which is a stumbling block to mitigating the efficacy of chemotherapy [Citation27]. Moreover, the cytotoxicity of DDP remains a familiar side effect of DDP [Citation28]. Hence, the study of the sensitivity mechanism of LC cells to DDP is vital for the combination of drugs on the grounds of specific molecular mechanisms to lighten the chemotherapy efficacy of LC.
DDP is a first-line chemotherapy drug in the clinical therapy of NSCLC [Citation29], but its efficacy is seriously limited owing to DDP resistance in NSCLC cells. Hence, at present, researchers are concentrating on the latent mechanism of DDP resistance, for developing promising therapeutic drugs to rescue the chemotherapy sensitivity of NSCLC cells to DDP [Citation30,Citation31]. In this study, it was discovered that a brand-new circRNA, Circ CLU2, was reduced in NSCLC tissues and cells. Circ CLU2 was available to repress cell advancement via miR-888-5p/RB1CC1 axis with depressive DDP resistance in NSCLC.
CircRNAs and miRNAs refer to key tumor phenotypes, like proliferation, apoptosis and chemotherapy resistance, and CircRNAs act biologically via disparate molecular mechanisms. A familiar molecular mechanism is that circRNA controls cell phenotypes via spongy miRNAs [Citation32–34]. In this study, detection of Circ-CUL2 was in NSCLC tissues and cells, clarifying the decline. Patients with elevated cirC-CUL2 manifested a better prognosis. Meanwhile, discussion of the impacts of Circ-CUL2 was on A549/DDP cell advancement with DDP resistance, affirming that Circ-CUL2 repressed cell advancement with enhancing sensitivity of A549/DDP cells to DDP.
MiRNAs are crucial in a great many biological processes, consisting of cell proliferation [Citation35], and in LC advancement and metastasis [Citation36]. In this study, a focus was on the downstream candidate miRNA of Circ-CUL2 (miR-888-5p), which has been manifested the elevation in hepatocellular carcinoma and acceleration of cancer cell metastasis [Citation18]. But its character in LC has not been explored. Here, via experimental detection was discovered that reduced miR-888-5p restrained A549/DDP cell advancement with enhancing the sensitivity to DDP.
Moreover, a recent study has clarified that RB1CC1 is available to repress RCC cell development [Citation21]. Similarly, RB1CC1 curbs prostate cancer [Citation37]. Reduced RB1CC1 forecasts unpleasing prognosis of breast cancer [Citation38]. In this study, it was discovered that RB1CC1 was the direct downstream target of miR-888-5p, and repressive RB1CC1 turned around the impacts of elevated Circ-CUL2 on A549/DDP cells. It was proved that Circ-CUL2 exerted its tumor suppressive influence via miR-888-5p/RB1CC1 axis.
5 Conclusion
Anti-tumor CircCUL2 is reduced in NSCLC tissues and cells. In the meantime, CircCUL2 represses DDP resistance of NSCLC via miR-888-5p/RB1CC1 axis. These results manifest that Circ-CUL2 is supposed to be a functional marker for NSCLC and a latent therapeutic target for NSCLC.
Ethics statement
Approval of the study was via the Ethics Committee of Cancer Hospital Chinese Academy of Medical Sciences with written informed consent of all patients
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Jonna S, Subramaniam D. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med. 2019;27(148):167–170.
- Tian LJ, Wu, YP, Wang, D, et al. Upregulation of Long Noncoding RNA (lncRNA) X-Inactive Specific Transcript (XIST) is Associated with Cisplatin Resistance in Non-Small Cell Lung Cancer (NSCLC) by Downregulating MicroRNA-144-3p. Med Sci Monit. 2019;25:8095–8104.
- Herbst R, Morgensztern D, Boshoff C. The biology and management of non-small cell lung cancer. Nature. 2018;553(7689):446–454.
- Lee S, Cheah Y. The Interplay between MicroRNAs and Cellular Components of Tumour Microenvironment (TME) on Non-Small-Cell Lung Cancer (NSCLC) Progression. J Immunol Res. 2019;2019:3046379.
- Chen Z, Fillmore CM, Hammerman PS, et al. Non-small-cell lung cancers: a heterogeneous set of diseases. Nat Rev Cancer. 2014;14(8):535–546.
- Fujimoto J, Wistuba I. Current concepts on the molecular pathology of non-small cell lung carcinoma. Semin Diagn Pathol. 2014;31(4):306–313.
- Fadejeva I, Olschewski H, Hrzenjak A. MicroRNAs as regulators of cisplatin-resistance in non-small cell lung carcinomas. Oncotarget. 2017;8(70):115754–115773.
- MacDonagh L, Gray SG, Breen E, et al. BBI608 inhibits cancer stemness and reverses cisplatin resistance in NSCLC. Cancer Lett. 2018;428:117–126.
- Panda A. Circular RNAs Act as miRNA Sponges. Adv Exp Med Biol. 2018;1087:67–79.
- Liu Z, Yu Y, Huang Z, et al. CircRNA-5692 inhibits the progression of hepatocellular carcinoma by sponging miR-328-5p to enhance DAB2IP expression. Cell Death Dis. 2019;10(12):900.
- Fu Y, Huang L, Tang H, et al. hsa_circRNA_012515 Is Highly Expressed in NSCLC Patients and Affects Its Prognosis. Cancer Manag Res. 2020;12:1877–1886.
- Xiao G, Huang W, Zhan Y, et al. CircRNA_103762 promotes multidrug resistance in NSCLC by targeting DNA damage inducible transcript 3 (CHOP). J Clin Lab Anal. 2020;34(6):e23252.
- Zhang Z, Gao X-H, Ma M-Y, et al. CircRNA_101237 promotes NSCLC progression via the miRNA-490-3p/MAPK1 axis. Sci Rep. 2020;10(1):9024.
- Zhu X, Han J, Lan H, et al. A novel circular RNA hsa_circRNA_103809/miR-377-3p/GOT1 pathway regulates cisplatin-resistance in non-small cell lung cancer (NSCLC). BMC Cancer. 2020;20(1):1190.
- Zhao Y, Zheng R, Chen J, et al. CircRNA CDR1as/miR-641/HOXA9 pathway regulated stemness contributes to cisplatin resistance in non-small cell lung cancer (NSCLC). Cancer Cell Int. 2020;20(1):289.
- Yoshida K, Yokoi A, Yamamoto Y, et al. ChrXq27.3 miRNA cluster functions in cancer development. J Exp Clin Cancer Res. 2021;40(1):112.
- Xu K, Zhan Y, Yuan Z, et al. Hypoxia Induces Drug Resistance in Colorectal Cancer through the HIF-1α/miR-338-5p/IL-6 Feedback Loop. Mol ther. 2019;27(10):1810–1824.
- Hao E, Yu J, Xie S, et al. Up-regulation of miR-888-5p in hepatocellular carcinoma cell lines and its effect on malignant characteristics of cells. J Biol Regul Homeost Agents. 2017;31(1):163–169.
- Li S, Qiang, Q, Shan, H, et al. MiR-20a and miR-20b negatively regulate autophagy by targeting RB1CC1/FIP200 in breast cancer cells. Life Sci. 2016;147:143–152.
- Ikebuchi K, Chano T, Ochi Y, et al. RB1CC1 activates the promoter and expression of RB1 in human cancer. Int J Cancer. 2009;125(4):861–867.
- Chen P, Duan, Y, Lu, X, et al. RB1CC1 functions as a tumor-suppressing gene in renal cell carcinoma via suppression of PYK2 activity and disruption of TAZ-mediated PDL1 transcription activation. Cancer Immunol Immunother. 2021;70(11):3261–3275 .
- Torre L, Bray F, Siegel RL, et al. Global cancer statistics, 2012. CA: a cancer journal for clinicians. 2015;65(2):87–108.
- Dougan M, Li D, Neuberg D, et al. A dual role for the immune response in a mouse model of inflammation-associated lung cancer. J Clin Invest. 2011;121(6):2436–2446.
- Meng YB, He, X, Huang, YF, et al. Long Noncoding RNA CRNDE Promotes Multiple Myeloma Cell Growth by Suppressing miR-451. Oncol Res. 2017;25(7):1207–1214.
- Didkowska J, Wojciechowska, U, Mańczuk, M, Łobaszewski, J, et al. Lung cancer epidemiology: contemporary and future challenges worldwide. Ann Transl Med. 2016;4(8):150.
- Lv X, Liu L, Cheng C, et al. SUN2 exerts tumor suppressor functions by suppressing the Warburg effect in lung cancer. Sci Rep. 2015;5(1):17940.
- Lin Y, Wang, Z, Liu, L, Chen, L . Akt is the downstream target of GRP78 in mediating cisplatin resistance in ER stress-tolerant human lung cancer cells. Lung Cancer. 2011;71(3):291–297.
- Fuertes M, Alonso C, Pérez J. Biochemical Modulation of Cisplatin Mechanisms of Action: enhancement of Antitumor Activity and Circumvention of Drug Resistance. Chem Rev. 2003;103(3):1207–1214.
- Xing Y, Liu, Y, Liu, T, et al. TNFAIP8 promotes the proliferation and cisplatin chemoresistance of non-small cell lung cancer through MDM2/p53 pathway. Cell Commun Signal. 2018;16(1):43.
- Cruz-Bermúdez A, Laza-Briviesca R, Vicente-Blanco RJ, et al. Cisplatin resistance involves a metabolic reprogramming through ROS and PGC-1α in NSCLC which can be overcome by OXPHOS inhibition. Free Radic Biol Med. 2019;135:167–181.
- Watanabe S, Yoshioka H, Sakai H, et al. Necitumumab plus gemcitabine and cisplatin versus gemcitabine and cisplatin alone as first-line treatment for stage IV squamous non-small cell lung cancer: a phase 1b and randomized, open-label, multicenter, phase 2 trial in Japan. Lung Cancer. 2019;129:55–62.
- Liu J, Xue N, Guo Y, et al. CircRNA_100367 regulated the radiation sensitivity of esophageal squamous cell carcinomas through miR-217/Wnt3 pathway. Aging (Albany NY). 2019;11(24):12412–12427.
- He J, Huang Z, He M, et al. Circular RNA MAPK4 (circ-MAPK4) inhibits cell apoptosis via MAPK signaling pathway by sponging miR-125a-3p in gliomas. Mol Cancer. 2020;19(1):17.
- Luo Y, Fu Y, Huang R, et al. CircRNA_101505 sensitizes hepatocellular carcinoma cells to cisplatin by sponging miR-103 and promotes oxidored-nitro domain-containing protein 1 expression. Cell Death Discov. 2019;5(1):121.
- Feliciano A, Garcia-Mayea Y, Jubierre L, et al. miR-99a reveals two novel oncogenic proteins E2F2 and EMR2 and represses stemness in lung cancer. Cell Death Dis. 2017;8(10):e3141.
- Wang H, Deng Q, Lv Z, et al. N6-methyladenosine induced miR-143-3p promotes the brain metastasis of lung cancer via regulation of VASH1. Mol Cancer. 2019;18(1):181.
- Suraneni M, Moore J, Zhang D, et al. Tumor-suppressive functions of 15-Lipoxygenase-2 and RB1CC1 in prostate cancer. Cell Cycle. 2014;13(11):1798–1810.
- Chano T, Ikebuchi, K, Tomita, Y, et al. RB1CC1 together with RB1 and p53 predicts long-term survival in Japanese breast cancer patients. PloS one. 2010;5(12):e15737.
