ABSTRACT
Long non-coding RNA (lncRNA) breast cancer-associated transcript 54 (BRCAT54) and microRNA-1269b (miR-1269b) are two critical ncRNAs in cancer biology, while their roles in hemangioma are unknown. Our preliminary sequencing data revealed their altered expression in hemangioma and predicted they could interact with each other. This study was therefore carried out to investigate the roles of BRCAT54 and miR-1269b in hemangioma, with a focus on their interaction. In this study, hemangioma samples donated by 20 infantile hemangioma patients at proliferating-phase and 20 infantile hemangioma patients at involuting-phase were used. The expression of BRCAT54 and miR-1269b in hemangioma samples, as well as hemangioma-derived endothelial cells (HDECs) and human umbilical vein endothelial cells (HUVECs) were detected by RT-qPCR. IntaRNA 2.0 was applied to predict the interaction between BRCAT54 and miR-1269b, which was then confirmed by RNA-RNA pulldown assay. Accumulation of BRCAT54 in the subcellular location of HDECs was detected by subcellular fractionation assay. The role of BRCAT54 and miR-1269b in cell proliferation has been explored by the BrdU assay. Compared to proliferating-phase tissues, involuting-phase tissues exhibited decreased expression levels of BRCAT54 and increased expression levels of miR-1269b. HDECs had decreased expression levels of BRCAT54 and increased expression levels of miR-1269b compared to that of HUVECs. In HDECs, BRCAT54, which was detected in both nuclear and cytoplasm fractions, directly interacted with miR-1269b. BRCAT54 and miR-1269b did not affect the expression of each other, while BRCAT54 suppressed the role of miR-1269b in enhancing the proliferation of HDECs. BRCAT54 may sponge miR-1269b to suppress the proliferation of HDECs.
Introduction
As the most common type of tumor originates from the soft tissues in childhood, infantile hemangioma (IH) is also called hemangiomas of infancy [Citation1]. IHs are caused by abnormally increased proliferation of endothelial cells, which is commonly observed in about 1 out of 10 children younger than 12 months old [Citation2]. IHs have the ability to involute after proliferation [Citation3–5]. Therefore, in the past, primary care providers usually assumed patients with IHs will recover without intervention [Citation5]. Unfortunately, without proper treatment, some patients with IHs will develop complications, which may cause chronic pain, or even permanent disfigurement in extreme cases. Therefore, timely treatment is still needed [Citation6]. Patients with IHs are usually treated with medicines, such as beta blockers [Citation7]. However, serious neurological disorders may still develop after active treatment [Citation8]. Therefore, novel therapies are of great importance.
The development of treatment approaches targeting IHs is limited due to the unclear molecular mechanisms [Citation9,Citation10]. Molecular targeted therapy is an emerging novel approach to treat human diseases. Some molecular pathways, such as the NF-κB signaling, have been characterized as potential targets to treat IHs [Citation11]. However, molecular targeted therapy is still under research. The development of IHs involves altered expression of non-coding RNAs (ncRNAs), such as long ncRNAs (lncRNAs) and microRNAs (miRNAs) [Citation12,Citation13]. Some of these ncRNAs exert critical functions in IHs and therefore could be targeted to treat IHs. For instance, lncRNA CASC2 suppresses cell proliferation in IHs [Citation14], while lncRNA OIP5-AS1 promotes cell IH cell proliferation [Citation15]. Previous studies have characterized lncRNA breast cancer-associated transcript 54 (BRCAT54) and miR-1269b as two critical players in cancer biology [Citation16–18], while their roles in hemangioma are unknown. Our preliminary sequencing data revealed their altered expression in hemangioma and predicted that they could interact with each other. Therefore, we speculated that BRCAT54 and miR-1269b may interact with each other to participate in hemangioma. The interaction between lncRNAs and miRNAs in IHs has rarely been studied. Increased elucidation of the molecular mechanisms of hemangioma may boost the development of novel approaches to treat this disease. The present study was then carried out to investigate the role of BRCAT54 and miR-1269b in hemangioma and the crosstalk between them.
Materials and methods
Patient samples
Hemangioma samples were donated by 20 IH patients at proliferating-phase (15 females and 5 males 5.6 ± 1.3 months old) and 20 IH patients at involuting-phase (15 females and 5 males, 5.7 ± 1.2 months old) who received surgical resection at Nanyang Central Hospital in Henan Province from May 2018 to August 2020. This study was approved by the Ethics Committee of this hospital. Patients’ parents signed the informed consent.
Cells and transient transfection
Hemangioma-derived endothelial cells (HDECs) and human umbilical vein endothelial cells (HUVECs) were bought from the Cell Bank of Chinese Academy Sciences (Shanghai, China) and were cultivated in DMEM (Gibco) containing fetal bovine serum (10%) and penicillin–streptomycin (1%). Cell culture was carried out in an incubator with 5% CO2 and with temperature and humidity set to 37 ºC and 95%, respectively.
To overexpress BRCAT54 and miR-1269b, cells were transfected with miR-1269b mimic and/or pcDNA 3.1- BRCAT54 vector through transient transfections achieved with Neon Electroporation Transfection (Thermo Fisher Scientific). In each transfection, 106 cells were transfected with 30 mM vector and/or 80 mM miRNA. The confirmation of overexpression (compared to NC mimic or empty vector transfection group) was performed every 24 h until the end of in vitro cell experiments.
RNA isolation and sample processing
The MagNA Pure LC RNA Isolation Kit (Roche) was used to isolate total RNAs. Briefly, 107 cultivated cells or 0.05 g tissue samples (ground in liquid nitrogen) were incubated with 10 volumes of lysis buffer containing Proteinase K. Magnetic glass particles were used to bind RNA, while DNase I digestion was performed to remove genomic DNA. RNA samples were subjected to Bioanalyzer analysis to determine RNA concentration and integrity. RNA concentrations higher than 1,000 ng/μl and RIN values higher than 9 were achieved in all samples.
Reverse transcriptions (RTs)-qPCR
The synthesis of complementary DNA (cDNA) samples was performed with 2,000 ng total RNA sample through RTs using the qScript cDNA Synthesis Kit (Quantabio). All cDNA samples were tested by amplifying actin genes prior to subsequent assays. The expression of BRCAT54 and miR-1269b was measured by qPCR with 18S rRNA and U6 as the internal control, respectively. The 2−ΔΔCT method [Citation19] was applied to normalize Ct values to their corresponding internal controls. Primer sequences were as follows: 5’-GATATCCCAGAAGGTTATGC-3’ (forward) and 5’-AGGCTCCACCTCCAAACACC-3’ (reverse) for BRCAT54; 5’-CTACCACATCCAAGGAAGCA-3’ (forward) and 5’-TTTTTCGTCACTACCTCCCCG-3’ for 18S rRNA; 5’-CGGCAGCACATATACTAAAAT-3’ (forward) and 5’-GCTTCACGAATTTGCGTGTC-3’ (reverse) for U6; 5’-CTGGATGAGCCATGCTAC-3’ (forward) and poly (T) (reverse) for miR-1269b.
Biotinylated RNAs for RNA-RNA pull-down assay
A T7 promoter vector expression BRCAT54 or NC RNA was used to prepare in vitro transcripts of both BRCAT54 and NC RNA. All in vitro transcriptions were performed using the HiScribe™ T7 High Yield RNA Synthesis Kit (NEB). The 3’-end of both RNA were biotinylated using the Biotin 3’ End DNA Labeling Kit (Life Technologies). Cells were then transfected with these two biotinylated RNAs (Bio-NC and Bio-BRCAT54) through the methods mentioned above. Cells were harvested 6 h later, followed by cell lysis on ice using lysis buffer. Centrifugations were performed on lysates, and the supernatants were collected and incubated with M-280 streptavidin magnetic beads (Sigma) coated with yeast tRNA and BSA (RNase-free). After that, beads were collected and washed with ice-cold PBS. Next, Trizol was used to purify the pulldown samples, followed by RT-qPCR to determine the expression of miR-1269b. Data normalizations were performed using the method mentioned above.
Analysis of BRCAT54 expression in subcellular locations
The preparation of both cytoplasm and nuclear fractions from cells was performed using the cell fractionation kit (Cat #9038, Cell Signaling Technology). This kit separates cells into cytoplasmic, membrane/organelle, and nuclear/cytoskeletal fractions, while only cytoplasm and nuclear fractions were used in this study. The separation of the cytoplasm fraction was performed through a simple centrifugation at 800 g for 15 min on cell lysates. Nuclear fraction, which was the cell pellets after centrifugation, was collected for further nuclear lysis. Both fractions were used for RNA isolation through the methods mentioned above. After the synthesis of cDNA samples, PCR was performed to amplify BRCAT54. The PCR products were subjected to 1.5% agarose gel separations, followed by EB staining and MyECL image analysis.
BrdU cell proliferation assay
DNA synthesis, which can be reflected by the incorporation of BrdU, was determined to reflect cell proliferation. Transfected cells were harvested at 48 h post-transfection. Cell culture was performed in a fresh medium for another 24 h, and then BrdU was added to reach a final concentration of 10 μM. Incubation with BrdU was performed for 2 h. After that the medium was removed, and cells were collected to incubate with peroxidase-coupled anti-BrdU antibody (Sigma–Aldrich). After incubation for 1 h, the cells were further incubated with a peroxidase substrate. Cell proliferation analysis was performed after 30 min of incubation by determining OD values at 450 nm.
Statistics analysis
SPSS 17.0 statistical software (SPSS) was used for the analyses of all datasets. Student’s t-test was used to compare two groups, while multiple independent groups were compared by ANOVA Tukey’s test. The differences were statistically significant when p value < 0.05.
Results
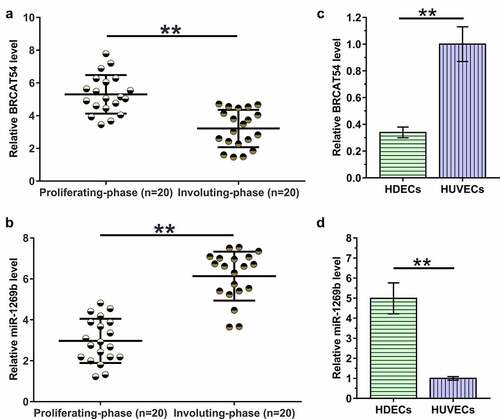
The expression of BRCAT54 and miR-1269b in hemangioma samples, HDECs and HUVECs
To analyze the differential expression of BRCAT54 and miR-1269b in IH, hemangioma samples from 20 IH patients at proliferating-phase and 20 IH patients at involuting-phase were subjected to RNA isolation and RT-qPCR. Compared to proliferating-phase tissues, involuting-phase tissues exhibited decreased expression levels of BRCAT54 (, p < 0.01) and increased expression levels of miR-1269b (, p < 0.01). The expression of BRCAT54 and miR-1269b in HDECs and HUVECs were also detected. The results showed decreased expression levels of BRCAT54 (, p < 0.01), and increased expression levels of miR-1269b (, p < 0.01) in HDECs compared to that in HUVECs. Therefore, BRCAT54 and miR-1269b may participate in the involuting-phase of IH.
Figure 1. Analysis of BRCAT54 and miR-1269b expression in hemangioma samples, HDECs and HUVECs.
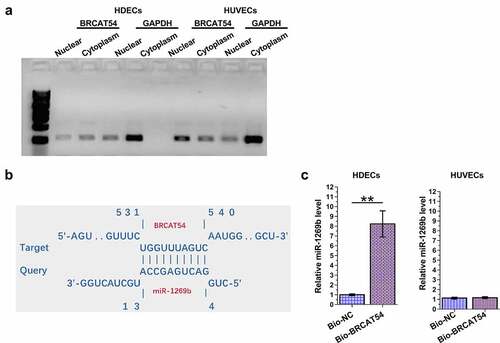
The subcellular location of BRCAT54 in HDECs and HUVECs, and its interaction with miR-1269b
The subcellular location determines the function. The subcellular location of BRCAT54 in HDECs and HUVECs was evaluated by subcellular fractionation assay. The data revealed that BRCAT54 can be detected in both cytoplasm and nuclear samples of both HDECs and HUVECs (). The online program IntaRNA 2.0 has been applied to predict the direct interaction between BRCAT54 and miR-1269b. It was predicted that BRCAT54 and miR-1269b could form multiple-base pairs (). RNA-RNA pulldown assay was performed to detect the direct interaction between BRCAT54 and miR-1269b, compared to Bio-NC group, Bio-BRCAT54 group showed significantly increased expression levels of miR-1269b in HDECs (p < 0.01), but not in HUVECs (). Therefore, BRCAT54 and miR-1269b may directly interact with each other in HDECs.
Figure 2. Analysis of the subcellular location of BRCAT54 in HDECs and HUVECs, and its interaction with miR-1269b.
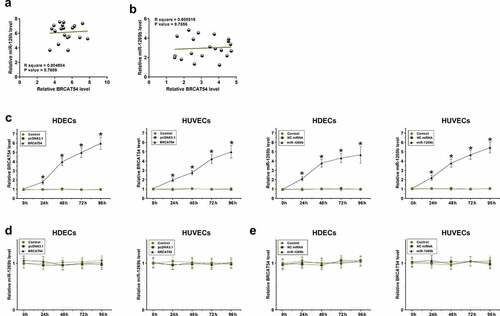
The role of BRCAT54 and miR-1269b in regulating the expression of each other
Correlations indicate possible interactions. The correlations between BRCAT54 and miR-1269b across both proliferating-phase and involuting-phase hemangioma samples were analyzed with Pearson’ correlation coefficient. It was observed that BRCAT54 and miR-1269b were not significantly correlated with each other across proliferating-phase () or involuting-phase () hemangioma samples. BRCAT54 and miR-1269b were overexpressed in HDECs and HUVECs, and the overexpression was confirmed every 24 h until 96 h (, p < 0.05). BRCAT54 did not affect the expression of miR-1269b in both cell types () and miR-1269b showed no role in regulating the expression of BRCAT54 (). Therefore, BRCAT54 was unlikely to be a target of miR-1269b.
Figure 3. Analysis of the role of BRCAT54 and miR-1269b in regulating the expression of each other.
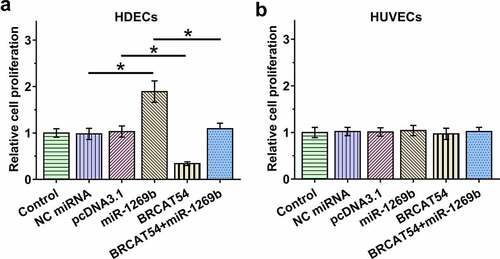
The role of BRCAT54 and miR-1269b in mediating the proliferation of both HDECs and HUVECs
Cell proliferation determines the progression of IH. A BrdU assay was performed to explore the role of BRCAT54 and miR-1269b in regulating the proliferation of both HDECs and HUVECs. BRCAT54 inhibited the proliferation of HDECs, and it also suppressed the role of miR-1269b in enhancing the proliferation of HDECs (, p < 0.05). In contrast, BRCAT54 and miR-1269b showed no role in mediating the proliferation of HUVECs (). Therefore, BRCAT54 may regulate the proliferation of HDECs through miR-1269b.
Discussion
The present study explores the involvement of BRCAT54 and miR-1269b and the direct interaction between them in IHs. Our results show that the expression of BRCAT54 and miR-1269b were altered in IHs. BRCAT54 may sponge miR-1269b to regulate the proliferation of HDECs.
BRCAT54 is a recently characterized lncRNA in non-small cell lung cancer [Citation16]. It was observed that BRCAT54 was downregulated in lung cancer and it regulates the expression of genes involved in the JAK-STAT and calcium pathways by binding to RPS9, thereby suppressing tumorigenesis [Citation16]. However, the involvement of BRCAT54 in other types of cancer is unclear. Our results revealed decreased expression levels of BRCAT54 in hemangioma-derived HDECs compared to that in normal HUVECs. IHs have three development stages, including proliferation, plateau, and involution [Citation20]. Our study also reported decreased expression levels of BRCAT54 in involution stage compared to that in the proliferation stage. Therefore, BRCAT54 is involved in IHs. In addition, the inhibitory effects of BRCAT54 on the proliferation of hemangioma-derived HDECs were observed. Therefore, BRCAT54 may serve as a target to treat IHs by suppressing cell proliferation.
MiR-1269b has been demonstrated to be an oncogenic miRNA in both liver cancer and lung cancer [Citation17,Citation18]. MiR-1269b is highly upregulated in these two types of malignancy, and it suppresses liver cancer growth and metastasis, and reduces the sensitivity of lung cancer cells to cisplatin by regulating the PTEN/PI3K/AKT signaling pathway [Citation17,Citation18]. In this study, we reported the upregulation of miR-1269b in IHs. Moreover, miR-1269b also increased the proliferation of hemangioma-derived HDECs. Therefore, miR-1269b may promote IHs by stimulating cell proliferation.
The present study reported the direct interaction between BRCAT54 and miR-1269b in HDECs, but not in HUVECs, and detected BRCAT54 in both cytoplasm and nuclear of both HDECs and HUVECs. In addition, BRCAT54 suppressed the role of miR-1269b in promoting the proliferation of HUVECs, while these two ncRNAs showed no role in mediating the proliferation of HUVECs. Therefore, BRCAT54 may sponge miR-1269b only in hemangioma-derived HDECs, but not in normal HUVECs, to regulate IHs. Therefore, the overexpression of BRCAT54 may serve as a target to suppress cell proliferation in IHs. However, novel techniques are needed to deliver BRCAT54 into vascular tissues.
Interestingly, BRCAT54 is an inhibitor of cell proliferation, but its accumulation was increased during the proliferation stage. In contrast, the accumulation of miR-1296b, which was an activator of cell proliferation, was decreased in proliferation stage. Studies have shown that cell proliferation inhibitors are usually downregulated in IHs [Citation14], while cell proliferation activators are usually upregulated in IHs [Citation15]. It is speculated that there are multiple inhibitors and activators in IHs, and they could interact to regulate the expression of each other. Our findings reveal the complexity of the molecular mechanism of IHs.
Conclusion
BRCAT54 is downregulated in IHs, and miR-1269b is highly upregulated in IHs. BRCAT54 may sponge miR-1269b to regulate the proliferation of HDECs, thereby participating in IHs. The exogenous application of BRCAT54 may serve as a potential target to treat IHs. However, novel RNA delivery techniques are needed to efficient deliver BRCAT54 to target sites.
Highlights
Involuting-phase tissues exhibited decreased expression levels of BRCAT54 and increased expression levels of miR-1269b;
BRCAT54 was detected in both nuclear and cytoplasmic fractions, and it interacted with miR-1269b;
BRCAT54 suppressed the role of miR-1269b in enhancing the proliferation of HDECs.
Authors’ contributions
Ya Wang designed the study. Zhonjun Lv carried out experiments and wrote the manuscript, Ya Wang revised the paper, Ke Yang collected patient specimens and related information. Zhonjun Lv and Ke Yang contributed to analyzing the data. All authors reviewed the results and approved the final version of the manuscript.
Availability of data and material
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Ethics approval and consent to participate
The Ethics Committee of Nanyang Central Hospital approved this study. Patients’ parents signed informed consent.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Holland KE, Drolet BA. Infantile hemangioma. Pediatr Clin North Am. 2010 Oct;57(5):1069–1083.
- Seiffert A, Schneider M, Roessler J, et al. Incidence, treatment patterns, and health care costs of infantile hemangioma: results of a retrospective German database analysis. Pediatr Dermatol. 2017 Jul;34(4):450–457.
- Chen ZY, Wang QN, Zhu YH, et al. Progress in the treatment of infantile hemangioma. Ann Transl Med. 2019 Nov;7(22):692.
- Chang L, Lv D, Yu Z, et al. Infantile hemangioma: factors causing recurrence after propranolol treatment. Pediatr Res. 2018 Jan;83(1–1):175–182.
- Darrow DH, Greene AK, Mancini AJ, et al. Diagnosis and management of infantile hemangioma. Pediatrics. 2015 Oct;136(4):e1060–104.
- Tiemann L, Hein S. Infantile hemangioma: a review of current pharmacotherapy treatment and practice pearls. J Pediatr Pharmacol Ther. 2020;25(7):586–599.
- Koh SP, Leadbitter P, Smithers F, et al. β-blocker therapy for infantile hemangioma. Expert Rev Clin Pharmacol. 2020 Aug;13(8):899–915.
- Thai T, Wang CY, Chang CY, et al. Central nervous system effects of oral propranolol for infantile hemangioma: a systematic review and meta-analysis. J Clin Med. 2019 Feb 22;8(2):268.
- Boye E, Olsen BR. Signaling mechanisms in infantile hemangioma. Curr Opin Hematol. 2009 May;16(3):202–208.
- Ji Y, Chen S, Li K, et al. Signaling pathways in the development of infantile hemangioma. J Hematol Oncol. 2014 Jan 31;7:13.
- Greenberger S, Adini I, Boscolo E, et al. Targeting NF-κB in infantile hemangioma-derived stem cells reduces VEGF-A expression. Angiogenesis. 2010 Dec;13(4):327–335.
- Liu X, Lv R, Zhang L, et al. Long noncoding RNA expression profile of infantile hemangioma identified by microarray analysis. Tumour Bio. 2016 Oct 5;37(12):15977–15987.
- Li J, Li Q, Chen L, et al. Competitive endogenous RNA networks: integrated analysis of non-coding RNA and mRNA expression profiles in infantile hemangioma. Oncotarget. 2018 Feb 23;9(15):11948–11963.
- Dai Y, Qiu M, Wang S, et al. lncRNA CASC2 suppresses the growth of hemangioma cells by regulating miR-18a-5p/FBXL3 axis. J Biol Regul Homeost Agents. 2020;34(1):49–56.
- Zhang J, Zhao T, Tian L, et al. LncRNA OIP5-AS1 promotes the proliferation of hemangioma vascular endothelial cells via regulating miR-195-5p/NOB1 axis. Front Pharmacol. 2019;10:449.
- Yang W, Qian Y, Gao K, et al. LncRNA BRCAT54 inhibits the tumorigenesis of non-small cell lung cancer by binding to RPS9 to transcriptionally regulate JAK-STAT and calcium pathway genes. Carcinogenesis. 2021 Feb 11;42(1):80–92.
- Chen L, Liu D, Yi X, et al. The novel miR-1269b-regulated protein SVEP1 induces hepatocellular carcinoma proliferation and metastasis likely through the PI3K/Akt pathway. Cell Death Dis. 2020 May 5;11(5):320.
- Yang W, Xiao W, Cai Z, et al. miR-1269b drives cisplatin resistance of human non-small cell lung cancer via modulating the PTEN/PI3K/AKT signaling pathway. Onco Targets Ther. 2020;13:109–118.
- Livak K, Schmittgen T. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25(4):402–408.
- Smith CJF, Friedlander SF, Guma M, et al. Infantile hemangiomas: an updated review on risk factors, pathogenesis, and treatment. Birth Defects Res. 2017 Jul 3;109(11):809–815.