ABSTRACT
As a common intraocular malignancy in pediatrics, retinoblastoma (RB) has high prevalence worldwide. We conducted this study, aiming to explore the molecular mechanism of Krüppel-like transcription factor 16 (KLF16)/cellular retinoic acid-binding proteins-2 (CRABP2) in regulating the invasion and migration and apoptosis of RB cells via integrin-β1/focal adhesion kinase (FAK)/extracellular signal-regulated kinase (ERK) pathway. With the adoption of real-time quantitative polymerase chain reaction (RT-qPCR) and Western blot, the mRNA and protein expression of CRABP2 and KLF16 were measured. In addition, the proliferation, clone formation ability and migration were detected with methyl thiazolyl tetrazolium (MTT), clone formation and wound healing assays, respectively. Furthermore, the invasion and apoptosis of transfected WERI-RB1 cells were evaluated with transwell and Tunel assays. With the application of Western blot, the expressions of proliferation-, apoptosis- and pathway-related proteins were assayed. The combination of KLF16 and CRABP2 was confirmed by dual-luciferase reporter assay and chromatin immunoprecipitation (ChIP). In this study, we found that CRABP2 gained a huge growth in RB cells and its silence promoted apoptosis but suppressed the proliferation, migration and invasiveness of WERI-RB1 cells. In addition, KLF16 could bind to CRABP2. It was also found that KLF16 overexpression reversed the effects of CRABP2 silence on the proliferation, migration and apoptosis of WERI-RB1 cells. What is more, CRABP2 silence blocked integrin-β1/FAK/ERK signaling pathway. In conclusion, KLF16 transcriptional up-regulation of CRABP2 promoted proliferation, invasion and migration but inhibited apoptosis of RB cells by activating integrin-β1/FAK/ERK pathway.
Graphical abstract
Background
Retinoblastoma (RB) originates from retinal embryonic nuclear layer cells. It is recognized that RB is due to heterozygous pathogenic variants in RB1 gene and that amplification of MYCN is the cause of RB in the absence of RB1 pathogenic variants in about 1.5% of individuals with isolated unilateral RB [Citation1]. Among all malignant tumors in children, the incidence of RB is second only to leukemia, with an annual incidence of 1/18,000-1/16,000, showing a genetic tendency [Citation2]. About 95% of the children were under five at onset. About 75% of the children were monocular RB, with the onset age of 2~3 years old, and the onset of binocular RB is earlier [Citation2–4]. There are 7000–8000 new cases in the world every year [Citation5] and 3000–4000 deaths [Citation6], and about 1100 new cases in China every year [Citation7,Citation8]. RB not only harms the visual function of children but even threatens their lives. In developing countries, most children with RB are found in advanced stages, and the survival rate is much lower than that in developed countries [Citation9,Citation10]. Early detection and treatment of RB help to enhance cure rate and reduce case fatality rate [Citation11]. Therefore, in-depth study on the mechanism of RB can provide scientific basis for the early diagnosis, treatment and prognosis evaluation of RB.
Compared with normal retina, 27 different differentially expressed proteins were identified in RB cells [Citation12]. Moreover, mRNA expressions of apolipoprotein A1, transferrin, cellular retinoic acid-binding proteins-2 (CRABP2), and α-crystalloprotein A were also observed to be higher in RB cells [Citation12]. CRABP2 is a member of intracellular lipid-binding proteins (iLBPs). CRABP2 transits retinoic acid (RA) into the nucleus where it binds to the homodimer formed by retinoic acid nuclear receptors (RARs) and retinoic acid X receptors (RXRs), resulting in subsequent RA-induced gene transcription to inhibit cell growth and promote differentiation [Citation13,Citation14]. A study indicated that CRABP2 level in plasma was obviously upregulated in non-small cell lung cancer (NSCLC) patients compared with that in control people, and NSCLC patients with higher CRABP2 level in plasma had poor survival [Citation15]. As tumor stage advances, CRABP2 expression level in HCC tissues and in HCC cell lines were increased. CRABP2 silence exhibited inhibitory effects on cell proliferation, invasiveness and expressions of extracellular signal-regulated kinase (ERK)/vascular endothelial growth factor (VEGF) pathway-related proteins but upregulated expression of apoptosis-related proteins [Citation16]. However, whether CRABP2 acts as an important player in the advancement of RB, its clear functional and molecular mechanisms have not been reported.
JASPAR predicts that the transcription factor Krüppel-like transcription factor 16 (KLF16) binds to the CRABP2 promoter region [Citation17]. KLF16 is a member of the Krüppel family of transcription factors involved in cell cycle and promoter-dependent transcriptional regulation [Citation18]. KLF16 played a tumor suppressive role in lung adenocarcinoma by inhibiting tumor cell proliferation and inducing apoptosis [Citation19]. Zhang et al. indicated that KLF16 was obviously increased in RB tissues and cells, and its overexpression enhanced the growth and migration of RB cells [Citation20]. CRABP2 silence suppressed the migration and invasive rates of lung cancer cells as well as metastasis in vivo, which might be partly dependent on HuR-mediated integrin β1/focal adhesion kinase (FAK)/ERK signaling [Citation21]. Axed Mucin 4 (MUC4/X) promoted tumorigenesis of pancreatic cancer through the activation of integrin-β1/FAK/ERK signaling pathway [Citation22].
The present study aimed to explore the molecular mechanism of KLF16/CRABP2 in regulating the invasion and migration and apoptosis of RB cells via integrin-β1/FAK/ERK pathway.
Materials and methods
Cell culture
American Type Culture Collection (Manassas, VA, USA) is the provider of human retinal pigment epithelial cells (ARPE-19). RB cells Y79 and SO-RB50 cells were provided from BioVector NTCC Inc. (Beijing, China), and WERI-RB1 cells were provided from Procell Life Science&Technology Co., Ltd (Wuhan, China). All the RB cell lines used in this study are RB1 variant. The incubation of cells was performed in Roswell Park Memorial Institute (RPMI)-1640 culture medium (HyClone, USA) with the addition of 10% FBS and 1% penicillin/streptomycin solution (Corning, Cambridge, USA) at 37°C in a humid incubator containing 5% CO2.
Cell transfection
Short hairpin (sh) RNA targeting CRABP2 (sh-CRABP2-1/2; 500 ng/µl), shRNA-KLF16-1/2 (500 ng/µl), and the negative control (sh-NC; 500 ng/µl), KLF16 overexpression vector (Oe-KLF16; 15 nM) and Oe-NC (15 nM) and were synthesized from GenePharma (Shanghai, China) and transfected to WERI-RB1 cells with the application of Lipofectamine 2000 (Thermo Fisher Scientific). The transfection effects were verified by real-time quantitative polymerase chain reaction (RT-qPCR) analysis and Western blot assays after 48 h following the cell transfection.
RT-qPCR analysis
Total RNA was isolated from the cells by TRIzol (Invitrogen) and converted to cDNA with ReverTra Ace® qPCR RT Master Mix (TOYOBO, Japan). The amplification and quantification of complementary DNA (cDNA) were performed using TransStart Top Green qPCR SuperMix (TransGen Biotech) on the Bio-rad machine. The primers used were synthesized from Sangon Biotech (Shanghai, China) and mRNA levels of KLF16 and CRABP2 were quantified using the 2−ΔΔCq method [Citation23].
Western blot
Proteins extract from the cells was obtained using radioimmunoprecipitation assay (RIPA) buffer (Beyotime), and BCA kit was used to determine the protein concentration. Separated by 12% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), the proteins were transferred onto polyvinylidene fluoride (PVDF) membranes. After the procedure of blocking membranes with 5% skim milk, primary antibodies were utilized to incubate membranes at 4°C overnight. The membranes were blocked with 5% skim milk and incubated with primary antibodies against CRABP2 (ab181255, 1:10,000; Abcam), Ki67 (ab16667, 1:1,000; Abcam), proliferating cell nuclear antigen (PCNA; ab18197, 1:1,000; Abcam), Bcl-2 (ab32124, 1:1,000; Abcam), Bax (ab32503, 1:10,000; Abcam), cleaved caspase 3 (#9661, 1:1,000; Cell Signaling Technology), cleaved caspase 9 (#20,750, 1:1,000; Cell Signaling Technology), KLF16 (orb39548, 1:1,000; Biorbyt), integrinbeta1 (ITGB1; #9699, 1:1,000; Cell Signaling Technology), p-FAK (#3284, 1:1,000; Cell Signaling Technology), FAK (#3285, 1:1,000; Cell Signaling Technology), p-ERK (#4370, 1:2,000; Cell Signaling Technology), ERK (#4695, 1:1,000; Cell Signaling Technology), and glyceraldehyde-phosphate dehydrogenase (GAPDH; ab9485, 1:2,500; Abcam) at 4°C overnight. Membranes were washed with Tris buffered saline Tween (TBST) for three times, after which horseradish peroxidase (HRP)-conjugated secondary antibody (ab6721, 1:2,000, Abcam) was employed to incubate membranes at room temperature for 2 h. Finally, the protein bands were imaged by enhanced chemiluminescence (ECL) reagents.
Methyl Thiazolyl Tetrazolium (MTT) assay
After indicated transfection, a total of 2000 WERI-RB1 cells were seeded in triplicate into a 96-well plate and cultured in a humid incubator at 37°C containing 5% CO2. Then, each well was added with 10 μl of MTT solution, and the cells were incubated for another 4 h. The proliferation detected by MTT was recorded at 24, 48 and 72 h. The cell proliferation reflected by absorbance was detected by microplate spectrophotometer at 490 nm.
Clone formation assay
After indicated transfection, total WERI-RB1 cells (5x104) were added to 6-well plates and incubated for two weeks at 37°C. Thereafter, 4% formaldehyde and GIMSA were applied to fix and counterstain the cells, respectively. The number of colonies (a diameter ≥ 100 μm) were counted under a digital camera.
Transwell assay
After indicated transfection, WERI-RB1 cells were suspended with FBS and serum-free RPMI-1640 medium and 200 µl cell suspension was added to the upper chamber of transwell plates (Corning). 600 µl RPMI-1640 culture medium containing 10% FBS was added into the lower chambers. After 24 h, the invading cells were fixed with methanol and stained with 0.5% crystal violet for 30 min. The area occupied by migrated cells was photographed with the application of a light microscope (Olympus Corporation).
Wound healing assay
WERI-RB1 cells were seeded in cell-culture dishes until the confluence has reached 90–100%. A 10 μl pipette tip was applied to make a scratch, followed by the phosphate buffer saline (PBS) washing for two times to remove the float. Then, the cells were incubated with fresh serum-free medium in an incubator at 37°C for 24 h. The width of the wound was photographed at 0 and 24 h using a light microscope (Olympus Corporation).
Tunel assay
The effects of CRABP2-2 silence and KLF16 overexpression on the apoptosis of WERI-RB1 cells were evaluated with TUNEL assays. In brief, WERI-RB1 cells were fixed and permeabilized with 4% paraformaldehyde and 0.3% Triton X-100, respectively. After TUNEL labeling (Beyotime Institute of Biotechnology), the nucleus was labeled with 4, 6-diamidino-2-phenylindole (DAPI; Beijing Solarbio Science & Technology Co., Ltd.). The images of positive cells were visualized under a fluorescence microscope.
Dual luciferase reporter assay
WERI-RB1 cells (1x105) were co-transfected with CRABP2-wide type (WT) or CRABP2-mutant type (MUT) vectors and Oe-KLF16 or Oe-NC using Lipofectamine 2000 reagent for 24 h. With the adoption of a dual luciferase reporter gene assay kit (Promega), the luciferase activity was assessed. The relative activity of the reporter gene was calculated by dividing the firefly luciferase activity by the renilla luciferase activity.
Chromatin Immunoprecipitation (ChIP)
The binding of KLF16 to the promoter of CRABP2 gene was confirmed by ChIP. WERI-RB1 cells were cross-linked in 1% formaldehyde for 10 min at 37°C and then incubated with anti-KLF16 or anti-IgG antibody overnight at 4°C. The next day, magnetic beads were added to the above mixture for 2 h at 4°C. Then, protein/DNA complexes were eluted and reverse cross-linked for the enrichment of DNA which was analyzed by RT-qPCR.
Statistical analysis
All data collected from experiments were displayed as the mean ± SD and analyzed with the GraphPad Prism 8 (GraphPad Software, Inc., La Jolla, CA, USA). Student’s t-test was employed for comparisons between two groups. One-way ANOVA was used for multiple comparisons, followed which Tukey’s post hoc test was applied. P less than 0.05 was viewed to be of statistically significance.
Results
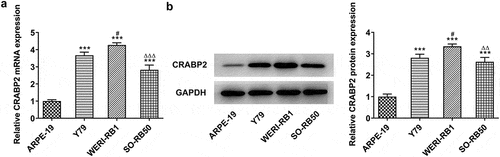
CRABP2 gained a huge growth in RB cells
To ensure the role of CRABP2 in RB, its expression level was detected. As () depicted, CRABP2 gained a huge growth in RB cells, including Y79, WERI-RB1 and SO-RB50. It is noted that CRABP2 has the highest expression in WERI-RB1 cells; therefore, we adopted WERI-RB1 cells for the following experiment.
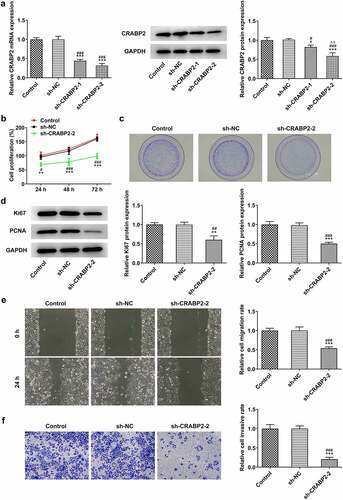
CRABP2 knockdown inhibited proliferation, invasion and migration of RB cells
In order to figure out the effects of CRABP2 knockdown on the proliferation, invasion and migration of RB cells, sh-CRABP2 plasmids were used to transfect WERI-RB1 cells. According to , CRABP2 expression was hugely diminished, especially in sh-CRABP2-2 group. CRABP2 knockdown exhibited suppressive effects on the proliferation (), clone formation ability () of WERI-RB1 cells as well as the expressions of proliferation-related proteins, such as Ki67 and PCNA (). In addition, the invasion and migration of WERI-RB1 cells was also suppressed after sh-CRABP2-2 transfection ().
Figure 2. CRABP2 knockdown inhibits proliferation, invasion and migration of RB cells. (a) CRABP2 mRNA and protein expression in WERI-RB1 transfected with sh-CRABP2-1/2 were respectively detected by RT-qPCR analysis and Western blot. *P < 0.05 and ***P < 0.001 vs. control group. #P < 0.05 and ###P < 0.001 vs. sh-NC group. ∆∆P < 0.01 vs. sh-CRABP2-1 group. (b) The proliferation and (c) colon formation ability of WERI-RB1 transfected with sh-CRABP2-2 was analyzed by CCK-8 and colon formation assay. (d) The expression of proliferation related proteins (Ki67 and PCNA) in WERI-RB1 transfected with sh-CRABP2-2 was determined by Western blot. (e) The migration and (f) invasion of WERI-RB1 transfected with sh-CRABP2-2 was analyzed by wound healing and transwell assay. **P < 0.01 and ***P < 0.001 vs. control group. #P < 0.05, ##P < 0.01 and ###P < 0.001 vs. sh-NC group.
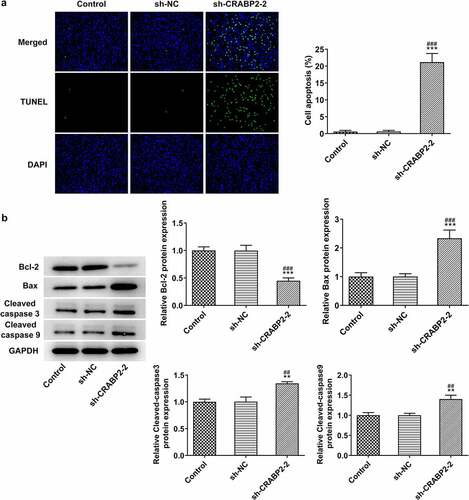
CRABP2 knockdown induced the apoptosis of RB cells
More experiments were conducted to figure out the effects of CRABP2 knockdown on the apoptosis of RB cells. The apoptosis of WERI-RB1 cells transfected with sh-CRABP2-2 was significantly increased (). Correspondingly, the Bcl-2 expression was decreased and the expression of Bax, cleaved-caspase 3 and cleaved-caspase 9 was increased by CRABP2 knockdown in WERI-RB1 cells ().
Figure 3. CRABP2 knockdown aggravates apoptosis of RB cells. (a) The apoptosis of WERI-RB1 transfected with sh-CRABP2-2 was analyzed by Tunel assay. (b) The expression of apoptosis related proteins (Bcl-2, Bax, cleaved-caspase 3 and cleaved-caspase 9) in WERI-RB1 transfected with sh-CRABP2-2 was determined by Western blot. **P < 0.01 and ***P < 0.001 vs. control group. ##P < 0.01 and ###P < 0.001 vs. sh-NC group.
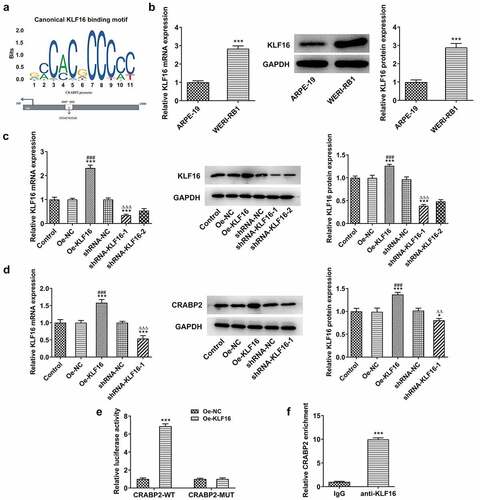
KLF16 positively modulated CRABP2 transcription in RB
To explore the potential mechanism of the regulatory role of CRABP2 in RB, a connection between KLF16 and CRABP2 was predicted by JASPAR, and the binding sites between KLF16 and CRABP2 are shown as . KLF16 mRNA and protein expression in WERI-RB1 cells were increased compared with that in ARPE-19 cells (). The KLF16 mRNA and protein expression were upregulated in WERI-RB1 cells transfected with Oe-KLF16 and downregulated in WERI-RB1 cells transfected with shRNA-KLF16-1/2. KLF16 mRNA and protein expression were lower in shRNA-KLF16-1 group than that in shRNA-KLF16-2 group (). KLF16 overexpression upregulated the CRABP2 expression and KLF16 knockdown downregulated the CRABP2 expression (). The luciferase activity was increased in WERI-RB1 cells co-transfected CRABP2-WT and Oe-KLF16 and was not changed obviously in WERI-RB1 cells co-transfected CRABP2-MUT and Oe-KLF16 (). The result of confirmed the combination of KLF16 and CRABP2 ().
Figure 4. KLF16 positively modulates CRABP2 transcription in RB. (a) The binding sites between KLF16 and CRABP2. (b) KLF16 mRNA and protein expression in WERI-RB1 cells and ARPE-19 cells were respectively detected by RT-qPCR analysis and Western blot. ***P < 0.001 vs. ARPE group. (c) KLF16 mRNA and protein expression in WERI-RB1 transfected with Oe-KLF16 or sh-KLF16-1/2 were respectively detected by RT-qPCR analysis and Western blot. (d) CRABP2 mRNA and protein expression in WERI-RB1 transfected with Oe-KLF16 or sh-KLF16-1 were respectively detected by RT-qPCR analysis and Western blot. *P < 0.05 and ***P < 0.001 vs. control group. ###P < 0.001 vs. Oe-NC group. ∆∆P < 0.01 and ∆∆∆P < 0.001 vs. shRNA-NC group. (e) The interaction between KLF16 and CRABP2 was analyzed by dual-luciferase reporter assay. ***P < 0.001 vs. Oe-NC group. (f) The binding ability of KLF16 to CRABP2 promoter was detected by ChIP. ***P < 0.001 vs. IgG group.
KLF16 overexpression reversed the effects of CRABP2 knockdown on proliferation, invasion, migration and apoptosis of RB cells
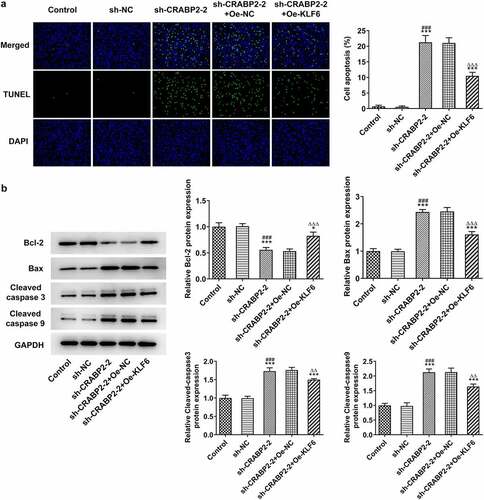
Next, rescue experiments were conducted to ensure the critical role of KLF16 underlying the regulatory effects of CRVBP2 on RB cells. KLF16 overexpression improved the proliferation and clone formation ability of sh-CRABP2-2 transfected WERI-RB1 cells () as well as increased Ki67 and PCNA expressions (). Furthermore, KLF16 overexpression enhanced the invasion and migration of sh-CRABP2-2 transfected WERI-RB1 cells (). As demonstrated, KLF16 overexpression suppressed the apoptosis of sh-CRABP2-2 transfected WERI-RB1 cells, and KLF16 overexpression increased the Bcl-2 expression and decreased the expression of Bax, cleaved-caspase 3 and cleaved-caspase 9 ().
Figure 5. KLF16 overexpression reverses the effects of CRABP2 knockdown on proliferation, invasion and migration of RB cells. (a) The proliferation and (b) colon formation ability of WERI-RB1 transfected with sh-CRABP2-2 and Oe-KLF16 was analyzed by CCK-8 and colon formation assay. (c) The expression of proliferation related proteins (Ki67 and PCNA) in WERI-RB1 transfected with sh-CRABP2-2 and Oe-KLF16 was determined by Western blot. (d) The migration and (e) invasion of WERI-RB1 transfected with sh-CRABP2-2 and Oe-KLF16 was analyzed by wound healing and transwell assay. *P < 0.05, **P < 0.01 and ***P < 0.001 vs. control group. ##P < 0.01 and ###P < 0.001 vs. sh-NC group. ∆∆P < 0.01 and ∆∆∆P < 0.001 vs. sh-CRABP2-2+ Oe-NC group.
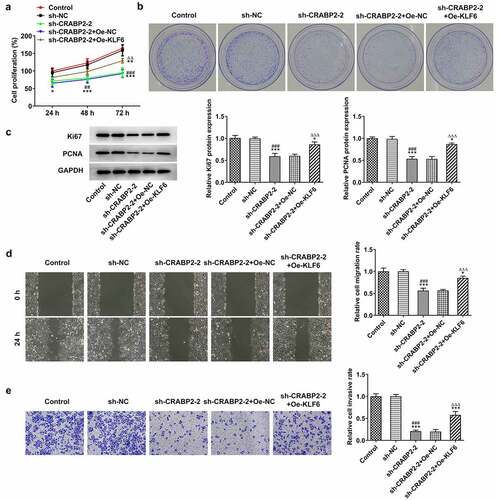
Figure 6. KLF16 overexpression reverses the effects of CRABP2 knockdown on apoptosis of RB cells. (a) The apoptosis of WERI-RB1 transfected with sh-CRABP2-2 and Oe-KLF16 was analyzed by Tunel assay. (b) The expression of apoptosis related proteins (Bcl-2, Bax, cleaved-caspase 3 and cleaved-caspase 9) in WERI-RB1 transfected with sh-CRABP2-2 and Oe-KLF16 was determined by Western blot. *P < 0.05 and ***P < 0.001 vs. control group. ###P < 0.001 vs. sh-NC group. ∆∆P < 0.01 and ∆∆∆P < 0.001 vs. sh-CRABP2-2+ Oe-NC group.
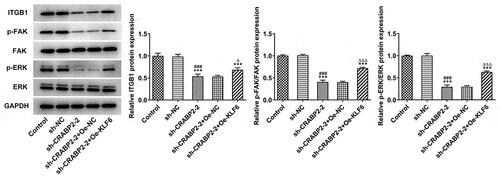
CRABP2 knockdown suppressed the integrin-β1/FAK/ERK pathway
Finally, a potential regulatory pathway was detected to further explore the potential mechanism. CRABP2 knockdown exerted suppressive effects on the expressions of ITGB1, p-FAK/FAK and p-ERK/ERK, which was reversed by KLF16 overexpression in WERI-RB1 cells ().
Discussion
As a primary intraocular malignant tumor, RB manifests leukocoria and strabismus, accounting for about 3% of children’s cancers. It is prone to intracranial and distant metastasis, posing severe threats for children’s life and health [Citation24]. Despite that there are many effective methods to treat RB, the mortality rate and recurrence rate are still high and the prognosis is poor [Citation25]. The emergence of gene therapy provides a new way for the treatment of RB.
CRABP2 is an intracellular lipid junction protein that binds to RA and is thought to be capable of regulating the RA pathway in cells [Citation26]. The specific combination of RA and its related molecules with some distinct cytoplasmic proteins will promote the RA pathway, which acts as a vital player in the progression of differentiation, apoptosis and proliferation [Citation27]. CRABP2 was found to exist in numerous tumors, but varies in the level of expression and function in different types of tumors. Studies [Citation28–30] have found that the decreased expression of CRABP2 in prostate cancer, head and neck squamous cell carcinoma, breast cancer and other tumors can lead to tumor genesis and development. Nevertheless, other studies [Citation31,Citation32] have found that elevated CRABP2 expression is a promoter of tumor development in pancreatic ductal carcinoma and nephroblastoma. Our study indicates that CRABP2 expression was highly expressed in RB cells and CRABP2 knockdown could suppress the proliferation, invasion and migration but promote the apoptosis of RB cells.
HuR is a member of the RNA-binding protein family. A study has found that in the case of RA deficiency, CRABP2 binds to HuR in the cytoplasm to improve the stability and transcription level of its target genes, thereby increasing the transcription level of downstream target genes and increasing protein translation [Citation33]. It has been reported in literatures that there is an interaction between CRABP2 and HuR in tumor cells [Citation34–36]. Integrin β1 is a known downstream of HuR in Jurkat T cells [Citation37], and molecules such as FAK and ERK in integrin signaling pathway are crucial to migration and anoikis resistance of cancer cells [Citation38–41]. Wu et al. revealed that CRABP2 enhanced the migration and anoikis resistance of metastatic lung cancer cells via activating integrin β1/FAK/ERK signaling [Citation21]. Integrin-β1 is essential to cell adhesion, proliferation and metastasis [Citation42–44]. Furthermore, cell to matrix adhesion promotes proliferation and invasion by integrin-β1/FAK/ERK signaling [Citation45]. Here, we also found that CRABP2 knockdown suppressed the integrin β1/FAK/ERK pathway. The proliferation, invasion and migration of RB cells were suppressed, and apoptosis of RB cells was promoted by CRABP2 knockdown maybe related to the inactivation of integrin β1/FAK/ERK pathway.
Previous study indicated that overexpression of KLF16 enhanced the proliferation, growth and migration of RB cells [Citation20]. In this study, KLF16 was demonstrated to positively modulate CRABP2 transcription in RB. Moreover, KLF16 overexpression reversed the effects of CRABP2 knockdown on the proliferation, invasion and migration and apoptosis of RB cells, and inactivation of integrin β1/FAK/ERK pathway.
However, there are still some limitations in the present study. Firstly, examining the expression level of CRABP2 and KLF16 in human samples, as well as analyzing the correlation among CRABP2, KLF16 and clinicopathological parameters, is beneficial to enrich our study goal. Secondly, confirmation of the mechanism in the animal model can further validate our results, which will be conducted in our future work.
Conclusion
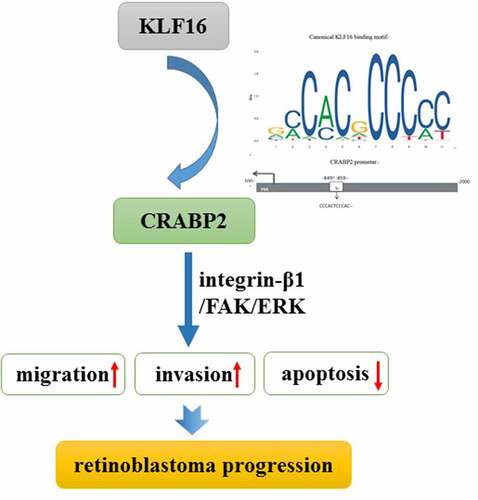
In conclusion, this study demonstrated the role of KLF16/CRABP2 in regulating the biological progress of RB cells. KLF16 transcriptional up-regulation of CRABP2 promoted the invasion and migration and inhibited apoptosis of RB cells by activating integrin-β1/FAK/ERK pathway. KLF16/CRABP2 might serve as a potential prognostic biomarker and a therapeutic target for RB.
Declarations
Availability of Data and Material
The experimental data will be available on the request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Dimaras H, Corson TW, Cobrinik D, et al. Retinoblastoma. Nat Rev Dis Primers. 2015;1(1):15021.
- Abramson DH, Schefler AC. Update on retinoblastoma. Retina (Philadelphia, Pa). 2004;24(6):828–848.
- Knudson AG Jr. Mutation and cancer: statistical study of retinoblastoma. Proc Natl Acad Sci U S A. 1971;68(4):820–823.
- Broaddus E, Topham A, Singh AD. Incidence of retinoblastoma in the USA: 1975-2004. Br J Ophthalmol. 2009;93(1):21–23.
- Leal-Leal C, Flores-Rojo M, Medina-Sansón A, et al. A multicentre report from the Mexican retinoblastoma group. Br J Ophthalmol. 2004;88(8):1074–1077.
- Naseripour M, Nazari H, Bakhtiari P, et al. Retinoblastoma in Iran: outcomes in terms of patients’ survival and globe survival. Br J Ophthalmol. 2009;93(1):28–32.
- Seregard S, Lundell G, Svedberg H, et al. Incidence of retinoblastoma from 1958 to 1998 in Northern Europe: advantages of birth cohort analysis. Ophthalmology. 2004;111(6):1228–1232.
- Kivelä T. The epidemiological challenge of the most frequent eye cancer: retinoblastoma, an issue of birth and death. Br J Ophthalmol. 2009;93(9):1129–1131.
- Chantada GL, Qaddoumi I, Canturk S, et al. Strategies to manage retinoblastoma in developing countries. Pediatr Blood Cancer. 2011;56(3):341–348.
- Canturk S, Qaddoumi I, Khetan V, et al. Survival of retinoblastoma in less-developed countries impact of socioeconomic and health-related indicators. Br J Ophthalmol. 2010;94(11):1432–1436.
- Golabchi K, Soleimani-Jelodar R, Aghadoost N, et al. MicroRNAs in retinoblastoma: potential diagnostic and therapeutic biomarkers. J Cell Physiol. 2018;233(4):3016–3023.
- Mallikarjuna K, Sundaram CS, Sharma Y, et al. Comparative proteomic analysis of differentially expressed proteins in primary retinoblastoma tumors. Proteomics Clin Appl. 2010;4(4):449–463.
- Sessler RJ, Noy N. A ligand-activated nuclear localization signal in cellular retinoic acid binding protein-II. Mol Cell. 2005;18(3):343–353.
- Delva L, Bastie JN, Rochette-Egly C, et al. Physical and functional interactions between cellular retinoic acid binding protein II and the retinoic acid-dependent nuclear complex. Mol Cell Biol. 1999;19(10):7158–7167.
- Kim DJ, Kim WJ, Lim M, et al. Plasma CRABP2 as a novel biomarker in patients with non-small cell lung cancer. J Korean Med Sci. 2018;33(26):e178.
- Chen Q, Tan L, Jin Z, et al. Downregulation of CRABP2 inhibit the tumorigenesis of hepatocellular carcinoma in vivo and in vitro. Biomed Res Int. 2020;2020:3098327.
- Fornes O, Castro-Mondragon JA, Khan A, et al. JASPAR 2020: update of the open-access database of transcription factor binding profiles. Nucleic Acids Res. 2020;48(D1):D87–d92.
- Daftary GS, Lomberk GA, Buttar NS, et al. Detailed structural-functional analysis of the Krüppel-like factor 16 (KLF16) transcription factor reveals novel mechanisms for silencing Sp/KLF sites involved in metabolism and endocrinology. J Biol Chem. 2012;287(10):7010–7025.
- Huang ZH, Luo WZ, Cai XD. Clinical significance of KLF16 expression in patients with lung adenocarcinoma. Chin J Physiol. 2013;29(11):1978–1983.
- Zhang JX, Yan XJ, Wu S, et al. KLF16 overexpression deleteriously affects the proliferation and migration of retinoblastoma by transcriptionally repressing BCL2L15. Biochem Biophys Res Commun. 2020;529(4):977–983.
- Wu JI, Lin YP, Tseng CW, et al. Crabp2 promotes metastasis of lung cancer cells via HuR and Integrin β1/FAK/ERK signaling. Sci Rep. 2019;9(1):845.
- Jahan R, Macha MA, Rachagani S, et al. Axed MUC4 (MUC4/X) aggravates pancreatic malignant phenotype by activating integrin-β1/FAK/ERK pathway. Biochim Biophys Acta Mol Basis Dis. 2018;1864(8):2538–2549.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods (San Diego. Calif). 2001;25(4):402–408.
- Yang XL, Hao YJ, Wang B, et al. Long noncoding RNA NORAD promotes the progression of retinoblastoma by sponging miR 136-5p/PBX3 axis. Eur Rev Med Pharmacol Sci. 2020;24(8):4055.
- Sun X, Shen H, Liu S, et al. Long noncoding RNA SNHG14 promotes the aggressiveness of retinoblastoma by sponging microRNA‑124 and thereby upregulating STAT3. Int J Mol Med. 2020;45(6):1685–1696.
- Percicote AP, Mardegan GL, Gugelmim ES, et al. Tissue expression of retinoic acid receptor alpha and CRABP2 in metastatic nephroblastomas. Diagn Pathol. 2018;13(1):9.
- McGrane MM. Vitamin A regulation of gene expression: molecular mechanism of a prototype gene. J Nutr Biochem. 2007;18(8):497–508.
- Okuducu AF, Janzen V, Ko Y, et al. Cellular retinoic acid-binding protein 2 is down-regulated in prostate cancer. Int J Oncol. 2005;27(5):1273–1282.
- Calmon MF, Rodrigues RV, Kaneto CM, et al. Epigenetic silencing of CRABP2 and MX1 in head and neck tumors. Neoplasia. 2009;11(12):1329–1339.
- Yang Q, Wang R, Xiao W, et al. Cellular retinoic acid binding protein 2 is strikingly downregulated in human esophageal squamous cell carcinoma and functions as a tumor suppressor. PLoS One. 2016;11(2):e0148381.
- Xiao W, Hong H, Awadallah A, et al. CRABP-II is a highly sensitive and specific diagnostic molecular marker for pancreatic ductal adenocarcinoma in distinguishing from benign pancreatic conditions. Hum Pathol. 2014;45(6):1177–1183.
- Takahashi M, Yang XJ, Lavery TT, et al. Gene expression profiling of favorable histology Wilms tumors and its correlation with clinical features. Cancer Res. 2002;62(22):6598–6605.
- Abdelmohsen K, Gorospe M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip Rev RNA. 2010;1(2):214–229.
- Vreeland AC, Levi L, Zhang W, et al. Cellular retinoic acid-binding protein 2 inhibits tumor growth by two distinct mechanisms. J Biol Chem. 2014;289(49):34065–34073.
- Vreeland AC, Yu S, Levi L, et al. Transcript stabilization by the RNA-binding protein HuR is regulated by cellular retinoic acid-binding protein 2. Mol Cell Biol. 2014;34(12):2135–2146.
- Wu JI, Lin YP, Tseng CW, et al. Crabp2 promotes metastasis of lung cancer cells via HuR and Integrin beta1/FAK/ERK signaling. Sci Rep. 2019;9(1):845.
- Mukherjee N, Lager PJ, Friedersdorf MB, et al. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol Syst Biol. 2009;5(1):288.
- Frisch SM, Vuori K, Ruoslahti E, et al. Control of adhesion-dependent cell survival by focal adhesion kinase. J Cell Biol. 1996;134(3):793–799.
- Collins NL, Reginato MJ, Paulus JK, et al. G1/S cell cycle arrest provides anoikis resistance through Erk-mediated Bim suppression. Mol Cell Biol. 2005;25(12):5282–5291.
- Miyazaki T, Kato H, Nakajima M, et al. FAK overexpression is correlated with tumour invasiveness and lymph node metastasis in oesophageal squamous cell carcinoma. Br J Cancer. 2003;89(1):140–145.
- Guo W, Giancotti FG. Integrin signalling during tumour progression. Nat Rev Mol Cell Biol. 2004;5(10):816–826.
- Grzesiak JJ, Tran Cao HS, Burton DW, et al. Knockdown of the β(1) integrin subunit reduces primary tumor growth and inhibits pancreatic cancer metastasis. Int J Cancer. 2011;129(12):2905–2915.
- Bouchard V, Harnois C, Demers MJ, et al. B1 integrin/Fak/Src signaling in intestinal epithelial crypt cell survival: integration of complex regulatory mechanisms. Apoptosis. 2008;13(4):531–542.
- Hazlehurst LA, Damiano JS, Buyuksal I, et al. Adhesion to fibronectin via beta1 integrins regulates p27kip1 levels and contributes to cell adhesion mediated drug resistance (CAM-DR). Oncogene. 2000;19(38):4319–4327.
- Reddig PJ, Juliano RL. Clinging to life: cell to matrix adhesion and cell survival. Cancer Metastasis Rev. 2005;24(3):425–439.