ABSTRACT
Circular RNAs (circRNAs) have been confirmed to be associated with the progression of various cancers, including hepatocellular carcinoma (HCC). However, the role and mechanism of circHIPK3 in HCC are still unclear. To investigate its function, circHIPK3 expression was first determined by RT–qPCR in HCC tissues or cells. Functionally, cell proliferation and invasion were investigated by CCK-8, EdU, or Transwell assays. In terms of understanding the mechanism, the interaction of the circRNA HIPK3/micro RNA 124 (miRNA 124) or micro RNA 506 (miRNA506) /PDK2 regulatory loop was verified by dual-luciferase reporter gene assay. In addition, a xenograft tumor model was established to confirm the impact of circHIPK3 on the growth of HCC cells in vivo. We found that circHIPK3 was upregulated in HCC patients and associated with clinical characteristics, while miR-124 and miR-506 were downregulated in HCC patients. Additionally, we proved that knock down of circHIPK3 remarkably suppressed the proliferation and invasion of HCC cells. Mechanistically, circHIPK3 directly bound to miR-124 or miR-506 and inhibited their expression, and PDK2 was a target gene of miR-124 or miR-506. Moreover, circHIPK3 overexpression reversed the inhibitory effect of miR-124 or miR-506 on HCC progression. miR-124 or miR-506 could also suppress tumorigenesis of HCC cells by PDK2. Furthermore, in vivo evidence confirmed that knock down of circHIPK3 inhibited tumor formation. We suggest that circHIPK3 can accelerate the proliferation and invasion of HCC cells by sponging miR-124 or miR-506 to upregulate PDK2, which is the underlying mechanism of circHIPK3-induced HCC progression.
Introduction
Liver cancer is the third leading cause of cancer-related death worldwide after lung and colorectal cancer [Citation1]. Hepatocellular carcinoma (HCC) accounts for 85% to 90% of primary liver cancer and is a highly invasive malignant tumor [Citation2]. Risk factors for HCC mainly include chronic hepatitis B and C, alcoholic liver disease, metabolic liver disease, exposure to dietary toxins, etc [Citation3,Citation4]. Statistically, approximately 750,000 patients die of HCC each year globally [Citation5]. The incidence of HCC is geographically heterogeneous, with approximately 85% of HCC occurring in developing countries and underdeveloped areas [Citation6]. Currently, liver transplantation and liver resection are two effective treatments for HCC [Citation7]. Due to the high degree of malignancy and poor prognosis of HCC, most patients do not have the opportunity for surgical treatment since most cases are found in an advanced stage [Citation8]. HCC has a high degree of malignancy and poor prognosis. Most patients are first diagnosed at an advanced stage and have lost the opportunity for surgical treatment [Citation9]. Consequently, there is an urgent need to find potential therapeutic targets for HCC therapy.
CircRNAs are a group of RNAs that regulate transcriptional and posttranscriptional gene expression and are produced from the back-splicing of RNA [Citation10,Citation11]. Due to the specificity and stability of circRNAs, they play a key role in the development and metastasis of different cancers. Recently, many circRNAs have been proven to be relevant to the progression of HCC, such as circ_101280 [Citation12], circ_0051443 [Citation13], circ_0000517 [Citation14], circ_0000517 [Citation15], Circ_0001955 [Citation16], and circ_0000285 [Citation17]. CircHIPK3 dysregulation has also been discovered as an oncogenic mechanism in multiple cancers, including lung cancer [Citation18], renal cancer [Citation19], gastric cancer [Citation20], bladder cancer [Citation21], prostate cancer [Citation22], pancreatic cancer [Citation23], and thyroid cancer [Citation24]. However, the function and possible mechanism of circHIPK3 in HCC progression are largely unknown.
It has been reported that circRNAs possess microRNA (miRNA) response elements (MREs) [Citation25]. CircRNAs, as competitive endogenous RNAs (ceRNAs), can competitively bind miRNAs and block their binding to targets, thereby affecting the expression of target genes [Citation26]. CircRNAs can act as sponges to absorb miRNAs and regulate their functions, which is a key mechanism by which circRNAs affect cancer progression [Citation27,Citation28]. Recent investigations have reported that miR-124-3p acts as an important tumor-suppressive miRNA to suppress HCC carcinogenesis through targeting CRKL [Citation29]. To investigate the possible mechanism of circHIPK3 in HCC, we also performed bioinformatics analysis to predict the targeted miRNAs of circHIPK3. In accordance with the predicted results, we unexpectedly discovered that there were latent binding sites between circHIPK3 and miR-124-3p or miR-506-3p. Therefore, we speculated that circHIPK3 might alter HCC progression by targeting miR-124-3p and miR-506-3p.
In this study, we aimed to determine the expression and roles of circHIPK3, miR-124-3p and miR-506-3p in HCC patients and HCC cells. In addition, we explored the possible regulatory mechanism of circHIPK3 on miR-124-3p and the miR-506-3p axis in HCC. Results from this current study reveal the potential value of the circHIPK3/miR-124-3p or miR-506-3p axis in the diagnosis, treatment and prognosis of HCC.
Materials and methods
Clinical specimens
Liver cancer and tumor-adjacent tissues were collected from 30 HCC patients who were admitted to Zhongshan Hospital. The clinical information of the 30 patients with HCC was as follows: sex (male 17; female, 13), age(≥55, 16; <55, 14), tumor size (≥4 cm, 14; <4 cm, 16), lymph node metastasis (positive, 13; negative, 17), and TNM stage (I–II, 16; III–IV, 14). Permission was obtained from the ethics committee of Zhongshan Hospital, and all participants signed written informed consent.
Cell lines
Liver cancer cell lines (HepG2, SMMC-7721, Bel-7402 and Huh-7) and a human hepatic cell line (HL7702) were purchased from ATCC. SMMC-7721, Bel-7402, and HL7702 cells were grown in RPMI 1640 medium (Gibco); HepG2 cells were cultured in MEM (Gibco, Cat. No. 41,500,034); Huh-7 cells were maintained in DMEM (Sigma–Aldrich, Cat. No. D6429). All media were supplemented with 10% fetal bovine serum (Gibco), and all the cells were grown at 37°C and 5% CO2.
Cell transfection
Negative control (NC), circHIPK3 siRNAs (si-circHIPK3), NC mimics, miR-124 mimics, and miR-506 mimics were chemically synthesized by GenePharma (Shanghai, China). Empty vector, circHIPK3 overexpression plasmid and PDK2 overexpression plasmid were obtained from HanBio Biotechnology (Shanghai, China). Based on the experimental purpose, HepG2 and SMMC-7721 cells were transfected with these plasmids or oligonucleotides using Lipofectamine 3000 (Invitrogen) for 48 h by referring to the reagent instructions [Citation30]. This assay was repeated three times.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Total RNA from HCC tissues and cells was extracted with TRIzol (Invitrogen), and the concentration was calculated by measuring the absorbance at A260/280. After incubation with or without 3 U/μg RNase R, an RNeasy MinElute cleaning kit was used to purify the resulting RNA. RNA (1.5 μg) was reverse transcribed into cDNA with a BestarTM qPCR RT kit (DBI; 2220). PCR amplification was conducted using SYBR Green Master Mix (Roche) on an Applied Biosystems 7500 H system (Foster City, CA, USA) with specific primers. The PCR conditions were as follows: denaturation at 95°C for 20s, 30 cycles of denaturation at 95°C for 30s, annealing at 55°C for 30s and extension at 72°C for 30s [Citation31]. This assay was repeated three times. All primer sequences are displayed in .
Table 1. The sequences of primers in RT-qPCR
Cell counting Kit-8 (CCK-8)
Treated HepG2 or SMMC-7721 cells were seeded in 96-well plates overnight at a concentration of 2 × 104/well. Twenty microliters of CCK-8 solution (Dojindo) were added to each well and incubated at 37°C. The optical density (OD) values were measured at 450 nm every 24 hours by a scanning multiwell spectrophotometer [Citation32]. This assay was repeated three times.
Cell proliferation assay using 5-ethynyl-2ʹ-deoxyuridine (EdU)
Cell viability was tested with a Cell-LightTM EdU kit (Cat. No. C10311). Transfected HepG2 or SMMC-7721 cells in a 24-well plate were fixed using 4% paraformaldehyde for 20 min, treated with 2 mg/mL glycine for 10 min, and cleared with 0.5% Triton X-100. Then, the cells were processed with 100 μL Apollo solution for 30 min and 100 μL Hoechst 33,342 for 20 min. After washing and permeating, the results were viewed with a fluorescence microscope [Citation33]. This assay was repeated three times.
Transwell assay
The bottom membrane of the Transwell chamber (Costar; MA, USA) was coated with diluted Matrigel (dilution ratio, 1:8, 50 μL) for 30 min at 37°C. Then, the transfected HepG2 or SMMC-7721 cells were suspended in serum-free medium and inoculated in the upper Transwell chamber, and medium with 20% FBS was added to the lower Transwell chamber. After incubation for 24 h at 37°C, the invaded cells were fixed, stained with 0.1% crystal violet, and photographed with a light microscope (Nikon, Japan) [Citation34]. Five random fields per group were photographed under an optical microscope and the number of cells was counted. This assay was repeated three times.
Dual luciferase reporter assay
The sites of circHIPK3 and miR-124 or miR-506 were predicted with Starbase (https://starbase.sysu.edu.cn/index.php), and the putative target genes associated with miR-124 or miR-506 were predicted by the TargetScan database (www.targetscan.org/vert_72). Wild-type (WT) or mutant (MUT) circHIPK3 or PDK2 were synthesized and then cloned into pis-Check 2 dual-luciferase reporter vectors (Promega). HepG2 cells were seeded in 24-well plates and cotransfected with miR-124 mimics or miR-506 mimics and WT- or MUT-circHIPK3 or PDK2 using Lipofectamine 2000 (Invitrogen). The relative luciferase activities were determined by the Dual-Luciferase Reporter Assay System (Promega) after 48 h of transfection [Citation35]. This assay was repeated three times.
Western blot
Proteins extracted from HepG2 or SMMC-7721 cells were separated by SDS–PAGE based on the molecular weight of the proteins. Protein from the gels was transferred to PVDF membranes and incubated with primary antibody (PDK2, abcam, UK) overnight at 4°C after blocking with 5% skim milk. The membranes were subsequently washed with 0.1 M phosphate‐buffered saline (PBS) with Tween‐20 and then incubated with HRP-conjugated secondary antibodies (abcam, UK). Finally, ECL (KeyGen) was adopted to test the immunoreactivities [Citation36]. This assay was repeated three times.
Tumor xenograft model
SPF experimental animals (BALB/c male nude mice, 4 weeks, 20 ± 2 g) were purchased from Laboratory Animal Center of Sun Yat-sen University. The mice were raised in a specific environment (temperature, 22°C-25°C; humidity, 45%-55%, 12 h light/12 h dark, and freely available food and water). This study was approved by the IACUC, and the experiment was strictly conducted based on IACUC. The transfected HepG2 cell suspension (1 × 106 cells) was inoculated subcutaneously into nude mice. We recorded the longest diameter (L) and shortest diameter (W) of the tumors every 7 days for 28 days. At 28 days, the mice were anesthetized, and the tumors were removed and photographed [Citation37].
Statistical analysis
The data were analyzed by Student’s t test and One-way analysis of variance using SPSS 22.0 software (SPSS, Chicago, IL, USA). All results are expressed as means ±SD. The statistical significance was set at P < 0.05.
Results
Silencing of circHIPK3 notably restrained the proliferation and invasion of HCC cells
To investigate the expression and function of circHIPK3 in HCC, we first collected HCC and tumor-adjacent tissues. RT–qPCR data showed that circHIPK3 expression was significantly increased in HCC compared with tumor-adjacent tissues (). In addition, we discovered that circHIPK3 expression was positively related to tumor size, lymph node metastasis and TNM stage, while there was no significant difference in circHIPK3 expression and sex or age in HCC patients (). Next, our results showed that circHIPK3 was noticeably upregulated in HepG2, SMMC-7721, Bel-7402 and Huh-7 cells relative to that in HL7702 cells, and the level of circHIPK3 was relatively high in HepG2 and SMMC-7721 cells and relatively low in Bel-7402 and Huh-7 cells (). Thus, we selected HepG2 and SMMC-7721 cells with higher circHIPK3 expression for circHIPK3 silencing through transfection with si-circHIPK3. RT–qPCR results showed that circHIPK3 expression was significantly lower in the si-circHIPK3 group than in the si-NC group, indicating the successful knock down of circHIPK3 in HepG2 and SMMC-7721 cells (). Moreover, the results from CCK-8 and EdU assays showed that circHIPK3 knockdown prevented the proliferation of HepG2 and SMMC-7721 cells ( and e). Additionally, Transwell data showed that circHIPK3 knockdown caused a remarkable suppression in the invasion capacity of HepG2 and SMMC-7721 cells (). Overall, our data confirmed that knock down of circHIPK3 contributed to the prevention of HCC progression.
Table 2. The correlation between circHIPK3 expression and clinicopathologic characteristics in liver cancer
Figure 1. Silencing of circHIPK3 notably restrained the proliferation and invasion of HCC cells. (a) RT–qPCR assay showed the change in circHIPK3 in HCC and tumor-adjacent tissues (n = 30). (b) RT–qPCR was conducted to confirm circHIPK3 expression in HL7702, HepG2, SMMC-7721, Bel-7402 and Huh-7 cells. (c) The change in circHIPK3 expression was monitored by RT–qPCR in HepG2 and SMMC-7721 cells transfected with si-circHIPK3. (d and e) CCK-8 and EdU assays were performed to assess cell proliferation in circHIPK3-silenced HepG2 and SMMC-7721 cells. (f) Cell invasion was tested by Transwell assay in HepG2 and SMMC-7721 cells after circHIPK3 knockdown, and the number of invaded cells was counted. *P< 0.05, ***P< 0.001.
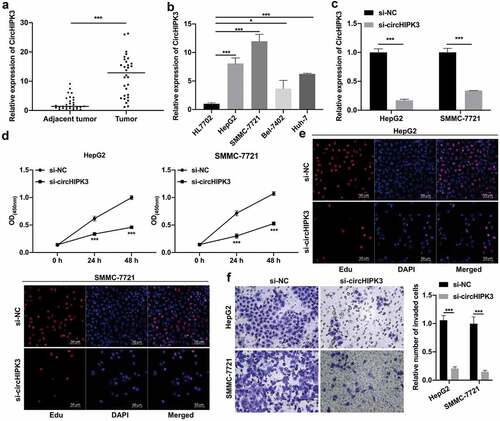
CircHIPK3 overexpression prominently reversed the inhibitory effects of miR-124 on the proliferation and invasion of HCC cells
Through bioinformatics prediction, we unexpectedly discovered a miRNA (miR-124) that might interact with circHIPK3. Moreover, our results also showed that miR-124 expression was markedly decreased in HCC versus tumor-adjacent tissues (). We further determined the binding ability between circHIPK3 and miR-124, and the data showed that miR-124 mimics could notably decrease the luciferase activity of WT-circHIPK3 but had no effect on the luciferase activity of MUT-circHIPK3 (). Moreover, RT–qPCR data revealed that miR-124 expression was significantly increased in miR-124 mimic-transfected HepG2 and SMMC-7721 cells, while this increased expression of miR-124 was markedly weakened by circHIPK3 overexpression (). Next, CCK-8 and EdU data revealed that miR-124 overexpression significantly diminished cell proliferation, which could also be partially reversed by circHIPK3 overexpression in HepG2 and SMMC-7721 cells ( and e). Transwell results revealed that miR-124 overexpression also prominently repressed the invasion of HepG2 and SMMC-7721 cells, which were also dramatically attenuated by circHIPK3 overexpression (). In summary, we discovered that circHIPK3, as a sponge for miR-124, could induce the proliferation and invasion of HCC cells by targeting miR-124.
Figure 2. CircHIPK3 overexpression prominently reversed the inhibitory effects of miR-124 on the proliferation and invasion of HCC cells. (a) RT–qPCR analysis of miR-124 expression in HCC and tumor-adjacent tissues (n = 30). (b) The interaction between circHIPK3 and miR-124 was analyzed via dual luciferase reporter gene assay in HCC cells. (c) The change in miR-124 expression was confirmed by RT–qPCR in HepG2 and SMMC-7721 cells cotransfected with miR-124 mimics and circHIPK3 plasmids. (d and e) The impact of miR-124 and circHIPK3 on cell proliferation was tested through CCK-8 and EdU assays in HepG2 and SMMC-7721 cells. (f) After cotransfection with miR-124 mimics and circHIPK3 plasmids, a Transwell assay was utilized to determine the invasion capacity of HepG2 and SMMC-7721 cells, and the number of invaded cells was calculated in each region. ***P< 0.001.
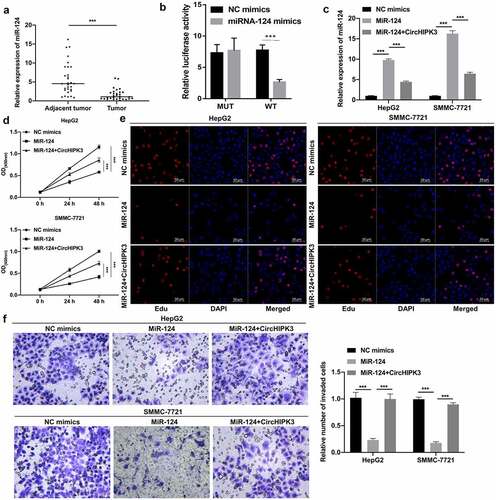
CircHIPK3 overexpression markedly attenuated the inhibitory action of miR-506 on the proliferation and invasion of HCC cells
Furthermore, we also discovered that miR-506 might be another underlying target of circHIPK3. The RT–qPCR results also indicated that the level of miR-506 was notably reduced in HCC tissues compared to tumor-adjacent tissues (). Moreover, our results showed that the luciferase activity of WT-circHIPK3 was noticeably reduced by miR-506 mimics, while the luciferase activity of MUT-circHIPK3 was unaffected by miR-506 mimics; thus, these results indicated that circHIPK3 could target miR-506 (). The RT–qPCR results showed that miR-506 expression was markedly heightened in the miR-506 group compared with the NC group, while this elevation of miR-506 expression was dramatically weakened by circHIPK3 overexpression (). Functionally, we first demonstrated that overexpression of miR-506 notably suppressed cell proliferation, while this suppression mediated by miR-506 overexpression could also be prominently reduced by circHIPK3 overexpression in HepG2 and SMMC-7721 cells ( and e). Transwell data also confirmed that miR-506 overexpression could dramatically prevent cell invasion, which was also markedly reversed by circHIPK3 overexpression in HepG2 and SMMC-7721 cells (). Generally, the data proved that circHIPK3 could also promotes HCC progression by targeting miRNA-506.
Figure 3. CircHIPK3 overexpression markedly attenuated the inhibitory action of miR-506 on the proliferation and invasion of HCC cells. (a) RT–qPCR analysis revealed a change in miR-506 expression in HCC and tumor-adjacent tissues (n = 30). (b) The interaction between circHIPK3 and miR-506 was evaluated using a dual luciferase reporter gene assay. (c) RT–qPCR was performed to confirm the expression change of miR-506 in miR-506- and circHIPK3-overexpressing HepG2 and SMMC-7721 cells. (d and e) CCK-8 and EdU assays revealed a change in cell proliferation of HepG2 and SMMC-7721 cells cotransfected with miR-506 mimics and/or circHIPK3 plasmids. (f) Transwell assays were performed to monitor the influences of circHIPK3 and miR-506 on the invasion ability of HepG2 and SMMC-7721 cells, and the invaded cells were also counted. **P< 0.01, ***P< 0.001.
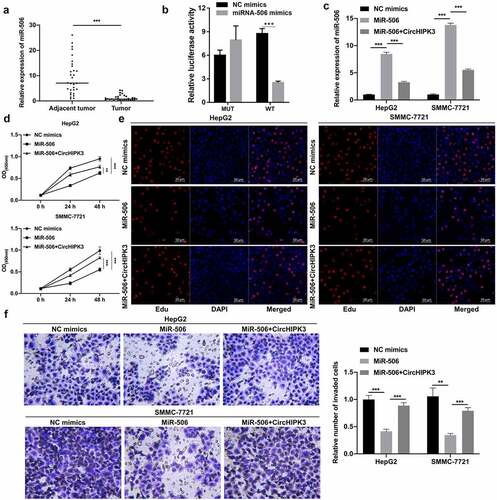
Overexpression of PDK2 markedly enhanced tumorigenesis of HCC cells mediated by miR-124 or miR-506
Subsequently, we further predicted the latent target gene of miR-124 or miR-506. First, Western blotting data showed that PDK2 was significantly downregulated in miR-124- or miR-506-transfected HepG2 and SMMC-7721 cells, while the miR-124- or miR-506 overexpression-mediated decrease in PDK2 expression was observably reversed by circHIPK3 overexpression (). Second, the data proved that miR-124 or miR-506 overexpression could markedly reduce the luciferase activity of WT-PDK2 but had no effect on the luciferase activity of MUT-PDK2 ( and c). Third, we cotransfected the PDK2 plasmid and miR-124 or miR-506 into HepG2 and SMMC-7721 cells, and our data verified that PDK2 overexpression could notably upregulate PDK2 in HepG2 and SMMC-7721 cells, which was dramatically reduced by miR-124 or miR-506, especially miR-124 ( and e). In addition, our results indicated that miR-124 or miR-506 overexpression prominently reversed the inhibitory action of PDK2 overexpression on the proliferation of HepG2 and SMMC-7721 cells ( and g). Additionally, miR-124 or miR-506 overexpression markedly attenuated the promoter role by PDK2 on the invasion of HepG2 and SMMC-7721 cells (). Thus, these results revealed that miR-124 or miR-506 could block the malignant biological properties of HCC cells through PDK2.
Figure 4. Overexpression of PDK2 markedly enhanced tumorigenesis of HCC cells mediated by miR-124 or miR-506. (a) Western blotting analysis revealed the change in expression of PDK2 in HepG2 cells after transfection with miR-124 or miR-506 and circHIPK3 plasmids. (b and c) The interaction between PDK2 and miR-124 or miR-506 was also determined with a dual luciferase reporter assay in HepG2 cells. (d and e) RT–qPCR was conducted to examine PDK2 expression in HCC cells after transfection with the PDK2 plasmid and miR-124 or miR-506. The effects of PDK2 and miR-124 or miR-506 on cell proliferation and invasion were tested with CCK-8 (f), EdU (g) and Transwell assays (h). *P< 0.05, **P< 0.01, ***P< 0.001.
Knock down of circHIPK3 dramatically suppressed growth and upregulated miR-124 and miR-506 in HCC xenograft tumors
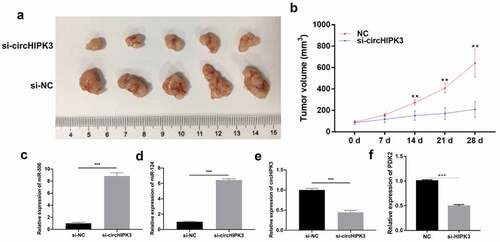
Based on the role and mechanism of circHIPK3 in HCC cells, we further verified the effect of circHIPK3 in HCC xenograft tumors. As shown in , tumors in nude mice were observably diminished in the si-circHIPK3 group compared with the si-NC group. Silencing circHIPK3 also significantly reduced the volume of xenograft tumors (). Additionally, RT–qPCR results revealed that knock down of circHIPK3 could prominently upregulate miR-124 and miR-506 and downregulate PDK2 and circHIPK3 (). Overall, these data revealed that circHIPK3 knockdown could also prevent the growth of HCC in nude mice and regulate miR-124, miR-506, and PDK2.
Figure 5. Knock down of circHIPK3 dramatically suppressed growth and upregulated miR-124 and miR-506 in HCC xenograft tumors. (a) After 4 weeks, groups of tumors were present in nude mice as observed by photography and comparison. (b) The length and width of the tumors were measured, and their volume was calculated at 0, 7, 14, 21 and 28 days. RT–qPCR assays revealed the expression changes of miR-506 (c), miR-124 (d), circHIPK3 (e) and PDK2 (f) in xenograft tumors. *P< 0.05, **P< 0.01, ***P< 0.001.
Discussion
HCC is one of the most universal malignant tumors of the digestive system worldwide and has not been fully and effectively controlled [Citation38]. Currently, finding new molecular targets to explore more effective therapeutic strategies has become an urgent challenge for HCC therapy. CircRNAs, as endogenous noncoding RNAs, have a covalently circular closed single-stranded RNA structure [Citation39]. Recently, researchers have gradually recognized the crucial role of circRNAs in the life sciences [Citation40]. CircHIPK3 (hsa_circ_0000284) is located on chr11:33,307,958–33,309,057 and is derived from the HIPK3 gene [Citation41]. In the literature, circHIPK3 has been shown to contribute to the acceleration of the progression of multiple cancers. For instance, loss of circHIPK3 could accelerate autophagy of lung cancer cells [Citation18]. Furthermore, circHIPK3 could induce tumorigenesis of renal cancer cells [Citation19], enhance invasion of thyroid cancer [Citation24], and also facilitate growth, metastasis and oxaliplatin-resistance of colorectal cancer cells [Citation42,Citation43]. The biological function of circHIPK3 in HCC has been reported in present studies, suggesting that circHIPK3 may contribute to HCC [Citation44–46]. In our study, we demonstrated that circHIPK3 was significantly upregulated in HCC tissues. Knock down of circHIPK3 also notably prevented the proliferation and invasion of HCC cells. Thus, we speculated that circHIPK3 could also accelerate the tumorigenesis of HCC cells.
CircRNAs act a miRNA sponges and play a crucial role in translation regulation [Citation10,Citation47]. A close association between circRNAs and miRNAs has been confirmed in various cancers [Citation48]. miRNAs are a type of endogenous, short and highly conserved small noncoding RNA [Citation49,Citation50]. miRNAs can regulate gene expression at the posttranscriptional stage by binding to the 3’-UTR of mRNAs [Citation51]. Many studies have shown that abnormal expression of miRNAs is associated with cancer processes, including proliferation, metastasis and apoptosis [Citation52,Citation53]. Through bioinformatics analysis, we unexpectedly discovered that there were potential binding sites between circHIPK3 and miR-124 or miR-506, indicating that miR-124 or miR-506 might be targeted by circHIPK3. miR-124 has also been reported to inhibit several cancer processes, including breast cancer [Citation54], gastric cancer [Citation55], and bladder cancer [Citation56]. Furthermore, miR-506 has also been proven to act as a tumor suppressor in multiple cancers, such as thyroid cancer [Citation57], cervical cancer [Citation58],colorectal cancer [Citation59] and HCC [Citation60]. Moreover, we discovered that miR-124 could delay hepatic carcinogenesis by interacting with miR-506 [Citation61]. Previous study revealed that circHIPK3 regulates AQP3 expression via miR-124 [Citation46]. However, it is not known whether circHIPK3 can accelerate the process of HCC through miR-124 and miR-506. In our study, we further confirmed that circHIPK3 can act as a sponge by regulating the expression of miR-124 and miR-506. In addition, circHIPK3 could also induce the proliferation and invasion of HCC cells by targeting miR-124 and miR-506. Therefore, we proved that in the progression of liver cancer formation, miR-124 and miR-506 could act as tumor suppressors, and miR-124 and miR-506 were the downstream regulatory targets of circHIPK3 in HCC.
Research has also demonstrated that circRNAs can prevent the inhibitory effect of miRNAs on mRNAs by sponging miRNAs [Citation62]. Although the basic biological roles of the circHIPK3/miR-124 or miR-506 axis in HCC were preliminarily investigated in this study, the mechanism of miR-124 and miR-506 was not further studied. Through bioinformatics prediction, we further discovered that PDK2 might be a possible target gene of miR-124 and miR-506. Thus, we speculated that circHIPK3 could also regulate the target gene (PDK2) of miR-124 and miR-506 by acting as a competitive endogenous RNA (ceRNA). Related studies have verified that tumor cells also rely on the glycolysis pathway as their main energy source even under normal oxygen content [Citation63,Citation64]. Pyruvate dehydrogenase kinases (PDKs), as key regulators of the aerobic glycolysis pathway, can change cell metabolism by affecting aerobic glycolysis and participate in malignant cell transformation [Citation65]. Among them, PDK2 was most closely associated with tumors. PDK2 is abnormally overexpressed in multiple tumors, which is relevant to the malignant phenotype of tumors [Citation66,Citation67]. In addition, research confirmed that PDK2 is upregulated in HCC, and downregulation of PDK2 can suppress the proliferation and metastasis of HCC cells [Citation68].
In our current study, we further confirmed that as the target gene of miR-124 or miR-506, PDK2 can also be involved in the prevention of miR-124- or miR-506-mediated liver cancer cell proliferation and invasion. Therefore, we further proved the role of the miR-124 or miR-506/PDK2 axis in HCC cells.
Conclusions
Our study indicated a novel regulatory loop involving the circHIPK3/miR-124 or miR-506/PDK2 axis in HCC progression. These data suggested that circHIPK3 might be a novel therapeutic target for HCC therapy.
Article highlights
circHIPK3 inhibited proliferation and invasion of HCC
miRNA-124/506 directly target PDK2
circHIPK3 positively regulates PDK2 expression by sponging miRNA-124 and miRNA-506
Acknowledgements
This work was supported by the Natural Science Foundation of Fujian Province (2019J01551) and the Medical Innovation Project of Fujian Province (2019-CXB-36).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Data Availability Statement
The data and material are available from the corresponding author upon reasonable request.
Additional information
Funding
References
- Sung HA-O, Ferlay, J, Siegel, RA-O, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries, CA: A Cancer Journal for Clinicians, 2021;71(3): 209-249.
- Forner A, Reig M, Bruix J. Hepatocellular carcinoma. Lancet. 2018;391(10127):1301–1314.
- Kulik L, El-Serag HB. Epidemiology and management of hepatocellular carcinoma. Gastroenterology. 2019;156(2):477–491.e1.
- Villanueva A. Hepatocellular carcinoma. N Engl J Med. 2019;380(15):1450–1462.
- Yang JD, Hainaut P, Gores GJ, et al. A global view of hepatocellular carcinoma: trends, risk, prevention and management. Nat Rev Gastroenterol Hepatol. 2019;16(10):589–604.
- Tellapuri S, Sutphin PD, Beg MS, et al. Staging systems of hepatocellular carcinoma: a review. Indian J Gastroenterol. 2018;37(6):481–491.
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34(2):153–159.
- Piñero F, Dirchwolf M, Pessôa MG. Biomarkers in hepatocellular carcinoma: diagnosis, prognosis and treatment response assessment. Cells. 2020;9(6):1370.
- Jiří T, Igor K, Ba M. Hepatocellular carcinoma future treatment options. Klin Onkol. 2020;33(Supplementum 3):26–29.
- Hansen TB, Jensen TI, Clausen BH, et al. Natural RNA circles function as efficient microRNA sponges. Nature. 2013;495(7441):384–388.
- Jeck WR, Sorrentino JA, Wang K, et al. Circular RNAs are abundant, conserved, and associated with ALU repeats. Rna. 2013;19(2):141–157.
- Cao S, Wang G, Wang J, et al. Hsa_circ_101280 promotes hepatocellular carcinoma by regulating miR-375/JAK2. Immunol Cell Biol. 2019;97(2):218–228.
- Chen W, Quan Y, Fan S, et al. Exosome-transmitted circular RNA hsa_circ_0051443 suppresses hepatocellular carcinoma progression. Cancer Lett. 2020;475:119–128.
- He S, Yang J, Jiang S, et al. Circular RNA circ_0000517 regulates hepatocellular carcinoma development via miR-326/IGF1R axis. Cancer Cell Int. 2020;20(1):404.
- Wang X, Wang X, Li W, et al. Up-Regulation of hsa_circ_0000517 predicts adverse prognosis of hepatocellular carcinoma. Front Oncol. 2019;9:1105.
- Yao Z, Xu R, Yuan L, et al. Circ_0001955 facilitates hepatocellular carcinoma (HCC) tumorigenesis by sponging miR-516a-5p to release TRAF6 and MAPK11. Cell Death Dis. 2019;10(12):945.
- Zhang XJ, Cao G, Fu J, et al. The role of hsa_circ_0000285 in metastasis of hepatocellular carcinoma. Eur Rev Med Pharmacol Sci. 2020;24(13):7204.
- Chen X, Mao R, Su W, et al. Circular RNA circHIPK3 modulates autophagy via MIR124-3p -STAT3-PRKAA/AMPKα signaling in STK11 mutant lung cancer. Autophagy. 2020;16(4):659–671.
- Lai J, Xin J, Fu C, et al. CircHIPK3 promotes proliferation and metastasis and inhibits apoptosis of renal cancer cells by inhibiting MiR-485-3p. Cancer Cell Int. 2020;20(1):248.
- Li Q, Tian Y, Liang Y, et al. CircHIPK3/miR-876-5p/PIK3R1 axis regulates regulation proliferation, migration, invasion, and glutaminolysis in gastric cancer cells. Cancer Cell Int. 2020;20(1):391.
- Li Y, Zheng F, Xiao X, et al. Circ HIPK 3 sponges miR-558 to suppress heparanase expression in bladder cancer cells. EMBO Rep. 2017;18(9):1646–1659.
- Liu F, Fan Y, Ou L, et al. CircHIPK3 facilitates the G2/M transition in prostate cancer cells by sponging miR-338-3p. Onco Targets Ther. 2020;13:4545–4558.
- Liu Y, Xia L, Dong L, et al. CircHIPK3 Promotes Gemcitabine (GEM) resistance in pancreatic cancer cells by sponging miR-330-5p and targets RASSF1. Cancer Manag Res. 2020;12:921–929.
- Shu T, Yang L, Sun L, et al. CircHIPK3 promotes thyroid cancer tumorigenesis and invasion through the Mirna-338-3p/RAB23 AXIS. Med Princ Pract. 2020. DOI:10.1159/000512548.
- Lee WJ, Moon J, Jeon D, et al. Possible epigenetic regulatory effect of dysregulated circular RNAs in epilepsy. PLoS One. 2018;13(12):e0209829.
- Chan JJ, Tay Y. Noncoding RNA:RNA regulatory networks in cancer. Int J Mol Sci. 2018;19(5):1310.
- Xiong DD, Dang Y-W, Lin P, et al. A circRNA-miRNA-mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16(1):220.
- Tay Y, Rinn J, Pandolfi PP. The multilayered complexity of ceRNA crosstalk and competition. Nature. 2014;505(7483):344–352.
- Majid A, Wang J, Nawaz M, et al. miR-124-3p suppresses the invasiveness and metastasis of hepatocarcinoma cells via targeting CRKL. Front Mol Biosci. 2020;15(7):223.
- Dalby B, et al. Advanced transfection with Lipofectamine 2000 reagent: primary neurons, siRNA, and high-throughput applications. Methods. 2004; 33(2):95-103.
- Zhao W, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR-20a-5p/HMGA2 axis in breast cancer. Cancer Med. 2018; 7(3):842-855.
- Wang S, et al. Long non-coding RNA LOC285194 inhibits proliferation and migration but promoted apoptosis in vascular smooth muscle cells via targeting miR-211/PUMA and TGF-β1/S100A4 signal. Bioengineered. 2020;11(1):718-728.
- Li D, Cheng X, Zheng W, et al. Glucosamine-6-phosphate isomerase 1 promotes tumor progression and indicates poor prognosis in hepatocellular carcinoma. Cancer Manag Res. 2020;12:4923-4935.
- Shi Z, Shen C, Yu C, et al. Long non-coding RNA LINC00997 silencing inhibits the progression and metastasis of colorectal cancer by sponging miR-512-3p. Bioengineered. 2021;12(1):627-639 .
- Xu X, Zheng SA-O. MiR-887-3p negatively regulates STARD13 and promotes pancreatic cancer progression. (1179-1322 (Print)).
- Zhang M, Xu Y, Yin S, et al. YY1-induced long non-coding RNA PSMA3 antisense RNA 1 functions as a competing endogenous RNA for microRNA 214-5p to expedite the viability and restrict the apoptosis of bladder cancer cells via regulating programmed cell death-ligand 1. Bioengineered. 2021;12(2):9150-9161.
- Cai C, Zhang Y, Peng XA-O. Knocking down Sterol regulatory element binding protein 2 (SREBF2) inhibits the Serine Protease 8 (PRSS8) /sodium channel epithelial 1alpha subunit (SCNN1A) axis to reduce the cell proliferation, migration and epithelial-mesenchymal transformation of ovarian cancer. (2165-5987 (Electronic)).
- Frager SZ, Schwartz JM. Hepatocellular carcinoma: epidemiology, screening, and assessment of hepatic reserve. Curr Oncol. 2020;27(Suppl 3):S138–s143.
- Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Li H, Heng B, Ouyang P, et al. Comprehensive profiling of circRNAs and the tumor suppressor function of circHIPK3 in clear cell renal carcinoma. J Mol Histol. 2020;51(3):317–327.
- Zeng K, Chen X, Xu M, et al. CircHIPK3 promotes colorectal cancer growth and metastasis by sponging miR-7. Cell Death Dis. 2018;9(4):417.
- Zhang Y, Li C, Liu X, et al. circHIPK3 promotes oxaliplatin-resistance in colorectal cancer through autophagy by sponging miR-637. EBioMedicine. 2019;48:277–288.
- Li W, Xue H, Li Y, et al. HIPK3 Circular RNA promotes metastases of HCC through sponging miR-338-3p to Induce ZEB2 expression. Dig Dis Sci. 2021;66(10):3439-3447.
- Zhang H, Dai Q, Zheng L, et al. Knockdown of circ_HIPK3 inhibits tumorigenesis of hepatocellular carcinoma via the miR-582-3p/DLX2 axis. Biochem Biophys Res Commun. 2020;533(3):501-509.
- Chen G, Shi Y, Liu M, et al. circHIPK3 regulates cell proliferation and migration by sponging miR-124 and regulating AQP3 expression in hepatocellular carcinoma. Cell Death Dis. 2018;9(2):175.
- Li Z, Huang C, Bao C, et al. Exon-intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol. 2015;22(3):256–264.
- Dong Y, He D, Peng Z, et al. Circular RNAs in cancer: an emerging key player. J Hematol Oncol. 2017;10(1):2.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207.
- Qianqian Tang SW, Qiao X, Wang F, et al. MiR-29 promotes ovarian carcinoma cell proliferation through the PTEN pathway. Eu J Gynaecol Oncol. 2020;41(5):774–778.
- Tafrihi M, Hasheminasab E. MiRNAs: biology, biogenesis, their web-based tools, and databases. Microrna. 2019;8(1):4–27.
- Andersen GB, Tost J. Circulating miRNAs as Biomarker in Cancer. Recent Results Cancer Res. 2020;215:277–298.
- Shah V, Shah J. Recent trends in targeting miRNAs for cancer therapy. J Pharm Pharmacol. 2020;72(12):1732–1749.
- Cai WL, Huang W-D, Li B, et al. microRNA-124 inhibits bone metastasis of breast cancer by repressing Interleukin-11. Mol Cancer. 2018;17(1):9.
- Liang F, Zhang H, Qiu Y, et al. MiR-124-5p inhibits the progression of gastric cancer by targeting MIEN1. Technol Cancer Res Treat. 2020;19:1533033820979199.
- Zo RB, Long Z. MiR-124-3p suppresses bladder cancer by targeting DNA methyltransferase 3B. J Cell Physiol. 2018;234(1):464–474.
- Chen L, Wang X, Ji C, et al. MiR-506-3p suppresses papillary thyroid cancer cells tumorigenesis by targeting YAP1. Pathol Res Pract. 2020;216(12):153231.
- Gong M, Chen C, Zhao H, et al. miR-506 suppresses cervical cancer cell proliferation both in vitro and in vivo. Neoplasma. 2018;65(3):331–338.
- Huang M, Xie X, Song X, et al. MiR-506 suppresses colorectal cancer development by inhibiting orphan nuclear receptor NR4A1 expression. J Cancer. 2019;10(15):3560–3570.
- Chen MA-O, Hu G, Zhou X, et al. Hsa_circ_0016788 regulates hepatocellular carcinoma progression via miR-506-3p/poly-adenosine diphosphate-ribose polymerase. J Gastroenterol Hepatol. 2021;36(12):3457-3468. DOI:10.1111/jgh.15635.
- Xiang H, Luo M, Hou P, et al. miR-124-3p combined with miR-506-3p delay hepatic carcinogenesis via modulating sirtuin 1. Biomarkers. 2021;26(3):196–206.
- Meng S, Zhou H, Feng Z, et al. CircRNA: functions and properties of a novel potential biomarker for cancer. Mol Cancer. 2017;16(1):94.
- Abbaszadeh Z, Çeşmeli S, Biray Avcı Ç. Crucial players in glycolysis: cancer progress. Gene. 2020;726:144158.
- Wu Z, Wu J, Zhao Q, et al. Emerging roles of aerobic glycolysis in breast cancer. Clin Transl Oncol. 2020;22(5):631–646.
- Woolbright BL, Rajendran G, Harris RA, et al. Metabolic Flexibility in Cancer: targeting the Pyruvate Dehydrogenase Kinase:Pyruvate Dehydrogenase Axis. Mol Cancer Ther. 2019;18(10):1673–1681.
- Cui L, Cheng Z, Liu Y, et al. Overexpression of PDK2 and PDK3 reflects poor prognosis in acute myeloid leukemia. Cancer Gene Ther. 2020;27(1–2):15–21.
- Hu T, Yu S, Li Y, et al. PDK2 induces cisplatin-resistance in lung adenocarcinoma via transcriptional regulation of CNNM3. J Drug Target. 2019;27(4):460–465.
- Gong J, Du C, Sun N, et al. Circular RNA hsa_circ_0005397 promotes hepatocellular carcinoma progression by regulating the miR-326/PDK2 axis. J Gene Med. 2021;23(6):e3332.
