ABSTRACT
Non-small cell lung cancer (NSCLC) is still one of the most challenging malignant tumors. Deregulation of circular RNAs (circRNAs) is associated with NSCLC progression. However, the regulatory mechanism of circRNAs in NSCLC still needs to be studied. We selected a differentially expressed hsa_circ_0006677 (circ_0006677) in NSCLC through analyzing the GSE158695 and GSE112214 datasets. Expression of circ_0006677 was evaluated by real-time quantitative polymerase-chain reaction (RT-qPCR). Effects of circ_0006677 overexpression on NSCLC cell proliferation, apoptosis, migration, invasion, and stemness were determined by clonogenic, 5-ethynyl-2’-deoxyuridine (EdU), flow cytometry, transwell, and sphere formation assays. The regulatory mechanism of circ_0006677 was predicted by bioinformatics analysis and verified by dual-luciferase reporter and RIP assays. Animal experiments were carried out to validate the function of circ_0006677 in vivo. We observed the downregulation of circ_0006677 in NSCLC samples and cells. Functionally, circ_0006677 overexpression decreased xenograft tumor growth and restrained NSCLC cell proliferation, invasion, migration, stemness, and induced NSCLC cell apoptosis in vitro. Molecular mechanism experiments exhibited that circ_0006677 functioned as a miR-1245a sponge and mediated SATB2 expression through adsorbing miR-1245a. Either miR-1245a overexpression or SATB2 knockdown weakened circ_0006677 overexpression-mediated repression on proliferation, invasion, migration, and stemness. In conclusion, circ_0006677 regulated SATB2-mediated tumor-suppressive effect via acting as a miR-1245a sponge in NSCLC, providing a new mechanism for understanding the progression of NSCLC.
KEYWORDS:
Introduction
Non-small cell lung cancer (NSCLC), which makes up to 80% of lung cancer cases, is an aggressive disease with a poor prognosis [Citation1]. Approximately 25% of patients are diagnosed with NSCLC disease amenable to potentially radical surgery [Citation2]. Despite this, the five-year survival rate for stage I NSCLC patients undergoing radical resection is 50% to 70%, while that for stage IIIA patients is 10% to 30% [Citation3]. Agents targeting NSCLC-driven mutations have revolutionized the approach to patients with metastatic disease [Citation4]. Therefore, exploring the molecular mechanisms that drive NSCLC progression is essential for the development of new targeted therapeutic agents [Citation5].
Recent studies have shown the contribution of circular RNAs (circRNAs) in the developmental and pathological processes [Citation6]. CircRNAs, a novel class of transcripts, are characterized by covalently linked 3’-5’ ends. The biogenesis of circRNAs via the back-splicing mechanism differs from the standard splicing mechanism used to produce linear RNA [Citation7]. This closed structure endows them with resistance to exonuclease degradation and attracts researchers to them because of their potential to become targets for the treatment of diseases [Citation8,Citation9]. An important feature of circRNA is its microRNA (miRNA) sponge function, which can effectively inhibit the activity of miRNA by binding to miRNA, resulting in affecting the expression of downstream genes and ultimately participating in various diseases [Citation10]. For instance, circRNA-cRAPGEF5 exerts a suppressive effect on renal cell carcinoma growth and metastasis through elevating TXNIP expression via sequestering miR-27a-3p [Citation11]. Another example is circ-solute carrier family 8 member A1, which acts as an endogenous miR-130b/miR-494 sponge to inhibit bladder cancer progression by increasing phosphatase and tensin homolog expression [Citation12]. Researchers now recognize that circRNAs act as pivotal molecular regulators in NSCLC [Citation13]. Nevertheless, the regulatory mechanism of circRNAs in NSCLC needs to be further elucidated.
Here, circ_0006677, derived from the WD repeat domain 78 (WDR78) gene on chr1:67,356,836–67,371,058, was selected as a differentially expressed circRNA in NSCLC based on bioinformatics analysis (the GSE158695 and GSE112214 datasets). This study aims to explore whether circ_0006677 is involved in the progression of NSCLC by functioning as a sponge for miRNA. We identified a novel mechanism by which circ_0006677 repressed NSCLC cell malignancy and stemness via regulating Special AT-rich binding protein-2 (SATB2) expression through function as a miR-1245a sponge.
Materials and methods
Patient specimens
The NSCLC tumor samples and corresponding non-tumor samples (n = 55) obtained during tumor resection surgery in the Civil Aviation General Hospital were stored at −80°C until use. All sample collections were done following a protocol approved by the Ethics Committee of the Civil Aviation General Hospital. All the participants gave their written informed consent before they entered the study.
Cell culture
Human bronchial epithelial cell line 16 HBE (#CL-0249) and lung cancer cell lines H1299 (#CL-0165), A549 (#CL-0016), HCC827 (#CL-0094), and H460 (#CL-0299) were obtained from Procell (Wuhan, China). Except the A549 cell line was maintained in Ham’s F-12 K medium (Thermo, Waltham, MA, USA), other cell lines were cultured in RPMI-1640 medium (Thermo). The growth conditions for these cells were in an incubator with 5% carbon dioxide at 37°C. To maintain cell growth, these media need to be supplemented with 10% FBS (Thermo) and 1% Penicillin–Streptomycin (Sigma-Aldrich, St. Louis, MO, USA).
RNA preparation
The total RNA extraction kit (Solarbio, Beijing, China) was utilized for the extraction of total RNA from collected samples and cultured cells as per manufacturer’s instructions. The active motif’s nuclear extract kit (Carlsbad, CA, USA) was used for the fractionation of nuclear and cytoplasmic RNA from cultured cells following the manufacturer’s instructions.
RNase R digestion
RNase R digestion was performed as previously described [Citation14] using RNase R (Epicenter Technologies, USA) at 37°C for 30 min. Three independent experiments were done in triplicate.
Real-time quantitative polymerase chain reaction (RT-qPCR)
Generation of cDNA was conducted using the script RT reagent kit (Takara, Tokyo, Japan) for circRNA and mRNA. For miRNA, reverse transcription was done using the miRCURY LNA™ Universal RT microRNA PCR system (Exiqon, Aarhus, Denmark). Amplifications were carried out using the Lightcycler 480 (Roche, Basel, Switzerland) in triplicate with an SYBR Premix Ex Taq II (Takara). Relative fold change was evaluated using the equation 2−ΔΔCt [Citation15], and all genes were normalized to the housekeeping gene β-actin or U6. Primer sequences were listed in .
Table 1. Primers for RT-qPCR used in the study
Plasmid construction
To construct the circ_0006677 overexpression plasmid, the cDNA of circ_0006677 was cloned into the pLO5-ciR vector (Geneseed, Guangzhou, China). The lentivirus-containing short hairpin RNA (shRNA) targeting SATB2 (sh-SATB2) was generated using the pLKO.1 vector (Addgene, Cambridge, MA, USA), and sh-NC was used as a control. Production of lentiviral particles and transduction of NSCLC cells (H1299 and A549) were performed as described [Citation16,Citation17].
miRNA mimic and inhibitor
The Lipofectamine RNAiMAX Reagent (Thermo) was utilized for the transfection of NSCLC cells with miR-1245a mimic (5’-AAGUGAUCUAAAGGCCUACAU-3’) and miR-NC (5’-CGGUACGAUCGCGGCGGGAUAUC-3’) as per manufacturer’s instructions.
Clonogenic assay
Clonogenic assay was conducted as the report of Mehta et al. [Citation18]. The transfected NSCLC cells (5 × 102 cells/well) were allowed to grow on 6-well plates for 14 days, followed by washing with PBS (Thermo). After staining with 0.25% crystal violet (Solarbio), the colonies (more than 50 cells) were captured and counted by a microscope (Olympus, Tokyo, Japan).
5-ethynyl-2’-deoxyuridine (EdU) assay
The EdU incorporation assay was carried out using Yefluor 488 EdU Imaging Kit (Yesen, Shanghai, China) following the manufacturer’s instructions. The EdU-positive cells were captured using an Olympus fluorescence microscope.
Measurement of the caspase 3 activity
Detection of caspase-3 activity was performed using the caspase-3/CPP32 colorimetric analysis kit (Bio Vision, USA) following the manufacturer’s steps. In brief, the supernatant of the lysed cells was diluted and added to the reaction tube, followed by the addition of reaction buffer and cetyl-Asp-Glu-Val-Asp p-nitroanilide substrate (DEVD-pNA). After incubating for 1 h, the optical density at 405 nm was measured in a microtiter plate reader (BioTek, Winooski, VT, USA).
Flow cytometry assay
Detection of cell apoptosis was conducted using the Annexin V-fluorescein isothiocyanate/Prodium Iodide kit (Keygen, Nanjing, China) according to the producer’s guidelines. The flow cytometer (Becton Dickinson, NJ, USA) was used to detect the cells.
Transwell assay
Analysis of cell migration and invasion was performed using transwell assays as previously described [Citation19]. Harvested NSCLC cells (1 × 105/chamber) re-suspended in a 200 μL of cell medium-serum free were added into the upper compartment of 24-well transwell chambers (Corning, NY, USA) without or with Matrigel (Corning), and 600 μL of complete medium was added to the bottom compartment. The cells were allowed to grow for 24 h. Migrating and invading cells fixed with 4% paraformaldehyde (Solarbio) and stained with 0.5% crystal violet (Solarbio) were captured and counted with a microscope (Olympus).
Sphere formation assay
The experiment was performed as previously described [Citation20]. Single-cell suspensions (2.5 × 103/well) were seeded onto a 24-well ultra-low attachment plate (Corning) in serum-free medium supplemented with 20 ng/mL epidermal growth factor (Sigma-Aldrich), 20 ng/mL fibroblast growth factor (Sigma-Aldrich), 4 mg/mL heparin (Sigma-Aldrich), and 2% B27 (Sigma-Aldrich). Fourteen days later, the number of sphere formations was counted under a microscope (Olympus).
Western blotting
Isolation of total protein from collected samples and cultured cells was performed using radioimmunoprecipitation assay lysis with protease and phosphates inhibitor (Sigma-Aldrich). Western blotting was carried out as previously described [Citation21]. Equal amount of protein extracts was loaded into 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis, followed by transferring onto a polyvinylidene fluoride membrane (Millipore, Billerica, MA, USA), which was then blocked with 5% skim milk powder. The membrane was probed with antibodies against proliferating cell nuclear antigen (PCNA) (ab92552, Abcam, Cambridge, MA, USA), E-cadherin (E-ca) (ab40772, Abcam), N-cadherin (N-ca) (ab18203, Abcam), SATB2 (ab92446, Abcam), matrix metalloproteinase (MMP)-2 (ab92536, Abcam), MMP-9 (ab76003, Abcam), t-MEK (#4694, Cell Signaling Technology, MA, USA), p-MEK (#3958, Cell Signaling Technology), t-ERK (#9102, Cell Signaling Technology), p-ERK (#8544, Cell Signaling Technology), and β-actin (ab8224, Abcam). Membranes were then incubated with a secondary antibody (Abcam). Protein bands were viewed using an enhanced chemiluminescence detection system with a chemiluminescence horseradish peroxidase (HRP) Substrate (Millipore).
Dual-luciferase reporter assay
The experiment was done according to the report by Wei et al. [Citation22]. The recombinant luciferase reporters circ_0006677 wild type (WT), circ_0006677 mutant (MUT), SATB2 3’ untranslated region (UTR) WT, and SATB2 3ʹUTR MUT were constructed using the pmirGLO vector (Promega, Madison, WI, USA), respectively. NSCLC cells were co-transfected with pRL-TK, recombinant luciferase reporter, and miR-NC or miR-1245a mimic. Luciferase activities of firefly luciferase and Renilla luciferase were measured using the dual-luciferase reporter system (Promega) after 48 h of transfection.
RIP assay
The RIP assay was used to analyze the binding between circ_0006677 and miR-1245a using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) according to the manufacturer’s instructions. The co-precipitated RNA complex was extracted and purified with the RNeasy kit (Qiagen, Valencia, CA, USA), followed by subjecting to RT-qPCR analysis.
Xenograft assay
Animal experiments were approved by the Animal Ethics Committee of the Civil Aviation General Hospital. Xenograft assay was done as previously depicted [Citation23]. Twelve BALB/c male nude mice (4 weeks old, 15–20 g) (Vital River Laboratory, Beijing, China) were reared in a highly clean SPF environment. A549 cells (5 × 106) stably overexpression of circ_0006677 in 100 μL of PBS were subcutaneously injected into the back of each nude mouse (n = 6 mice per group), and A549 cells carrying sh-NC were used as controls. Tumor volume was recorded once a week [(length × width2)/2]. After 28 days, the mice were sacrificed and their xenograft tumors were stripped.
Immunohistochemical (IHC) staining
IHC assay was conducted based on the report of Xu et al. [Citation24]. Xenograft tumors fixed with 4% formaldehyde were embedded in paraffin and cut into 4 μm sections. The sections were deparaffinized, rehydrated, antigen retrieval, and blocked, followed by incubation of antibodies against SATB2 (ab92446, Abcam), PCNA (ab92552, Abcam), and Bax (ab32503, Abcam). After incubation with HRP-conjugated secondary antibody, the sections were stained with 3,3’-Diaminobenzidine and hematoxylin, followed by viewing under a microscope (Olympus).
Statistical analysis
All experiments were repeated at least three times. Results, which were presented as mean ± standard deviation, were analyzed using GraphPad Prism version 8 software (GraphPad, La Jolla, CA, USA). Comparisons of numerical values were performed using Student’s t-test or analysis of variance. A difference was considered statistically significant when P < 0.05.
Results
Circ_0006677 was prominently underexpressed in NSCLC
To explore the role of circRNAs in NSCLC, we first searched for differentially expressed circRNAs in NSCLC through analyzing the GSE158695 and GSE112214 datasets. Venn diagram showed that circ_0006677 was the only downregulated differentially expressed circRNA with a change greater than 4 in both two datasets (). And the expression of circ_0006677 in the GSE158695 and GSE112214 datasets was shown in . Circ_0006677 is derived from the WDR78 gene on chr1:67,356,836–67,371,058, resulting from back-splicing of exons 2, 3, and 4 (473 bp) (). We then verified that circ_0006677 was prominently underexpressed in NSCLC via detection of circ_0006677 expression in 55 NSCLC samples (). Compared to the 16HBE cell line, NSCLC cell lines exhibited an overt reduction in circ_0006677 expression (). Moreover, linear WDR78 was severely degraded after RNase R incubation, but circ_0006677 was hardly affected by this enzyme (). The subcellular localization of circ_0006677 in NSCLC cells was analyzed. Data in displayed that the majority of circ_0006677 localized to the cytoplasm. Collectively, low circ_0006677 expression might be related to NSCLC tumorigenesis.
Figure 1. The expression and characteristics of circ_0006677 in NSCLC. (a) Venn diagram showing the overlap of downregulated differentially expressed circRNA in the GSE158695 and GSE112214 datasets with a fold change of more than 4. (b and c) Relative expression of circ_0006677 in the GSE158695 and GSE112214 datasets. (d) Schematic illustration of circ_0006677. (e and f) RT-qPCR analysis of circ_0006677 expression in NSCLC samples and cell lines. (g and h) RT-qPCR analysis of the levels of circ_0006677 and WDR78 mRNA in total RNA with or without RNase R incubation. (i and j) The subcellular localization of circ_0006677 in NSCLC cells. *P < 0.05.
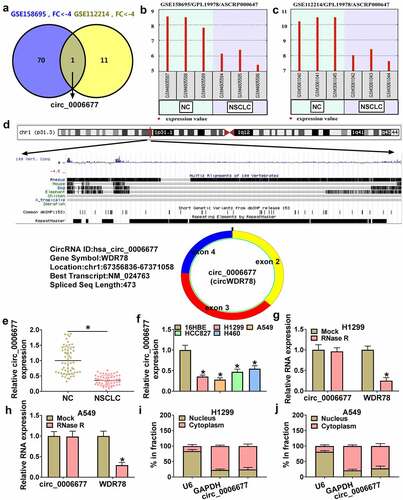
Circ_0006677 overexpression decreased NSCLC cell malignancy and stemness
Subsequently, we constructed the circ_0006677 overexpression plasmid to further analyze the function of circ_0006677 in NSCLC. RT-qPCR demonstrated the transfection efficiency of the circ_0006677-overexpressing plasmid (). Clonogenic and EdU assays showed that circ_0006677 overexpression decreased the proliferative capability of NSCLC cells (). In contrast, circ_0006677 overexpression significantly increased the caspase 3 activity in NSCLC cells and promoted the apoptosis of NSCLC cells (). Transwell assays exhibited that exogenetic expression of circ_0006677 restrained the migrating and invading capabilities of NSCLC cells (). Sphere formation assay displayed that the number of spheroids in the circ_0006677-overexpressing group was reduced significantly (). Overexpression of circ_0006677 reduced N-ca and PCNA protein levels and increased E-ca protein levels in NSCLC cells (). In addition, there were lower protein levels of MMP-2 and MMP-9 in circ_0006677-overexpressed NSCLC cells than the control cells (). Together, these results suggested that circ_0006677 overexpression decreased NSCLC cell malignancy and stemness.
Figure 2. Impacts of circ_0006677 overexpression on NSCLC cell malignancy and stemness. (a) RT-qPCR exhibiting the transfection efficiency of the circ_0006677 overexpression plasmid. (b-i) NCCLC cells transfected with circ_0006677 or vector were utilized. (b and c) Clonogenic and EdU assays detection of the proliferation of NCCLC cells. (d) Analysis of the caspase 3 activity in NSCLC cells. (e) Flow cytometry assay evaluation of the apoptosis of NCCLC cells. (f and g) Transwell assays analysis of the migration and invasion of NCCLC cells. (h) Sphere formation assay detection of the stemness of NCCLC cells. (i and j) Western blotting showing N-ca, PCNA, and E-ca protein levels in NSCLC cells. (k and l) Western blotting showing MMP-2 and MMP-9 protein levels in NSCLC cells. *P < 0.05.
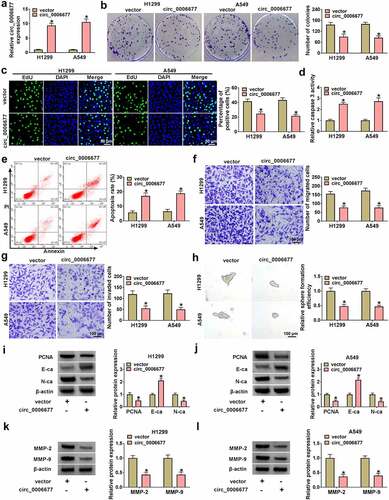
Circ_0006677 served as a miR-1245a sponge
Considering the importance of circRNAs as miRNA sponges, we conducted bioinformatics analysis (Circular RNA Interactome) [Citation25]. By consulting the literature [Citation26], it was found that circ_0006677 may be combined with miR-1245a (). To validate this prediction, we used miR-1245a mimic to overexpress miR-1245a in NSCLC (). Luciferase reporter assays showed that a remarkable reduction in luciferase reporter activity was detected in NSCLC cells co-transfected with miR-1245a mimic and the circ_0006677 WT reporter, but not that of the circ_0006677 MUT reporter (). RIP assay exhibited that circ_0006677 and miR-1245a could be amplified from the complex precipitated by the anti-Ago2 antibody, suggesting that circ_0006677 bound with miR-1245a via Ago2 (). Additionally, we also detected miR-1245a expression in NSCLC samples, and the results exhibited a prominent elevation in miR-1245a expression in NSCLC samples compared to corresponding non-tumor samples (). And Pearson’s correlation coefficient analysis exhibited a negative correlation between circ_0006677 and miR-1245a expression in NSCLC samples (). Homoplastically, miR-1245a was overexpressed in NSCLC cells (). Also, circ_0006677 overexpression decreased miR-1245a expression, but this decrease was weakened by co-transfection with circ_0006677 and miR-1245a mimic (). Together, these results suggested that circ_0006677 functioned as a miR-1245a sponge.
Figure 3. The function of circ_0006677 as a miR-1245a sponge. (a) Bioinformatics prediction of binding sites between circ_0006677 and miR-1245a. (b) The transfection efficiency of miR-1245a mimic. (c and d) The luciferase activity of the circ_0006677 WT/circ_0006677 MUT reporter in NSCLC cells with or without miR-1245a mimic. (e and f) Relative levels of circ_0006677 and miR-1245a in the complex precipitated by the anti-Ago2/anti-IgG antibody. (g) Relative expression of miR-1245a in NSCLC samples. (h) Pearson’s correlation coefficient analysis showing the correlation of miR-1245a and circ_0006677 in NSCLC samples. (i) Relative expression of miR-1245a in NSCLC cells. (j) Relative expression of miR-1245a in NSCLC cells after transfection with circ_0006677 combined with or without miR-1245a mimic. *P < 0.05.
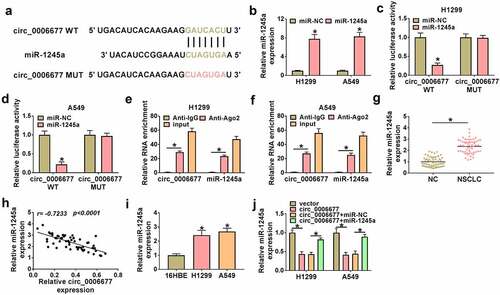
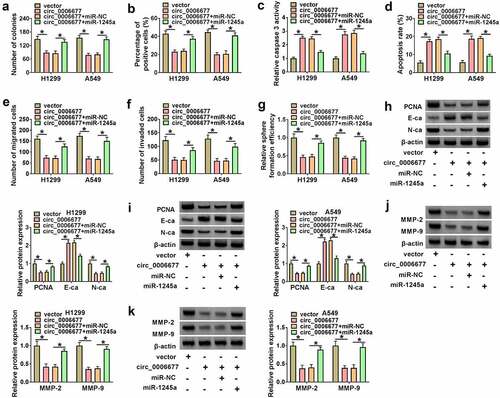
Circ_0006677 exerted its anti-tumor effect via repressing miR-1245a
Whether circ_0006677 functions as a tumor suppressor gene through sequestering miR-1245a was further validated. We co-transfected the circ_0006677 overexpression plasmid and miR-1245a mimic into NSCLC to determine whether the tumor-inhibiting effect of circ_0006677 could be impaired by miR-1245a overexpression. We observed that introduction of miR-1245a mimic partly overturned the decreased proliferation of NSCLC cells mediated by circ_0006677 overexpression (). Moreover, the elevated caspase 3 activity and apoptosis rate in circ_0006677-overexpressing NSCLC cells were partly antagonized by introduction of miR-1245a mimic (). Ectopic expression of miR-1245a also attenuated the inhibiting effects of circ_0006677 upregulation on NSCLC cell migration, invasion, and spheroid formation (). In addition, the decreased N-ca and PCNA protein levels and the elevated E-ca protein levels in circ_0006677-overexpressing cells were reversed by exogenous expression of miR-1245a (). In addition, the downregulated protein levels of MMP-2 and MMP-9 in circ_0006677-overexpressed NSCLC cells were impaired after miR-1245a upregulation (). Altogether, circ_0006677 repressed NSCLC cell malignancy and stemness via sequestering miR-1245a.
Figure 4. Circ_0006677 sponged miR-1245a to regulate NSCLC cell malignancy and stemness. (a-g) The proliferation (a and b), caspase 3 activity (c), apoptosis (d), migration (e), invasion (f), and sphere formation (g) of NSCLC cells after transfection with circ_0006677 combined with or without miR-1245a mimic. (h and i) Protein levels of E-ca, PCNA, and N-ca in NSCLC cells after transfection with circ_0006677 combined with or without miR-1245a mimic. (j and k) Protein levels of MMP-2 and MMP-9 in the above NSCLC cells. *P < 0.05.
Validation of SATB2 as a target of miR-1245a
To gain further insight into the mechanism by which miR-1245a regulates NSCLC cell malignancy and stemness, we predicted SATB2 was a target of miR-1245a using online prediction databases (TargetScan, miRDB, and miRWalk). Their binding sites were exhibited in . Luciferase assays exhibited that co-transfection of the SATB2 3ʹUTR WT reporter with miR-1245a mimic caused an overt decrease in luciferase activity, while co-transfecting the SATB2 3ʹUTR MUT reporter and miR-1245a mimic did not change (). We also observed an obvious reduction in SATB2 mRNA expression in NSCLC samples (), and its expression was negatively correlated with miR-1245a () and positively correlated with circ_0006677 (). Moreover, downregulated SATB2 protein levels were observed in NSCLC samples and cells (). As expected, SATB2 protein levels were also downregulated in miR-1245a mimic-transfected NSCLC cells (). Additionally, circ_0006677 overexpression led to a distinct elevation in SATB2 protein levels, but this effect was overturned by co-transfecting with miR-1245a mimic (). Collectively, these results suggested that miR-1245a directly targeted SATB2.
Figure 5. SATB2 acted as a miR-1245a target. (a) The possible binding sites between SATB2 and miR-1245a. (b and c) The luciferase activity of the SATB2 3ʹUTR WT/SATB2 3ʹUTR MUT reporter in NSCLC cells after transfection with miR-1245a mimic. (d) Relative expression of SATB2 mRNA in NSCLC samples. (e and f) Correlation of SATB2 mRNA and miR-1245a/circ_0006677 was analyzed. (g and h) Relative protein levels of SATB2 in NSCLC samples and cells. (i) Influence of miR-1245a mimic on SATB2 protein levels in NSCLC cells. (j) Relative protein levels of SATB2 in NSCLC cells after transfection with circ_0006677 combined with or without miR-1245a mimic. *P < 0.05.
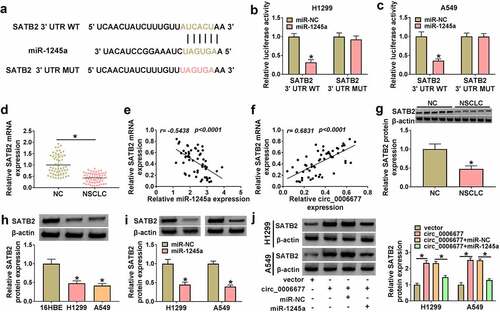
Circ_0006677 played its anti-tumor effect via upregulating SATB2
To confirm the SATB2-dependent effect of circ_0006677 on NSCLC cell malignancy and stemness, we designed a short hairpin RNA targeting SATB2 and its interference efficiency was demonstrated by Western blotting (). Indeed, SATB2 knockdown reversed circ_0006677 overexpression-mediated repression on cell proliferation (), elevation on cell caspase 3 activity (), and promotion on cell apoptosis in NSCLC cells (). Silencing of SATB2 was indeed able to re-establish the migrating and invading capacities (), as well as the spheroid formation capacity of circ_0006677-overexpressing NSCLC cells (). In addition, SATB2 inhibition overturned the changes in N-ca, PCNA, and E-ca protein levels in NSCLC cells mediated by overexpression of circ_0006677 (). And SATB2 silencing partly overturned the decreased protein levels of MMP-2 and MMP-9 in NSCLC cells prompted by circ_0006677 overexpression (). In sum, circ_0006677 restrained NSCLC cell malignancy and stemness by SATB2.
Figure 6. Circ_0006677 elevated SATB2 expression to restrain NSCLC cell malignancy and stemness. (a) Western blotting analysis of the interference efficiency of sh-SATB2. (b-h) The proliferation (b and c), caspase 3 activity (d), apoptosis (e), migration (f), invasion (g), and sphere formation (h) of NSCLC cells after transfection with circ_0006677 combined with or without sh-SATB2. (i and j) Relative protein levels of E-ca, PCNA, and N-ca in NSCLC cells after transfection with circ_0006677 combined with or without sh-SATB2. (k and l) Western blotting analysis of the protein levels of MMP-2 and MMP-9 in the above NSCLC cells. *P < 0.05.
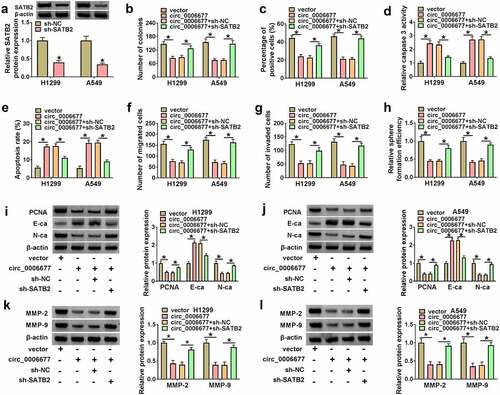
Circ_0006677 mediated the MEK/ERK via the miR-1245a/SATB2 axis
Because oncogenic KRAS mutations often occur in NSCLC and cause the activation of the MEK/ERK pathway, we explored whether circ_0006677 mediates the MEK/ERK pathway through the miR-1245a/SATB2 axis. Data in exhibited that circ_0006677 overexpression resulted in a reduction in the activity of MEK and ERK, but these changes were weakened by miR-1245a overexpression or SATB2 silencing. These results suggested that circ_0006677 overexpression could block the MEK/ERK pathway through the miR-1245a/SATB2 axis.
Figure 7. Circ_0006677 mediated the MEK/ERK via the miR-1245a/SATB2 axis. (a and b) Western blotting analysis of the protein levels of t-MEK, p-MEK, t-ERK, and p-ERK in NSCLC cells with vector, circ_0006677, circ_0006677+ miR-NC, circ_0006677+ miR-1245a, miR-1245a+sh-NC, or circ_0006677+ SATB2. *P < 0.05.
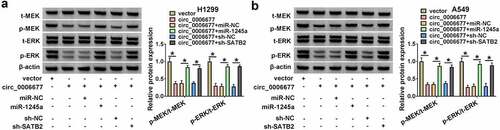
Circ_0006677 decreased xenograft tumor growth in vivo
We further ascertained the cancer-repressing role of circ_0006677 in NSCLC through constructing xenograft models in vivo. Mice injected with A549 cells stably overexpressing circ_0006677 had smaller tumor volume and lighter tumor weight compared with the control group (). Tumors derived from mice injected with A549 cells stably overexpressing circ_0006677 had higher levels of circ_0006677 and SATB2 and lower levels of miR-1245a compared with the control group (). Also, the tumors in the circ_0006677 group had higher SATB2 and E-ca protein levels and lower PCNA and N-ca protein levels compared with the vector group (). Furthermore, the protein levels of MMP-2 and MMP-9 were lower in tumor samples of the circ_0006677-overexpression group than those from the vector group (). IHC analysis showed a significant decrease in PCNA expression and an overt increase in SATB2 and Bax expression in the circ_0006677 group compared with the vector group (). Collectively, circ_0006677 decreased xenograft tumor growth in vivo.
Figure 8. Circ_0006677 reduced NSCLC cell growth in vivo. (a) Growth curves of tumor volume in different groups. (b) Representative images of xenograft tumors in different groups. (c) The tumor weight of xenograft tumors in different groups. (d) Relative levels of circ_0006677, miR-1245a, and SATB2 mRNA in xenograft tumors. (e) Relative protein levels of SATB2, E-ca, PCNA, and N-ca in xenograft tumors. (f) Relative protein levels of MMP-2 and MMP-9 in xenograft tumors. (g) IHC analysis of the expression of PCNA, SATB2, and Bax in xenograft tumors. *P < 0.05.
Discussion
It is becoming increasingly clear that circRNAs play significant roles in NSCLC. Nevertheless, their roles in NSCLC remain largely unknown. In this research, our findings demonstrated that reduced expression of circ_0006677 released miR-1245a with concomitant downstream downregulation of SATB2, resulting in the facilitation of NSCLC cell proliferation, migration, invasion, and stemness. Thus, circ_0006677 upregulation might represent a potential therapeutic strategy for NSCLC.
Here, this study selected a downregulated differentially expressed circRNA circ_0006677 with a change greater than 4 in both GSE158695 and GSE112214 datasets. A recent study reported the downregulation of circ_0006677 in NSCLC, and circ_0006677 could repress NSCLC cell glycolysis, proliferation, migration, and invasion via sequestering miR-578 with concomitant downstream upregulation of SOSC2 [Citation27]. Analogously, a distinct downregulation in circ_0006677 expression was observed in NSCLC samples and cells in this study. In line with the report of Yang et al. [Citation27], overexpression of circ_0006677 repressed NSCLC cell proliferation, migration, and invasion in vitro. In addition, our data also exhibited the repressive effect of circ_0006677 overexpression on NSCLC cell stemness. Furthermore, circ_0006677 overexpression decreased xenograft tumor growth, and PCNA and N-ca levels were lower in xenograft tumors derived from circ_0006677-overexpressing NSCLC cells, but E-cad and Bax levels were higher, suggesting that circ_0006677 could repress NSCLC cell growth in vivo. These results primarily suggested the inhibiting function of circ_0006677 in NSCLC.
Up to now, a series of studies have demonstrated that circRNAs can exert their function by acting as competing endogenous RNAs (ceRNAs) [Citation28]. miR-1245a has been reported to exert an oncogenic role in breast cancer [Citation29] and colon adenocarcinoma [Citation30] progression by downregulating encoded by breast-cancer susceptibility gene 2 (BRCA2). In NSCLC, circ-prolyl 4-hydroxylase subunit beta downregulation reduced BRCA2 expression via releasing miR-1245a, thus promoting NSCLC progression [Citation26]. By extracting the cytoplasm and nucleus of NSCLC cells, circ_0006677 was validated to be mainly distributed in the cytoplasm, indicating that circ_0006677 might be a ceRNA. Importantly, we identified that circ_0006677 could interact with miR-1245a in NSCLC through luciferase and RIP assays. Ectogenic expression of miR-1245a weakened the inhibiting effects of circ_0006677 overexpression on NSCLC cell proliferation, migration, invasion, and stemness. Our findings demonstrated that circ_0006677 exerted an anti-tumor function in NSCLC by repressing miR-1245a activity.
SATB2, a transcription factor, can regulate gene expression through serving as a transcriptional cofactor and modulating chromatin architecture [Citation31]. SATB2 has been uncovered as a tumor-inhibiting gene in colorectal cancer [Citation32], and clear cell renal cell cancer [Citation33], but it exerts an oncogenic effect on glioblastoma [Citation34], hepatocellular cancer [Citation35], oral squamous cell cancer [Citation36], and osteosarcoma [Citation37]. Also, SATB2 could repress epithelial-mesenchymal transition (EMT) through decreasing G9a and histone methylation in NSCLC [Citation38]. Moreover, SATB2 could inhibit NSCLC initiation by suppressing stemness marker genes such as SRY-Box Transcription Factor 2 [Citation39]. Our results exhibited low SATB2 levels in NSCLC samples and cells, and SATB2 was identified as a miR-1245a target. Interestingly, SATB2 could be regulated by circ_0006677, which functioned as a miR-1245a decoy. Importantly, SATB2 silencing whittled circ_0006677 overexpression mediated suppression on NSCCL cell proliferation, migration, invasion, and stemness. These findings prompted us to conclude that circ_0006677 exerts an anti-tumor effect in NSCLC at least in part through the ceRNA network, that is, circ_0006677 adsorbs miR-1245a to increase the expression of the anti-tumor gene SATB2.
Conclusion
This study demonstrated that circ_0006677 acts as an endogenous miR-1245a sponge and elevated SATB2 expression via adsorbing miR-1245a, leading to suppressing NSCLC cell malignancy and stemness. These findings provide a better understanding of the mechanism involved in NSCLC progression.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Liu JC, Narva S, Zhou K, et al. A review on the antitumor activity of various nitrogenous-based heterocyclic compounds as NSCLC inhibitors. Mini Rev Med Chem. 2019;19(18):1517–1530.
- Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594.
- Friedlaender A, Addeo A, and Russo A, et al. 2020. Targeted therapies in early stage NSCLC: hype or hope?. International journal of molecular sciences; 21(17):6329. https://doi.org/10.3390/ijms21176329.
- Naylor EC, Desani JK, Chung PK. Targeted therapy and immunotherapy for lung cancer. Surg Oncol Clin N Am. 2016;25(3):601–609.
- Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): an update. Discov Med. 2019;27(148):167–170.
- Kristensen LS, Andersen MS, Stagsted LVW, et al. The biogenesis, biology and characterization of circular RNAs. Nature Reviews. Genetics. 2019;20(11):675–691.
- Hsiao KY, Sun HS, Tsai SJ. Circular RNA - new member of noncoding RNA with novel functions. Exp Biol Med (Maywood). 2017;242(11):1136–1141.
- Zhang HD, Jiang LH, Sun DW, et al. CircRNA: a novel type of biomarker for cancer. Breast Cancer. 2018;25(1):1–7.
- Li J, Sun D, Pu W, et al. Circular RNAs in cancer: biogenesis, function, and clinical significance. Trends Cancer. 2020;6(4):319–336.
- Verduci L, Strano S, Yarden Y, et al. The circRNA-microRNA code: emerging implications for cancer diagnosis and treatment. Mol Oncol. 2019;13(4):669–680.
- Chen Q, Liu T, Bao Y, et al. CircRNA cRAPGEF5 inhibits the growth and metastasis of renal cell carcinoma via the miR-27a-3p/TXNIP pathway. Cancer Lett. 2020;469:68–77.
- Lu Q, Liu T, Feng H, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Molecular Cancer. 2019;18(1):111.
- Li C, Zhang L, Meng G, et al. Circular RNAs: pivotal molecular regulators and novel diagnostic and prognostic biomarkers in non-small cell lung cancer. Journal of Cancer Research and Clinical Oncology. 2019;145(12):2875–2889.
- Liu H, Bi J, Dong W, et al. Invasion-related circular RNA circFNDC3B inhibits bladder cancer progression through the miR-1178-3p/G3BP2/SRC/FAK axis. Mol Cancer. 2018;17(1):161.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta delta C(T)) method. Methods. 2001;25(4):402–408.
- Li X, Tian G, Wu J. Novel circGFRα1 promotes self-renewal of female germline stem cells mediated by m(6)A writer METTL14. Front Cell Dev Biol. 2021;9:640402.
- He SW, Xu C, Li YQ, et al. AR-induced long non-coding RNA LINC01503 facilitates proliferation and metastasis via the SFPQ-FOSL1 axis in nasopharyngeal carcinoma. Oncogene. 2020;39(34):5616–5632.
- Mehta AK, Hua K, Whipple W, et al. Regulation of autophagy, NF-κB signaling, and cell viability by miR-124 in KRAS mutant mesenchymal-like NSCLC cells. Sci Signal. 2017;10(496). DOI:10.1126/scisignal.aam6291.
- Zhang J, Wang G, Chu SJ, et al. Loss of large tumor suppressor 1 promotes growth and metastasis of gastric cancer cells through upregulation of the YAP signaling. Oncotarget. 2016;7(13):16180–16193.
- Sugano T, Seike M, Noro R, et al. Inhibition of ABCB1 overcomes cancer stem cell-like properties and acquired resistance to MET inhibitors in non-small cell lung cancer. Mol Cancer Ther. 2015;14(11):2433–2440.
- Yu M, Chen Y, Li X, et al. YAP1 contributes to NSCLC invasion and migration by promoting slug transcription via the transcription co-factor TEAD. Cell Death Dis. 2018;9(5):464.
- Wei S, Zheng Y, Jiang Y, et al. The circRNA circPTPRA suppresses epithelial-mesenchymal transitioning and metastasis of NSCLC cells by sponging miR-96-5p. EBioMedicine. 2019;44:182–193.
- Yan M, Sun L, Li J, et al. RNA-binding protein KHSRP promotes tumor growth and metastasis in non-small cell lung cancer. J Exp Clin Cancer Res. 2019;38(1):478.
- Xu S, Zhang H, Wang A, et al. Silibinin suppresses epithelial-mesenchymal transition in human non-small cell lung cancer cells by restraining RHBDD1. Cell Mol Biol Lett. 2020;25(1):36.
- Liang ZZ, Guo C, Zou MM, et al. circRNA-miRNA-mRNA regulatory network in human lung cancer: an update. Cancer Cell International. 2020;20:173.
- Yang L, Wang J, Fan Y, et al. Hsa_circ_0046264 up-regulated BRCA2 to suppress lung cancer through targeting hsa-miR-1245. Respir Res. 2018;19(1):115.
- Yang B, Zhao F, Yao L, et al. CircRNA circ_0006677 inhibits the progression and glycolysis in non-small-cell lung cancer by sponging miR-578 and regulating SOCS2 expression. Front Pharmacol. 2021;12:657053.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
- Song L, Dai T, Xie Y, et al. Up-regulation of miR-1245 by c-myc targets BRCA2 and impairs DNA repair. J Mol Cell Biol. 2012;4(2):108–117.
- Pan Z, Gan W, Liang C, et al. miR-1245a promotes the proliferation and invasion of colon adenocarcinoma by targeting BRCA2. Ann Transl Med. 2019;7(23):777.
- Britanova O, de Juan Romero C, Cheung A, et al. Satb2 is a postmitotic determinant for upper-layer neuron specification in the neocortex. Neuron. 2008;57(3):378–392.
- Mansour MA, Hyodo T, Ito S, et al. SATB2 suppresses the progression of colorectal cancer cells via inactivation of MEK5/ERK5 signaling. FEBS J. 2015;282(8):1394–1405.
- Sliwinska-Jewsiewicka A, Kowalczyk AE, Krazinski BE, et al. Decreased expression of SATB2 associates with tumor growth and predicts worse outcome in patients with clear cell renal cell carcinoma. Anticancer Res. 2018;38(2):839–846.
- Tao W, Zhang A, and Zhai K, et al. SATB2 drives glioblastoma growth by recruiting CBP to promote FOXM1 expression in glioma stem cells. EMBO molecular medicine. 2020;12(12):e12291. https://doi.org/10.15252/emmm.202012291.
- Wang Y, Li CF, Sun LB, et al. microRNA-4270-5p inhibits cancer cell proliferation and metastasis in hepatocellular carcinoma by targeting SATB2. EMBO Mol Med. 2020;33(4):1155–1164.
- Gao J, Meng Y, Ge X, et al. SATB2 overexpression promotes oral squamous cell carcinoma progression by up-regulating NOX4. Cell Signal. 2021;82:109968.
- Seong BK, Lau J, Adderley T, et al. SATB2 enhances migration and invasion in osteosarcoma by regulating genes involved in cytoskeletal organization. Oncogene. 2015;34(27):3582–3592.
- Ma YN, Zhang HY, Fei LR, et al. SATB2 suppresses non-small cell lung cancer invasiveness by G9a. Clin Exp Med. 2018;18(1):37–44.
- Kucuksayan H, Ozes ON, Akca H. Downregulation of SATB2 is critical for induction of epithelial-to-mesenchymal transition and invasion of NSCLC cells. Lung Cancer. 2016;98:122–129.
