ABSTRACT
SOCS3 is low-expressed in breast cancer and may be a potential target. Ultrasound targeted microbubble destruction (UTMD) improved the efficiency of gene transfection. We explored the effects of UTMD-mediated transfection of SOCS3 on the biological characteristics and epithelial-mesenchymal transition (EMT) of breast cancer stem cells (BCSCs). The expression of SOCS3 in breast cancer (BC) and its association with prognosis were evaluated by GEPIA and The Cancer Genome Atlas (TCGA) websites. BCSCs were sorted by flow cytometry and immunomagnetic bead method, followed by sphere formation, 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) and xenograft assays to test their effects in vitro and in vivo. The levels of SOCS3, EMT- and STAT3 pathway-related genes were determined by RT-qPCR and Western blot, respectively. The effects of liposome and UTMD on BCSCs and mice were compared by the gain-of-function experiments. Low expression of SOCS3 was associated with poor prognosis of BC patients, and found in BC and BCSCs. BCSCs were successfully sorted, with high viability and tumorigenicity. UTMD increased the transfection rate of SOCS3. Moreover, UTMD- and liposome-mediated SOCS3 reduced cell viability, proliferation, migration and invasion, blocked cell cycle, inhibited sphere formation in BCSCs, and retarded tumor growth in mice. Mechanistically, overexpressed SOCS3 inhibited the expressions of EMT-related genes and the activation of STAT3 pathway in BCSCs and mice. The regulatory effects of UTMD-mediated SOCS3 on the above-mentioned biological characteristics were better than liposome-mediated SOCS3. UTMD-mediated SOCS3 has a better therapeutic effect in BC, providing new experimental evidence for the treatment of BC.
Graphical abstract
Introduction
Breast cancer has been reported as the most prevalent malignant tumor in women, accounting for 11.6% of all cancer-related deaths [Citation1]. In 2012, almost 1.7 million people were diagnosed globally and about 50 thousands of people died from this disease [Citation2]. At present, the incidence of breast cancer worldwide is increasing at a rate of 3% per year, and this disease is becoming more prevalent among a younger age group [Citation3]. In addition, about 30% of breast cancer patients will relapse and metastasize in spite of early treatment [Citation4]. The effective rate and long-term survival rate of breast cancer treatment are extremely poor [Citation5], the reason of which lies in that the mechanism concerning the occurrence and development of breast cancer is not clear. Additionally, the known information is not adequate enough to explain the specific biological characteristics of breast cancer and the molecular signaling pathways involved [Citation6]. Therefore, an in-depth study on the mechanism implicated in the occurrence and development of breast cancer is significant so as to come up with new ideas and methods for the targeted therapy of breast cancer.
Cancer stem cells (CSCs) are considered to be a group of stem cell-like cells that exist in a very small proportion of tumors, with high tumorigenicity, expression of specific markers, self-renewal and other biological characteristics [Citation7,Citation8]. The presence of breast cancer stem cells (BCSCs) enhances the proliferation, invasion and metastasis of breast cancer [Citation9]. BCSCs could be recognized by activities of CSC markers (CD44+/CD24− and enzyme aldehyde dehydrogenase (ALDH+)) [Citation10,Citation11]. A previous study has reported that BCSCs with both these CSC markers could show the greatest tumor-initiating capacity [Citation12]. Additionally, breast cancers can be subdivided into luminal breast cancers, HER2+ breast cancers, and basal breast cancers with triple negative (TNBC). The common gene expression profiles in BCSCs across molecular subtypes of breast cancer have been evidenced, indicating that agents targeting BCSCs may have utility across molecular subtypes of breast cancer [Citation13]. In comparison with other breast cancer subtypes, the enrichment of CSCs in combination with the aggressive nature of TNBC, may contribute to the poor prognosis of TNBC patients [Citation14]. At the same time, BCSCs are usually resistant to conventional chemotherapy and are considered to account for the failure in the treatment [Citation15]. Therefore, how to eradicate CSCs has become a hot topic for scholars.
Existing data suggest that Janus kinase (JAK)/activator of transcription (STAT) signaling pathway and the process of epithelial-mesenchymal transition (EMT) are closely related to the occurrence of tumors [Citation16,Citation17]. The activity of JAK/STAT signaling pathway can be inhibited by SOCS protein family [Citation18]. SOCS3 is one of the most active members of the SOCS family and the most important inhibitor in the STAT signal transduction pathway [Citation19]. Analysis of human patient genome database reports that the expression of SOCS3 is positive prognostic marker of breast cancer [Citation20]. SOCS3 also participates in the development and metastasis of breast cancer [Citation21–23]. Besides, it has been reported that anti-miR-203 represses growth and stemness of estrogen receptor (ER)-positive luminal breast cancer cells via targeting SOCS3 [Citation24]. The activation of STAT3 is essential for the induction and maintenance of TNBC stem cell [Citation25]. In addition, with the rapid development of molecular biology and genetic engineering, gene therapy is very likely to become a new therapeutic method for humans to conquer many diseases difficult to cure and malignant tumors [Citation26]. Ultrasound targeted microbubble destruction (UTMD), as a new type of gene delivery method and targeted drug delivery system, can be combined with RNA interference technology and acoustic pulse ultrasound to effectively improve the efficiency and targeting of transfected genes into cells [Citation27,Citation28]. During UTMD, the gene is integrated into a microbubble and then locally released at the targeted tissue by ultrasound triggering [Citation29]. The destruction of microbubble by ultrasound triggering induces an increase in capillary permeability and induces irreversible holes in the membranes of target cells, which helps improve the transference of gene into the nucleus and promotes the expression and transfection of the interested gene [Citation30]. Additionally, UTMD has shown great potential in cancer immunotherapy as aiding method [Citation31], which is expected to become an efficient, safe, and targeted method for both gene transfection and gene therapy [Citation29].
Based on the reports above, we hypothesize that SOCS3-mediated EMT process of breast cancer may be associated with JAK/STAT signaling pathway, and that UTMD-mediated SOCS3 may enhance the effects of SOCS3 in breast cancer cells. Therefore, the aim of this study was to explore the effects and mechanism of UTMD-mediated transfection of SOCS3 on the biological characteristics and EMT as well as the tumorigenic ability of BCSCs. The goal of this work is to investigate the feasibility of UTMD-mediated SOCS3 therapy on breast cancer.
Materials and methods
Ethics statement and animals
All animal experiments were executed in accordance with the guidelines of the China Council on Animal Care and Use. The experiments with animals had been reviewed by the Institutional Animal Care and Use Committee of Nanfang Hospital (BD201910029). Food and water were provided ad libitum, and every effort was made to minimize any pain or discomfort to the animals.
Sixty-six female BALB/c nude mice (4–6 weeks old, 18–20 g) were obtained from Jiangsu ALF Biotechnology Co., LTD. (China) and raised in SPF-level animal laboratory. The conditions of feeding were adjusted to constant temperature (22–25°C) and humidity (55 ± 5%), and the food and water used were sterilized.
Bioinformatics assay
Based on the GEPIA (http://gepia.cancer-pku.cn/index.html) website, we analyzed and evaluated the expression of SOCS3 in the database of The Cancer Genome Atlas (TCGA)-BRCA (Breast Invasive Carcinoma, including 1085 tumor samples and 291 normal samples) (https://www.cancer.gov/) and the impact of SOCS3 expression on the survival of patients with breast cancer using Kaplan-Meier survival analysis. To construct the survival curve, patients were divided into two groups according to the median expression level of SOCS3.
Isolation and identification of BCSCs
The human breast cancer cell line (MCF-7) is a breast cancer-derived cell line widely used to study breast cancer. MCF-7 cell line (CRL-3435, American Type Culture Collection, USA) was adopted for our study and cultured in DMEM/F12 medium (ATCC, 30–2006) containing 10% fetal bovine serum (FBS, ATCC, 30–2021) and 100 U/ml penicillin-streptomycin (ATCC, 30–2300) at 37°C, 5% CO2 in an incubator (SCO6WE-2, SHELLAB, USA). The cells were detached using a mixture of EDTA and trypsin (T4049, Sigma-Aldrich, USA) to prepare a single cell suspension. For stem-like characteristics of the cells, 4 × 104 cells per well were seeded in an ultra-low adhesion 6-well culture plate (3471, Corning, USA) with DMEM/F12 serum-free culture medium (SFM) containing 20 μg/L EGF (SRP3027, Sigma-Aldrich), 20 μg/L b-FGF (450–33, Peprotech, USA) and 2% B27 (17504044, Gibco, USA). The cells were collected after 7–10 days.
CD44+CD24− BCSCs were sorted by fluorescence activated cell sorting (FACS) using a FACS Calibur flow cytometer (BD Biosciences, San Jose, CA, USA) or sorted by magnetic activated cell sorting (MACS). For FACS [Citation32], fluorescence-labeled antibodies against CD44 (PE, ab269300, Abcam, UK) and CD24 (FITC, ab30350) were added to the cells and incubated for 30 minutes (min) in the dark, and cells were then sorted by a flow cytometer to collect CD44+CD24− BCSCs and the rest of the non-stem cell population cells (non-BCSCs). For both FACS and flow cytometry analysis, gates were established and fluorescent compensation performed using single color stained samples and unstained control. All FACS data were analyzed by postcollection compensation with FowJo vv10.0.4 software (Tree Star Inc, Ashland, OR, USA).
For MACS, 10 μg of primary antibody of CD24 (ab202073) was added to the cell suspension and incubated at 4°C for 30 min. 40 μl of IgG immunomagnetic beads (130–095-951, Miltenyi Biotec) were added to the resuspended cells, followed by the mixture and incubation at 4°C for 30 min in the dark. 500 μl of cell suspension was poured into the sorting column (LS magnetic columns (130–042-401, Miltenyi Biotec in a MidiMACS Separator (130–042-302, Miltenyi Biotec) on the sorting rack to collect CD24− cells. Subsequently, the cells were added with 10 μg of primary antibody against CD44 (ab189524) and incubated at 4°C for 30 min. 40 μl of IgG immunomagnetic beads (130–095-194, Miltenyi Biotec) were added to the resuspended cells, mixed, and incubated at 4°C for 30 min. The new sorting column was put into the sorting rack, where 500 μl of cell suspension was poured to collect CD44+CD24− cells.
The fresh isolated cells were collected and cultured in the SFM. A testing diagram is presented in Supplementary Figure S1 to illustrate the experimental design.
Sphere formation experiment
Differently sorted cell populations were collected and seeded in ultra-low adhesion 6-well plates at 5000 cells/well, and 4 μg/ml heparin and 0.48 μg/ml hydrocortisone were added to MammoCult complete medium (05620, STEMCELL Technologies, Canada) and cells were cultured for 7–10 days. Mammary spheres with a diameter larger than 60 μm were observed and counted with an inverted microscope (ECLIPSE Ts2, Nikon, Japan) (magnification×200).
Cell viability assay
MTT assay kit was used for determining the viability of the BCSCs. BCSCs were seeded in 96-well plates containing SFM for 24, 48 or 72 h. Then MTT reagents (10 μl) were added and cells were further cultured for 4 h, following which 110 μl of formazan solution were added for a 10-min incubation. Thereafter, the OD value at an absorbance of 490 nm was read by a SpectraMax5 microplate reader (Molecular Devices, USA).
Transfection
The recombinant plasmid for SOCS3 overexpression (SOCS3, C05003) and its negative control (NC, A06001) were obtained from Shanghai GenePharma Company (China). The BCSCs were randomly divided into 5 groups: Control, liposome (LIP)-NC, LIP-SOCS3, UTMD-NC, and UTMD-SOCS3 groups. Control group: cells were cultured routinely; LIP-NC group: cells were transfected with liposome-mediated NC plasmid (1 μg plasmid + 2 μl Lipofectamine 2000 (11668019, Invitrogen, USA)); LIP-SOCS3 group: cells were transfected with liposome-mediated SOCS3 overexpression plasmid; UTMD-NC group: cells were transfected with UTMD-mediated NC plasmid; UTMD-SOCS3 group: cells were transfected with UTMD-mediated SOCS3 overexpression plasmid. Ultrasound contrast agent SonoVue was purchased from Bracco (Italy). After preheating for 30 min with the ultrasonic therapy device (Microwave25P, Fysiomed, Belgium), the cells of the UTMD group were subjected to ultrasonic treatment (frequency: 1 MHz, intensity: 0.75 W/cm2, time: 45 seconds (s)) [Citation33].
Cell cycle assay
The cell cycle of BCSCs in each group was detected with DNA Content Quantitation Assay (Cell Cycle) (CA1510, Solarbio, China). The BCSCs of each group were made into single cell suspension (1 × 106 cells/ml), then centrifuged to remove the supernatant and fixed with 70% pre-cooled ethanol for 2 h. BCSCs were subsequently resuspended in a water bath at 37°C for 30 min. Then, 400 µl of PI staining solution was added to the mix, which was incubated at 4°C for 30 min in the dark. A flow cytometer was used for the detection and recording.
Colony formation assay
BCSCs (1 × 103 cells/well) were seeded in 6-well culture plates and cultured for about 2 weeks. Afterward, the colonies formed were fixed with 4% paraformaldehyde (158,127, Sigma-Aldrich, USA), and stained with 0.1% crystal violet (C8470, Solarbio, China), followed by the calculation and analysis using Olympus IX73 microscope (Olympus, Japan).
Wound healing assay
The suspensions of BCSCs (about 1 × 105 cells/well) were seeded in 6-well plates and cultured. After BCSCs reached a confluence of 90%, a 200 μl pipette tip was used to scratch the cell layers to form wounded gaps. After 24 h of culture, the wounded gaps in cells of each group were photographed and calculated with Japan Nikon ECLIPSE Ts2 microscope (magnification × 100) and Adobe Illustrator software (USA).
Transwell assay
The invasion ability of BCSCs was assessed by Transwell assay using Transwell chamber (3422, Corning, USA) covered with Matrigel (354230, BD, USA). The suspension of BCSCs (1 × 105 cells/ml) in serum-free medium was added to the upper chamber, while the medium containing 10% FBS was put into the lower chamber. After being cultured 24 h, the invading cells were fixed with 4% paraformaldehyde, and stained with 0.1% crystal violet, and photographed under a microscope (magnification × 250).
RNA isolation and RT-qPCR
Total RNA of BCSCs or tumor tissues of mice was extracted with RNA Extraction Kit (R1200, Solarbio, China). Its integrity was detected by agarose gel and its quantity was determined in NanoDrop-1000 spectrophotometer (Thermo Scientific, Wilmington, DE, USA). The RNA was reverse-transcribed to cDNA with a RT-PCR kit (KR123, TIANGEN, China). The SuperReal PreMix Plus (FP205, TIANGEN, China) was utilized for the conduction of RT-qPCR in ABI Prism 7500 HT detection system (Applied Biosystems, China). The expression of RNA was quantified by the 2−ΔΔCt method [Citation11] and normalized to GAPDH. All the sequences of primers were listed as follows (5’-3’). SOCS3: CCTGCGCCTCAAGACCTTC, GTCACTGCGCTCCAGTAGAA; GAPDH: GCACCGTCAAGGCTGAGAAC, TGGTGAAGACGCCAGTGGA; E-Cadherin: CGAGAGCTACACGTTCACGG, GGGTGTCGAGGGAAAAATAGG; Vimentin: GACGCCATCAACACCGAGTT, CTTTGTCGTTGGTTAGCTGGT; N-Cadherin: TTTGATGGAGGTCTCCTAACACC, ACGTTTAACACGTTGGAAATGTG. For mice, the sequences of primers were additionally listed as follows (5’-3’). SOCS3: ATGGTCACCCACAGCAAGTTT,TCCAGTAGAATCCGCTCTCCT; GAPDH: AGGTCGGTGTGAACGGATTTG, TGTAGACCATGTAGTTGAGGTCA. The sequences of primers were obtained from PrimerBank (https://pga.mgh.harvard.edu/primerbank).
Western blot
Total protein in BCSCs or tumor tissues of mice was extracted with a total protein extraction kit (BC3711, Solarbio, China) and quantified with BCA kit (P0011, Beyotime, China). Proteins were detached by 12% SDS-PAGE gel, transferred onto the PVDF membrane (160–0184, Bio-Rad, USA) and blocked for 2 hours (h). Later, the sealed membranes were separately probed with primary antibodies and secondary antibodies. The primary antibodies used here including those against SOCS3 (ab3693, 1 µg/ml, 27 kDa), E-cadherin (ab1416, 1/50, 110 kDa), Vimentin (ab92547, 1/1000, 54 kDa), N-Cadherin (ab18203, 1 µg/ml, 130 kDa), p-STAT3 (ab76315, 1/2000, 88 kDa), STAT3 (ab119352, 1/5000, 88 kDa), cyclin D1 (ab134175, 1/10,000, 34 kDa), PCNA (ab92552, 1/1000, 29 kDa), GAPDH (ab181602, 1/10,000, 36 kDa), and the secondary antibodies were those against rabbit IgG (ab205718) and mouse IgG (ab205719). Afterward, BeyoECL Plus (P0018M, Beyotime) was employed for the visualization and exposure. The images were quantified by ImageJ software (http://imagej.nih.gov/ij/).(The expressions of proteins were normalized to GAPDH.
Tumorigenesis assay
Animal experiments were divided into two parts.
For the first part [Citation28], MCF-7 cells, non-BCSCs and BCSCs were resuspended into the suspension at the concentration of 1 × 103 cells/200 μl or 1 × 104 cells/200 μl for use. BALB/c nude mice were randomly divided into 3 groups (n = 6 for each group), and 1 × 103 or 1 × 104 cells were inoculated into the subcutaneous tissues of both hind limbs of BALB/c nude mice. The tumor formation of the BALB/c nude mice was observed every other day. Six weeks later, the BALB/c nude mice were deeply anesthetized by sodium pentobarbital [150 mg/kg, intraperitoneal injection, (B5646-50 mg, ApexBio, USA)] and sacrificed by cervical dislocation.
In the second part, the BCSCs of each group were collected after transfection and made into a cell suspension of 2 × 105 cells/ml. BALB/c nude mice were randomly divided into 5 groups (n = 6 for each group), and 200 μl of cell suspension were slowly injected subcutaneously. The tumor formation under the skin of BALB/c nude mice was observed every other day, and the length and width of the tumor were recorded with vernier calipers at 0, 15, 19, 27, and 35 days. After 35 days, the BALB/c nude mice were anesthetized with sodium pentobarbital and sacrificed by cervical dislocation. The transplanted tumor was carefully peeled off, the weight of which was calculated and the photo of which was taken and recorded. The tumor samples were quickly frozen for subsequent analyses with RT-qPCR and Western blot.
Data analysis
The statistical data were expressed by the mean ± SD from at least three experiments and analyzed using Graphpad prism 8.0. Kolmogorov-Smirnov test was used to verify normality. The one-way analysis of variance (ANOVA) or two-way ANOVA was used for the comparison, followed by the Tukey multiple-comparison test. P < 0.05 was considered as statistically significant.
Results
SOCS3 may be a potential target in breast cancer, and UTMD improved the efficiency of gene transfection. We hypothesize that SOCS3-mediated EMT process of breast cancer may be associated with JAK/STAT signaling pathway, and UTMD-mediated SOCS3 may enhance the effects of SOCS3. The goal of this work is to investigate the feasibility of UTMD-mediated SOCS3 therapy on breast cancer. This work showed that UTMD-mediated SOCS3 attenuated biological characteristics and EMT of BCSCs and tumorigenic ability of breast cancer than liposome-mediated SOCS3, which was related to JAK/STAT signaling pathway.
The expression of SOCS3 in breast cancer and its relationship with prognosis
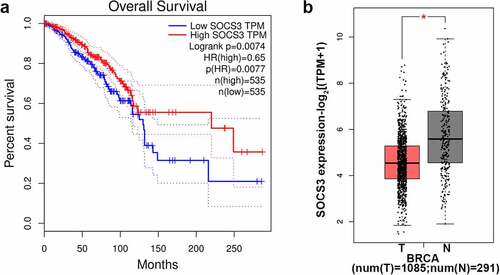
Based on the GEPIA website, we analyzed and evaluated the SOCS3 expression in the TCGA-BRCA database and the impact of SOCS3 expression on the survival of patients with breast cancer. The results showed that low expression of SOCS3 was significantly associated with the poor prognosis of patients with breast cancer (P = 0.0074, )). Meanwhile, when compared with the normal group, SOCS3 was evidently lower expressed in samples of BRCA (P < 0.05, )).
Figure 1. The expression of SOCS3 in breast cancer and its association with prognosis of patients with breast cancer. (a-b) Based on the GEPIA (http://gepia.cancer-pku.cn/index.html) website, we analyzed and evaluated the expression of SOCS3 in the TCGA-BRCA database (Breast Invasive Carcinoma, including 1085 tumor samples and 291 normal samples). (a) The impact of SOCS3 expression on the survival of patients with breast cancer. (b) The different expression of SOCS3 in BRCA and normal samples of people. *P < 0.05 vs. T. T, tumor samples in BRCA. N, normal samples.
Identification and biological characteristics of BCSCs
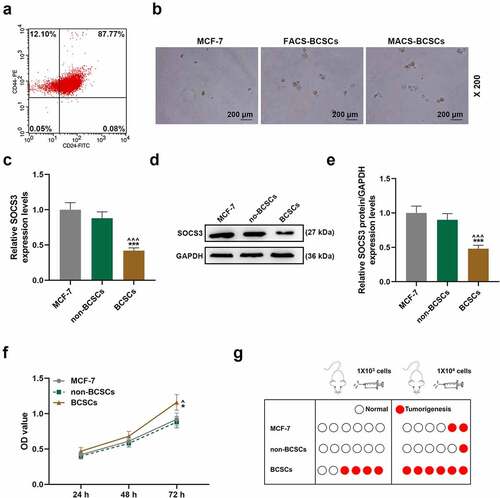
Next, we used FACS and MACS to sort out CD44+CD24− BCSCs and non-BCSCs ()). There were sorted BCSCs with 12.1% purity. No significant difference was found in the spheroidization rate of BCSCs obtained by the two sorting methods, but they were significantly higher than MCF-7 cells (magnification × 200, )). Subsequently, we detected the expression of SOCS3 in MCF-7 cells, non-BCSCs and BCSCs and found that the expression of SOCS3 in BCSCs was significantly lower than those in MCF-7 cells and non-BCSCs (P < 0.001, )). In addition, we verified the cell viability of different cells and their effects on the tumorigenicity in BALB/c nude mice. It was found that the viability of BCSCs was significantly higher than that of MCF-7 cells and non-BCSCs, and the tumorigenicity of BCSCs in BALB/c nude mice was significantly higher than that of MCF-7 cells and non-BCSCs (P < 0.05, )).
Figure 2. The isolation of breast cancer stem cell and the detection of biological characteristics. (a) Flow cytometry was used to sort CD44+/CD24- breast cancer stem cells (BCSCs). (b) The formation of mammary glands of MCF-7 cells and sorted BCSCs was measured by sphere formation assay (magnification × 200). (c-e) The expression of SOCS3 in MCF-7 cells, non-BCSCs and BCSCs were measured by RT-qPCR and Western blot, respectively. Each experiment was repeated three times independently. RT-qPCR: Real-time quantitative PCR. GAPDH was set as control. (f) MTT assay was constructed to detect the viability of each group of cells at 24 h, 48 h, 72 h. (g) The nude mouse tumor xenotransplantation experiment was used to detect the tumorigenic ability of cells in each group of within nude mice. ^P < 0.05, ^^^P < 0.001 vs. MCF-7; *P < 0.05, ***P < 0.001 vs. non-BCSCs.
Effects of SOCS3 transfection with liposomes and UTMD on the biological characteristics of BCSCs
UTMD is a novel gene delivery method and targeted drug delivery system, while liposome transfection is a conventional method of transfection. We found that the transfection of both UTMD and lipoplasmid can successfully up-regulate the expression of SOCS3, and the transfection of SOCS3 overexpression vector using UTMD significantly promotes the expression of SOCS3, the effects of which were better than the transfection using liposome (P < 0.01, )). Compared with their respective control groups, the viability of cells in UTMD-SOCS3 and LIP-SOCS3 groups were significantly reduced, and the inhibitory effect of UTMD-SOCS3 on cell viability was obviously stronger than that of LIP-SOCS3 (P < 0.01, )).
Figure 3. The comparison on the effects of liposome and UTMD transfection with SOCS3 on the viability, cell cycle and sphere formation of BCSCs. BCSCs were separately transfected with NC or SOCS3 overexpression plasmid using liposome (LIP) (1 μg plasmid + 2 μL Lipofectamine 2000) or ultrasound microbubbles (ultrasound radiation conditions: 1 MHz, 0.75 W/cm, 45 s). (a-c) RT-qPCR and Western blot were used to quantify the expression of SOCS3 in cells of each group after transfection. (d) The change of cell viability is detected at 48 h by MTT assay. (e-f) Flow cytometry was performed to detect the cell cycle. (g-i) The number and size of tumor spheres in cells of Control, LIP-NC, LIP-SOCS3, UTMD-NC, UTMD-SOCS3 groups were determined by the sphere formation assay (magnification × 100). *P < 0.05, **P < 0.01, ***P < 0.001 vs. LIP-NC; ^^^P < 0.001 vs. UTMD-NC; ##P < 0.01, ###P < 0.001 vs. LIP-SOCS3.
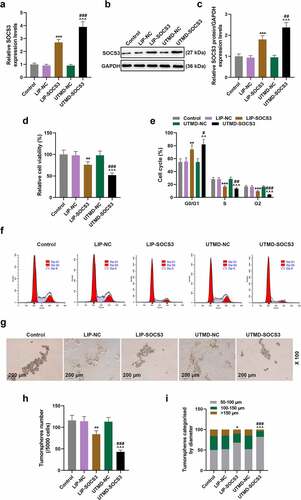
Subsequently, we found that the G0/G1 phase of cells in UTMD-SOCS3 and LIP-SOCS3 groups was prolonged, while the S and G2 phases were shortened, in addition to the discovery where the regulatory effect of UTMD-SOCS3 was significantly better than that of LIP-SOCS3 (P < 0.05, )). The number and size of tumor spheres in UTMD-SOCS3 and LIP-SOCS3 groups were significantly reduced, and UTMD-SOCS3 posed higher inhibitory effects on the sphere formation of BCSCs than LIP-SOCS3 group (P < 0.05, )). Additionally, the cell proliferation, migration and invasion abilities of cells in UTMD-SOCS3 and LIP-SOCS3 groups were significantly reduced, and the inhibitory effects of UTMD-SOCS3 on the above-mentioned biological characteristics were evidently stronger than those of LIP-SOCS3 (P < 0.001, )). The data indicated that both UTMD and liposome can transfect the plasmids promoting SOCS3 into cells and thereby inhibited the stemness, growth, migration, invasion of BCSCs, and the transfection using the method of UTMD was more efficient.
Figure 4. The effects of liposome and UTMD transfection of SOCS3 on the proliferation, migration and invasion of BCSCs. (a-c) The histogram represented the clones, migration and invasion rates of cells in the control, LIP-NC, LIP-SOCS3, UTMD-NC, UTMD-SOCS3 groups. (d) The clone formation of each group was evaluated by the clone formation assay. (e) The change of cell migration ability in each group was detected by wound healing assay (magnification × 100). (f) Transwell assay was performed to detect the invasion ability of cells in each group (magnification × 250). ***P < 0.001 vs. LIP-NC; ^^^P < 0.001 vs. UTMD-NC; ###P < 0.001 vs. LIP-SOCS3.
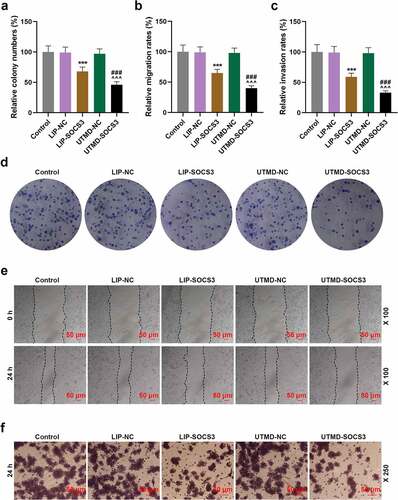
Effects of SOCS3 transfection with liposomes or UTMD on the expressions of EMT- and STAT3 pathway-related genes in BCSCs
In order to unveil the mechanism with regards to the effects of SOCS3 and the two transfection methods on BCSCs, we quantified the expressions of EMT- and STAT3 pathway-related genes. It was found that both UTMD-SOCS3 and LIP-SOCS3 could significantly inhibit the expressions of EMT-related genes Vimentin and N-Cadherin yet promote the expression of E-Cadherin, in addition to the discovery suggesting that the regulatory effects of UTMD-SOCS3 were significantly greater than those of LIP-SOCS3 (P < 0.01, )). Simultaneously, both UTMD-SOCS3 and LIP-SOCS3 effectively reduced the phosphorylation of STAT3 in cells, as well as the decreased expressions of c-Myc, CyclinD1 and PCNA, where we confirmed that the stronger inhibitory effects of UTMD-SOCS3 on the activation of STAT3 pathway as compared to LIP-SOCS3 (P < 0.05, )).
Figure 5. Effects of SOCS3 transfection with liposomes or UTMD on the expressions of EMT- and STAT3 pathway-related genes in BCSCs. (a-e) Effects concerning the transfection of SOCS3 into BCSCs with liposomes and UTMD on EMT-related molecules (E-Cadherin, N-Cadherin, Vimentin) and STAT3 pathway-related molecules (STAT3, CyclinD1, PCNA) were detected by RT-qPCR and Western blot as needed. Each experiment was repeated three times independently. GAPDH was set as control. *P < 0.05, ***P < 0.001 vs. LIP-NC; ^^^P < 0.001 vs. UTMD-NC; ##P < 0.01, ###P < 0.001 vs. LIP-SOCS3.
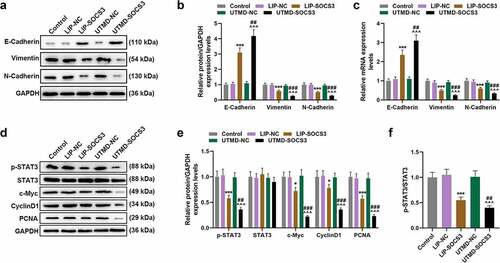
The effect of liposomes- or UTMD-mediated transfection of SOCS3 in vivo
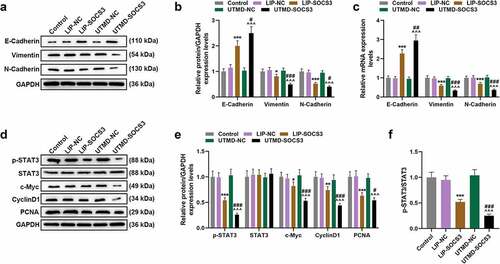
A nude mice subcutaneous tumor xenograft model was constructed to determine the effect of SOCS3 on the tumor growth in mice. As shown in ), the tumor volume and weight of mice in the UTMD-SOCS3 and the LIP-SOCS3 groups were both suppressed, and a greater inhibitory effect was evidenced in mice of the UTMD-SOCS3 group when compared to that of LIP-SOCS3 group (P < 0.001). Similarly, the promotive effect of UTMD-SOCS3 on the expression of SOCS3 was significantly greater than that of LIP-SOCS3 (P < 0.001, )). Mechanistically, we detected EMT- and STAT3 pathway-related genes in tumor tissues of mice and found that the results are consistent with experiments in vitro. Both UTMD-SOCS3 and LIP-SOCS3 significantly inhibited the expressions of Vimentin, N-Cadherin, c-Myc, CyclinD1 and PCNA as well as the phosphorylation of STAT3, while facilitating the expression of E-Cadherin. Furthermore, UTMD-SOCS3 posed a higher regulatory effect on EMT- and STAT3 pathway-related genes than LIP-SOCS3 in tumor tissues of mice (P < 0.05, )).
Figure 6. UTMD-mediated SOCS3 showed higher inhibition on tumor growth of BC mice than liposome-mediated SOCS3. (a-c) The tumor xenograft model was constructed to detect the effects of different transfection methods of SOCS3 on the volume and weight of tumors in nude mice. (d-f) The expression of SOCS3 in the tumor tissues of mice was calculated by RT-qPCR and Western blot. Each experiment was repeated three times independently. GAPDH was set as the control. ***P < 0.001 vs. LIP-NC; ^^^P < 0.001 vs. UTMD-NC; ###P < 0.001 vs. LIP-SOCS3.
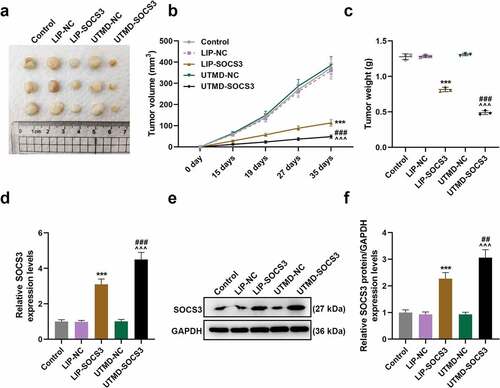
Figure 7. The effect of liposomes- or UTMD-mediated transfection of SOCS3 in EMT-related and STAT3 pathway-related molecules in BC mice in vivo. (a-e) The effects of liposome- and UTMD-mediated transfection with SOCS3 on the expressions of EMT-related molecules (E-Cadherin, N-Cadherin, Vimentin) and STAT3 pathway-related molecules (STAT3, CyclinD1, PCNA) in the tumor tissues of mice were measured by RT-qPCR and Western blot, as required. Each experiment was repeated three times independently. GAPDH was set as control. *P < 0.05, **P < 0.01, ***P < 0.001 vs. LIP-NC; ^^^P < 0.001 vs. UTMD-NC; #P < 0.05, ##P < 0.01, ###P < 0.001 vs. LIP-SOCS3.
Discussion
Multiple studies have indicated that BCSCs have self-renewal, multi-directional differentiation potential and high tumorigenicity [Citation15]. As BCSCs are closely related to tumor occurrence, invasion and metastasis, recurrence and resistance after treatment, people have gradually dedicated to proposing viable method to eradicate BCSCs and understanding their related biological characteristics [Citation9]. So far, the roles of SOCS3 or UTMD alone in breast cancer have been studied individually by scholars, but the combination of these two in breast cancer, BCSCs in particular, has not been discovered and addressed [Citation21,Citation34–36]. In this study, it was reported, for the first time, that UTMD-mediated SOCS3 might attenuate the malignant biological progression of BCSCs by inhibiting the activation of the STAT3 pathway.
CSCs can be identified by the cell surface markers they express [Citation7]. For example, Al-Hajj et al. discovered and isolated BCSCs in breast cancer specimens for the first time, and suggested that BCSCs have the characteristics of CD44+CD24− [Citation37]. In this experiment, two sorting methods, namely, FACS and MACS, were used to sort BCSCs with CD44+CD24− as surface markers as well. Both sorting methods were able to sort BCSCs successfully, and high sphere formation rates were depicted. Furthermore, SOCS3 has lower expression in BCSCs, higher cell viability and tumorigenesis of which are evidenced, not only proving that BCSCs have high tumorigenicity and strong self-renewal ability, but also implying that the silence of SOCS3 may promote the tumorigenesis of breast cancer.
UTMD is currently one of the most promising non-viral gene transfection methods [Citation38]. Its principle of action is to use a specific frequency of ultrasound irradiation at a specific site to rupture the microbubbles that reach that site, which increases permeability and makes it easier for foreign substances to enter the target cells and achieve the purpose of targeted therapy [Citation39]. For example, Liao et al. pointed out that UTMD-mediated transfection of HIF-1α shRNA can significantly silence HIF-1α and successfully inhibit tumor growth in rats with liver cancer [Citation40]. Zhao et al. found that UTMD enhanced the local accumulation of FOXA1 and showed an excellent therapeutic effect on ER-positive breast cancer [Citation41]. It was demonstrated in our study that the efficiency of UTMD-mediated transfection of SOCS3 into BCSCs was significantly higher than that mediated by liposome. In addition, when compared to the liposome-mediated SOCS3, the effects of UTMD-mediated SOCS3 on the biological functions of BCSCs are significantly better.
SOCS3 is an important regulatory molecule that regulates many cytokine signals in the body, and plays an important role in many physiological regulation processes [Citation42]. SOCS3 attenuates tumor growth in lung cancer xenograft models, and inhibits the proliferation and malignant transformation of lung adenocarcinoma cells in vitro [Citation43]. The expression of SOCS3 is reduced, and the high expression of SOCS3 induces cell apoptosis and reduces cell proliferation in pancreatic cancer [Citation44], the results of which are similar to those we proposed in our study [Citation44]. Specifically, we also found that SOCS3 is lower-expressed in BCSCs, and the high expression of SOCS3 causes cell cycle arrest, inhibits the viability of BCSCs, andreduces the sphere formation and malignant EMT.
SOCSs are induced by STATs, and in turn, SOCSs inhibit the cascade of JAK/STAT signaling [Citation45,Citation46]. SOCS3 overexpression inhibits the activity of JAK-STAT3 signaling pathway in breast cancer cells, inhibits cell proliferation, and improves the sensitivity to ADM-induced apoptosis [Citation47]. Similarly, in this study, we found that the overexpression of SOCS3 inhibited the activation of the STAT3 pathway in BCSCs and mice. The mechanism implicated in the action of SOCS3 may be associated with the usage of the SH2 domain, which is similar to STAT, to compete for the binding to the phosphorylated Tyr site in the cytoplasmic region of the cytokine receptor, thereby preventing the activation of the transcription factor STAT. Additionally, approximately two-thirds of all breast cancers diagnosed are classified as hormone-dependent, and growth hormone plays a significant role in the development, progression, and metastasis of breast cancer [Citation48]. The previous studies have shown the regulatory effect of SOCS3 on hormone signaling [Citation49,Citation50]. Barclay et al. [Citation51] showed the complexity of SOCS3 regulation and crosstalk in T47D cells in response to prolactin, a key mammotropic hormone. A gut-derived hormone, glucose-dependent insulinotropic polypeptide (GIP) increased proinflammatory-related factors such as SOCS3 in the hypothalamus [Citation52]. These reports also indicated the important role of SOCS3 in hormone-dependent breast cancer. However, further mechanism implicated required to be further investigated.
Conclusion
Taken together, we compared the effects of liposome- and UTMD-mediated SOCS3 on the proliferation, migration, and EMT of BCSCs. Then, further experiments in vivo were conducted to test the effects of the two methods on the xenograft formation using BCSCs. Finally, we propose that UTMD-mediated SOCS3 has a better therapeutic effect, and these results may provide new experimental evidence for the treatment of breast cancer.
Highlights
High viability and tumorigenicity were evidenced in BCSCs.
Under the mediation of UTMD and liposome, SOCS3 attenuates biological characteristics and EMT of BCSCs.
The regulatory effects of UTMD-mediated SOCS3 were better than liposome-mediated SOCS3.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Supplemental Material
Download Zip (225.6 KB)Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424.
- Harbeck N, Gnant M. Breast cancer. Lancet. 2017;389:1134–1150.
- Anastasiadi Z, Lianos GD, Ignatiadou E, et al. Breast cancer in young women: an overview. Updates Surg. 2017;69:313–317.
- Wormann B. Breast cancer: basics, screening, diagnostics and treatment. Med Monatsschr Pharm. 2017;40:55–64.
- Ganz PA, Goodwin PJ. Breast cancer survivorship: where are we today?. Adv Exp Med Biol. 2015;862:1–8.
- Sun YS, Zhao Z, Yang ZN, et al. Risk factors and preventions of breast cancer. Int J Biol Sci. 2017;13:1387–1397.
- Dawood S, Austin L, Cristofanilli M. Cancer stem cells: implications for cancer therapy. Oncology (Williston Park). 2014;28:1101–7, 10.
- Nassar D, Blanpain C. Cancer stem cells: basic concepts and therapeutic implications. Annu Rev Pathol. 2016;11:47–76.
- Luo M, Clouthier SG, Deol Y, et al. Breast cancer stem cells: current advances and clinical implications. Methods Mol Biol. 2015;1293:1–49.
- Bilek SL, Lay T. Variation of interplate fault zone properties with depth in the Japan subduction zone. Science (New York, NY). 1998;281:1175–1178.
- Zou Y, Chen Y, Yao S, et al. MiR-422a weakened breast cancer stem cells properties by targeting PLP2. Cancer Biol Ther. 2018;19:436–444.
- Xie G, Ji A, Yuan Q, et al. Tumour-initiating capacity is independent of epithelial-mesenchymal transition status in breast cancer cell lines. Br J Cancer. 2014;110:2514–2523.
- Liu S, Cong Y, Wang D, et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Reports. 2014;2:78–91.
- Sulaiman A, McGarry S, Han X, et al. CSCs in breast cancer-one size does not fit all: therapeutic advances in targeting heterogeneous epithelial and mesenchymal CSCs. Cancers (Basel). 2019;11:1128.
- Dittmer J. Breast cancer stem cells: features, key drivers and treatment options. Semin Cancer Biol. 2018;53:59–74.
- Groner B, von Manstein V. Jak stat signaling and cancer: opportunities, benefits and side effects of targeted inhibition. Mol Cell Endocrinol. 2017;451:1–14.
- Inan S, Hayran M. Cell signaling pathways related to epithelial mesenchymal transition in cancer metastasis. Crit Rev Oncog. 2019;24:47–54.
- Slattery ML, Lundgreen A, Kadlubar SA, et al. JAK/STAT/SOCS-signaling pathway and colon and rectal cancer. Mol Carcinog. 2013;52:155–166.
- Liu K, Wu Z, Chu J, et al. Promoter methylation and expression of SOCS3 affect the clinical outcome of pediatric acute lymphoblastic leukemia by JAK/STAT pathway. Biomed Pharmacother. 2019;115:108913.
- Li Y, Hodge J, Liu Q, et al. TFEB is a master regulator of tumor-associated macrophages in breast cancer. J Immunother Cancer. 2020;8:e000543.
- Xu JZ, Shao CC, Wang XJ, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175.
- Jiang M, Zhang W, Zhang R, et al. Cancer exosome-derived miR-9 and miR-181a promote the development of early-stage MDSCs via interfering with SOCS3 and PIAS3 respectively in breast cancer. Oncogene. 2020;39:4681–4694.
- Zhang KJ, Tan XL, Guo L. The long non-coding RNA DANCR regulates the inflammatory phenotype of breast cancer cells and promotes breast cancer progression via EZH2-dependent suppression of SOCS3 transcription. Mol Oncol. 2020;14:309–328.
- Muhammad N, Bhattacharya S, Steele R, et al. Anti-miR-203 suppresses ER-positive breast cancer growth and stemness by targeting SOCS3. Oncotarget. 2016;7:58595–58605.
- Thiagarajan PS, Zheng Q, Bhagrath M, et al. STAT3 activation by leptin receptor is essential for TNBC stem cell maintenance. Endocr Relat Cancer. 2017;24:415–426.
- Sun W, Shi Q, Zhang H, et al. Advances in the techniques and methodologies of cancer gene therapy. Discov Med. 2019;27:45–55.
- Wu J, Li RK. Ultrasound-targeted microbubble destruction in gene therapy: a new tool to cure human diseases. Genes Dis. 2017;4:64–74.
- Liu YM, Li XF, Liu H, et al. Ultrasound-targeted microbubble destruction-mediated downregulation of CD133 inhibits epithelial-mesenchymal transition, stemness and migratory ability of liver cancer stem cells. Oncol Rep. 2015;34:2977–2986.
- Chen H, Hwang JH. Ultrasound-targeted microbubble destruction for chemotherapeutic drug delivery to solid tumors. J Ther Ultrasound. 2013;1:10.
- Tinkov S, Bekeredjian R, Winter G, et al. Microbubbles as ultrasound triggered drug carriers. J Pharm Sci. 2009;98:1935–1961.
- Tu J, Zhang H, Yu J, et al. Ultrasound-mediated microbubble destruction: a new method in cancer immunotherapy. Onco Targets Ther. 2018;11:5763–5775.
- Tang W, Li M, Qi X, et al. β1,4-Galactosyltransferase V modulates breast cancer stem cells through Wnt/β-catenin signaling pathway. Cancer Res Treat. 2020;52:1084–1102.
- Xu J, Wang Y, Li Z, et al. Ultrasound-Targeted Microbubble Destruction (UTMD) combined with liposome increases the effectiveness of suppressing proliferation, migration, invasion, and Epithelial- Mesenchymal Transition (EMT) via Targeting Metadherin (MTDH) by ShRNA. Med Sci Monit. 2019;25:2640–2648.
- Zhang W, Jiang M, Chen J, et al. SOCS3 Suppression Promoted the Recruitment of CD11b(+)Gr-1(-)F4/80(-)MHCII(-) early-stage myeloid-derived suppressor cells and accelerated Interleukin-6-Related tumor invasion via affecting myeloid differentiation in breast cancer. Front Immunol. 2018;9:1699.
- Shi D, Guo L, Sun X, et al. UTMD inhibit EMT of breast cancer through the ROS/miR-200c/ZEB1 axis. Sci Rep. 2020;10:6657.
- Ji Y, Han Z, Shao L, et al. Ultrasound-targeted microbubble destruction of calcium channel subunit alpha 1D siRNA inhibits breast cancer via G protein-coupled receptor 30. Oncol Rep. 2016;36:1886–1892.
- Al-Hajj M, Wicha MS, Benito-Hernandez A, et al. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003;100:3983–3988.
- Chen S, Grayburn PA. Ultrasound-targeted microbubble destruction for cardiac gene delivery. Methods Mol Biol. 2017;1521:205–218.
- Geis NA, Katus HA, Bekeredjian R. Microbubbles as a vehicle for gene and drug delivery: current clinical implications and future perspectives. Curr Pharm Des. 2012;18:2166–2183.
- Liao Y, Luo H, He Z, et al. A combination of UTMD-Mediated HIF-1alpha shRNA transfection and TAE in the treatment of hepatic cancer. Biomed Res Int. 2019;2019:1937460.
- Zhao R, Liang X, Zhao B, et al. Ultrasound assisted gene and photodynamic synergistic therapy with multifunctional FOXA1-siRNA loaded porphyrin microbubbles for enhancing therapeutic efficacy for breast cancer. Biomaterials. 2018;173:58–70.
- Pedroso JAB, Ramos-Lobo AM, Donato J Jr. SOCS3 as a future target to treat metabolic disorders. Hormones (Athens). 2019;18:127–136.
- Speth JM, Penke LR, Bazzill JD, et al. Alveolar macrophage secretion of vesicular SOCS3 represents a platform for lung cancer therapeutics. JCI Insight. 2019;4. DOI:10.1172/jci.insight.131340.
- Wang J, Guo J, Fan H. MiR-155 regulates the proliferation and apoptosis of pancreatic cancer cells through targeting SOCS3. Eur Rev Med Pharmacol Sci. 2019;23:5168–5175.
- Gao Y, Zhao H, Wang P, et al. The roles of SOCS3 and STAT3 in bacterial infection and inflammatory diseases. Scand J Immunol. 2018;88:e12727.
- Sun D, Jiang Z, Chen Y, et al. MiR-455-5p upregulation in umbilical cord mesenchymal stem cells attenuates endometrial injury and promotes repair of damaged endometrium via Janus kinase/signal transducer and activator of transcription 3 signaling. Bioengineered. 2021;2:78–91.
- Li MZ, Lai DH, Zhao HB, et al. SOCS3 overexpression enhances ADM resistance in bladder cancer T24 cells. Eur Rev Med Pharmacol Sci. 2017;21:3005–3011.
- Subramani R, Nandy SB, Pedroza DA, et al. Role of growth hormone in breast cancer. Endocrinology. 2017;158:1543–1555.
- Carow B, Rottenberg ME. SOCS3, a major regulator of infection and inflammation. Front Immunol. 2014;5:58.
- Liu Z, Gan L, Zhou Z, et al. SOCS3 promotes inflammation and apoptosis via inhibiting JAK2/STAT3 signaling pathway in 3T3-L1 adipocyte. Immunobiology. 2015;220:947–953.
- Barclay JL, Anderson ST, Waters MJ, et al. SOCS3 as a tumor suppressor in breast cancer cells, and its regulation by PRL. Int J Cancer. 2009;124:1756–1766.
- Fu Y, Kaneko K, Lin HY, et al. Gut hormone GIP induces inflammation and insulin resistance in the hypothalamus. Endocrinology. 2020;46:161.
