ABSTRACT
The expression of METTL14 is significantly reduced in patients with retinitis pigmentosa (RP). To clarify the significance of the N6-methyladenosine (m6A) RNA modification in RP, we examined phagocytosis, apoptosis, and cell cycle distribution in a human RPE cell line, ARPE-19, following lentivirus-mediated knockdown of METTL14. Differentially expressed genes and changes in m6A level were evaluated by RNA sequencing (RNA-seq) and methylated RNA immunoprecipitation sequencing (MeRIP-seq), respectively. The results showed that phagocytosis and proliferation were decreased whereas apoptosis was increased in RPE cells by METTL14 silencing. We found that METTL14 directly regulated m6A level and the expression of MAP2, as determined by RNA-seq, MeRIP-seq, MeRIP quantitative PCR, and the RNA pull-down assay. Additionally, MAP2 could bind to neuronal differentiation (NEUROD)1, a pathogenic gene in RPE-associated diseases. A family member of the YTH domain, (YTHDF)2 was recognized as an m6A reader of MAP2 mRNA. MAP2 overexpression had the same effects as METTL14 knockdown in RPE cells. Thus, METTL14 regulates the expression of MAP2 via the modification of m6A, resulting in the dysregulation of NEUROD1 and pathologic changes in RPE cells. These findings suggest that therapeutic strategies targeting the m6A modification of MAP2 or the METTL14/YTHDF2/MAP2/NEUROD1 signaling axis may be effective in the treatment of RPE-associated ocular diseases.
1. Introduction
The RPE forms the outermost layer of the human retina. It is a monolayer of hexagonal cells in a polar arrangement located between the sensory layer and Bruch’s membrane. The RPE functions primarily in the phagocytosis of the outer segment of photoreceptor cells, providing nutrients for the outer cells of the sensory retina and regulating metabolism. Abnormal RPE structure and function leads to apoptosis and necrosis of retinal photoreceptor cells, resulting in severe RPE-associated ocular disorder including RP, uveitis, as well as age-related macular degeneration (AMD)[Citation1].
RP represents multiple inherited retinal diseases with variable incidence, progression, and severity that eventually result in vision loss. The major features of RP are progressive damage to the RPE and photoreceptors and a high degree of genetic heterogeneity [Citation2]. In many RP patients, phagocytosis is impaired and there is a loss of RPE cell tight junction integrity [Citation3,Citation4]. Uveitis is an intraocular inflammatory disease involving the uvea, retina, and vitreous body that is mainly the result of autoimmune dysfunction and can lead to blindness. AMD is an oculopathy involving the outer retinal layer, pigment epithelium, and choroid that is thought to result from a combination of genetic and environmental factors and is associated with progressive central vision impairment.
Many studies have investigated the pathogenesis of RPE-associated ocular diseases [Citation5–7]. NEUROD1 was identified as a pathogenic gene in RP, although its mechanism of action is not fully understood [Citation8]; and RP1 and RP1-like (RP1L)1 are also related to RP [Citation9]. MAP2 is highly expressed in the retina of AMD patients [Citation10]. Additionally, DNA methylation, histone modification, chromatin remodeling, and noncoding RNA regulation are epigenetic factors associated with the pathogenesis of RPE-associated ocular lesions [Citation11–14]. As a novel type of epigenetic modification, RNA methylation has been studied in the context of multiple diseases.
m6A is the main type of RNA methylation in eukaryotes, affecting approximately 80% of mRNAs in processes such as transcription, translation, and protein degradation [Citation15,Citation16]. The consensus m6A motif is RACH (R represents G or A; H represents U, A, or C). The modification levels of an m6A transcript can be mediated via a methyltransferase (writer), binding protein (reader), and demethylase (eraser) [Citation17]. The writer mediates the RNA methylation process [Citation18]. METTL3 and METTL14 are the most common writers, and their activities have been identified both in vitro and in vivo [Citation19]. The eraser (e.g., AlkB homolog 5, RNA demethylase [ALKBH]5, and fat mass and obesity-associated [FTO]) removes the RNA methylation mark [Citation20,Citation21]. The reader detects the RNA methylation mark and promotes RNA translation and degradation; m6A binding proteins in the cytoplasm include YTHDF1, YTHDF2, YTHDF3, and YTH domain-containing (YTHD)2 [Citation22,Citation23].
m6A methylation links to the pathogenesis of several diseases namely type 2 diabetes mellitus, neurologic disorders, mitochondria-related diseases, and various tumors [Citation24]. However, the significance of m6A methylation in RPE-associated ocular diseases has not been fully explored. We hypothesized that METTL14 may affect RPE cell phagocytosis, tight junction, apoptosis and cell cycle by regulating m6A methylation. In order to prove this hypothesis, RPE cells were used as materials in vitro. Our results demonstrated that METTL14 is of vital significance in development by regulating the expression of MAP2 via m6A modification and METTL14/YTHDF2/MAP2/NEUROD1 signaling pathway.
2. Materials and methods
2.1. Cell culture and identification
ARPE-19 human RPE cells (CRL-2302) obtained from ATCC (Manassas, USA), were cultured in Dulbecco’s Modified Eagle’s Medium/F-12 (Gibco, USA) by the addition of 100 U/ml penicillin (Beyotime, China), 100 μg/ml streptomycin (Beyotime, China), and 10% fetal bovine serum (Gibco, USA). Passage culture of cells was performed every 2–3 days. Isolation of human RPE cells was from the eye tissue of human cadavers in the eye bank of The First Affiliated Hospital of Chongqing Medical University. As per Helsinki Declaration, written consent of the experimental materials for medical research was gained from donors or their families. The method of isolation and culture of RPE cells have been previously described (29). Cells were grown at a constant temperature of 37°C inside a humidified 5% CO2 condition. The expression of RPE65 was detected by cell immunofluorescence and the cells were identified [Citation25]. RPE65 antibody (A9615, Abclonal, China) and fluorescent secondary antibody (AS011, Abclonal, China) were purchased from Abclonal, China.
2.2. Lentivirus preparation
Lentivirus harboring a short hairpin (sh)RNA against METTL14 for gene silencing experiments was packaged by Chongqing Biomedicine Biotechnology Co (Chongqing, China). Briefly, 293 T cells were incubated under a 5% CO2 atmosphere at 37°C and for 8–24 h. Transfection was carried out when the cells reached >50% confluence. A 400-μl mixture of core and packaging plasmids were supplied dropwise to a cell culture dish; the culture medium was mixed by gently rocking the dish and cell incubation was conducted at 37°C with 5% CO2. Following 12 h incubation, the culture medium was discarded and an appropriate quantity of complete medium prewarmed to 37°C was added. The supernatant containing lentivirus was collected 24 and 48 h following transfection, filtered by passing through a 0.45-μm mesh, and subsequently maintained under a condition of −80°C [Citation26].
2.3. RNA sequencing (RNA-seq) and methylated RNA immunoprecipitation sequencing (MeRIP-seq)
Annoroad (Beijing, China) performed RNA-seq and MeRIP-seq sample sequencing. For RNA-seq, the total RNA of samples was initially extracted and sample quality was evaluated. Enrichment and purification were performed using oligo(dT) magnetic beads. A commercial kit was used to remove rRNA and a fragmentation buffer was applied for mRNA purification to obtain shorter fragments that were adopted as a template for first-strand cDNA synthesis with random hexamer primers. A buffer solution containing RNase H, dNTPs, and DNA polymerase I was supplied for second-strand cDNA synthesis. A QIAquick PCR kit (Qiagen, Valencia, CA, USA) was used for purification. The eluted double-stranded cDNA was processed end repair, polyadenylation, and linker addition. Followed by verification of target fragments using agarose gel electrophoresis, PCR amplification was performed for library construction. Quantification was carried out by applying a Qubit 3.0 fluorometer (Thermo Fisher Scientific, USA), and the library was diluted to 1 ng/μl. The instrument 2100 Bioanalyzer (Agilent, USA) was applied to detect the insert size of the library; if it was smaller than expected, fluorescence quantitative (q)PCR was conducted via iQ SYBR Green Supermix on a CFX96 Touch PCR detection system (Bio-Rad, USA). The concentration of the library was > 10 nM indicating good quality. Based on a sequencing strategy of PE150, the library was sequenced on the Illumina platform (San Diego, USA).
For MeRIP-seq, mRNA was enriched with oligo d(T) magnetic beads and 100-bp fragments were obtained using divalent cations. An anti-m6A antibody was used for coimmunoprecipitation (co-IP). The cDNA synthesis template used IP extracted RNA with random hexamers. The splint-ligated products were purified with magnetic beads and amplified by PCR. After agarose gel electrophoresis ran for confirming the quality of the constructed library, the library was quantified with Qubit v3.0. Libraries with sufficiently high concentrations were sequenced on an Illumina sequencer based on target end reads [Citation27,Citation28].
2.4. Cell cycle analysis
ARPE-19 cells were initially rinsed using 3 ml phosphate-buffered saline (PBS), which was removed and replaced with 1 ml trypsin for 1–5 min. A 5-ml volume of PBS was added and followed by transference of cell suspension to a 15-ml centrifuge tube for centrifugation at 1500 g for 5 min. Following the removal of supernatant, the cells were resuspended using 500 μl PBS; 2 ml of 95% cold ethanol (20°C) was supplemented dropwise and the mixture was vortexed and fixed for 30 min. Centrifugation was performed at 1 500 g for 5 min by the addition of 5 ml PBS followed by the removal of supernatant. Of 5 ml PBS was supplied for cell resuspension and the supernatant was removed after centrifugation at 1500 g for 5 min. Propidium iodide (PI) staining solution (800 μl) was added, incubation was performed subsequently at room temperature away from light for 30 min. Flow cytometry analysis was ultimately conducted (CytoFLEX, Kraemer Boulevard Brea, CA, USA) [Citation29].
2.5. Cell apoptosis assay
ARPE-19 cells were firstly added to 10 ml capacity tubes; each sample contained 1–5 × 106 cells/ml. The tube was centrifuged at 1000 g for 5 min followed by the removal of supernatant. Following a cycle of washing with incubation buffer, the cells were centrifuged at 1000 g for 5 min, resuspended in 100 μl labeling solution and incubated in the dark for 10–15 min at room temperature; Centrifugation was conducted again at 1000 g for 5 min and followed by precipitation, incubation and a cycle of washing with the solution. Fluorescent solution (SA-FLOUS; Beyotime Biotechnology, Shanghai, China) was supplemented and incubated in the dark at 4°C for 20 min with occasional agitation. Cell apoptosis was subjected to flow cytometry at 488 nm excitation wavelength. Fluorescein isothiocyanate and PI fluorescence were detected at 515 and more than 560 nm, respectively [Citation30].
2.6. Cytophagocytosis assay
ARPE-19 cells were cultured in T-25 multiwell plates. At approximately 90% cell confluency, the solution was replaced with 3 ml of fresh complete medium. Each plate contained 30 μl microspheres. After incubation for 24 h, the cell medium was transferred to an appropriate centrifuge tube. Adherent cells were followed by three cycles of washing with PBS and then digested using trypsin at room temperature until the cells could be detached by gentle trituration. The trypsin was inactivated by adding a fresh complete culture medium. The culture supernatant was carefully pipetted to another tube for centrifugation at 1 000 g for 5 min, followed by the removal of the supernatant. The cells were rinsed and resuspended in PBS and maintained in the dark until they were counted. Flow cytometry was immediately performed [Citation31].
2.7. Co-IP
Co-IP was performed with a kit (Bes3011; Bersin, Guangzhou, China) as per manufacturer’s instructions. Briefly, ARPE-19 cells were cleaned twice by the addition of PBS and radioimmunoprecipitation assay (RIPA) buffer precooled at 4°C and subsequently centrifuged at 14,000 g for 15 min. The supernatant was obtained and for every 1 ml containing total protein, 100 μl Protein A agarose beads were provided, vortexed at 4°C for 10 min and centrifugated at 4°C and 14,000 g for 15 min. The supernatant was obtained and 500 μl was combined with 20 μl primary antibodies and after vortexing, the antigen–antibody mixture was incubated overnight at 4°C. Centrifugation was performed at 14,000 g for 5 s and agarose bead–antigen–antibody complex was harvested, resuspended in sample buffer, heated for 5 min in boiling water, and rapidly cooled on ice for Western blot analysis [Citation32].
2.8. m6A detection
m6A was detected using a kit (p-9005-48; EpiGen, Farmingdale, USA) based on manufacturer’s instructions. Generally, 2 μl negative control (NC), 2 μl positive control (PC), or 200 ng sample RNA (2 μl) were mixed with 80 μl binding solution in a tube, followed by incubation at 37°C for 90 min. The binding solution was removed and subsequently, a 50 μl capture antibody was supplied for incubation at room temperature for 60 min. Being washed and supplemented with 50 μl of detection antibody, the solution was incubated 30 min and washed a second time. Of 50 μl enhancer solution was provided for incubation 30 min and then being washed. Developer solution at 100 μl was added and incubated for 10 min in the dark. The reaction was terminated with a 100 μl stop solution. The optical density (OD) was measured using a microplate reader at 450 nm. The following formula was used to calculate m6A%: ([ODsample−ODNC]/S)/([ODPC−ODNC]/P), where S indicates the input amount of sample RNA in ng and P indicates the input amount of PC in ng [Citation33].
2.9. MeRIP-qPCR
MeRIP was performed using a kit (Bes5203; Bersin) as per the manufacturer’s instruction. Total RNA was extracted and combined with A/G immunomagnetic beads and anti-m6A antibody. The enriched RNA–antibody complex was digested with protease, and MAP2 mRNA in the enriched concentrate was detected by qPCR.
2.10. Western blotting
RIPA lysis buffer was supplemented to ARPE-19 cells. The mixed solution was vortexed for 1 min and maintained on ice for 3 min; this was repeated 10 times. Following ice incubation for 15 min until the proteins fully decomposed, the cells were centrifuged at 13,000 g at 4°C for 20 min. After centrifugation, the supernatant was pipetted to a precooled tube with 1.5 ml capacity and the protein concentration was determined subsequently. The samples were boiled at 100°C for 10 min, and 30 μg of protein was isolated by gel electrophoresis using 10% sodium dodecyl sulfate (SDS) polyacrylamide and transferred to a polyvinylidene difluoride membrane. The film was blocked with 5% skimmed milk in blocking buffer at room temperature for 1 h. After three repeated washing using Tris-buffered saline containing 0.1% Tween-20, each time 5 min. the membrane was incubated overnight at 4°C with primary antibody diluted 1:1000–1:2000 in 3% bovine serum albumin. After the addition of secondary antibody (1:1000), the membrane was incubated for 1 h at room temperature and immunoreactivity was visualized via enhanced chemiluminescence. Antibodies against the following proteins were applied: MAP2 (BM2572), NEUROD1 (BM1147), and METTL14 (BM8530) (all from IGEE, Chongqing, China).
2.11. Quantitative real-time (qRT-)PCR
Total RNA extraction was conducted via TRIzol reagent (Takara, Japan) as per manufacturer’s instructions. Reverse transcription used the PrimeScript RT Kit (Takara) to synthesize the first strand cDNA. CFX96 Real-Time system (Bio-Rad) was applied for qRT-PCR assays via TB Green Premix Ex Taq II (Takara). The primers were synthesized by Chongqing Biomedicine Biotechnology Co. Ltd. Relative gene expression was calculated using 2−ΔΔCt method (30). GAPDH was used as an internal control gene.
2.12. RNA pull-down
The RNA pull-down assay was performed through a pull-down kit (Bes51021; Bersin) as per manufacturer’s instructions. ARPE-19 cells were rinsed with PBS and digested with trypsin. The reaction was terminated by adding a complete medium. After centrifugation, the supernatant was removed. Following twice washes with ice-cold PBS, an addition of 500 μl lysis buffer was provided for incubation on ice for 20 min with occasional rocking. Centrifugation was conducted at 4°C 5000 g for 5 min; the supernatant was obtained and 500 μl was shifted to another tube, of which 50 μl was removed and maintained at 4°C as the input control (total RNA). A 100-μl volume of prepared magnetic bead suspension was added and subsequently incubated on a shaker at 4°C for 4 h. Following precipitation of magnetic beads, the supernatant was discarded. The beads had five cycles of washing with l ml lysis buffer and recovered in 100 μl lysis buffer and the input was removed as described above. A 500-μl volume of TRIzol LS was supplied and vortexed. The mixture was subsequently incubated at room temperature for 5 min, supplemented with 100 μl chloroform, vortexed, and centrifuged at 16,000 g 4°C for 15 min. The supernatant (250 μl) was shifted to a new tube, supplemented with 850 μl anhydrous ethanol, and stored overnight at −20°C; the following centrifugation at 16,000 g 4°C for 15 min, the supernatant was abandoned. The pellets were rinsed 3 times applying 75% ice-cold ethanol prepared with diethylpyrocarbonate (DEPC) water. The dried RNA pellets were dissolved by the addition of 200 μl DEPC water, extracted with 400 μl phenol: chloroform: isopropanol, agitated for 15s, and centrifuged at 14,000 g 25°C for 10 min. Of 350 μl aqueous supernatant was transferred to another Eppendorf tube, supplemented 5 μl precipitate enhancer for RNA precipitation, 50 μl salt solution I, 15 μl salt solution II, and 850 μl anhydrous ethanol and incubated for 1 h or overnight at −80°C. After discarding the supernatant, the samples were centrifuged at 4°C 14000 g for 15 min, washed with 80% ice-cold ethanol, and centrifuged at 14,000 g 4°C for 15 min. After the removal of the supernatant, the pellets were dried on ice. A 10 μl-volume of enzyme-free water was added and the pellets were dissolved on ice. The quality of the extraction was estimated based on the RNA measurement of the input sample [Citation6].
2.13. RNA immunoprecipitation (RIP)
RIP technique was adopted through the Magna RIP Kit (Bes5203; Bersin) based on manufacturer’s instructions. All of the cells were cross-linked for 8 min using 0.2% formaldehyde and kept in a neutralization buffer (containing 2.5 M glycine and 25 mM Tris [pH = 7.4]) for 5 min, followed by two washes with PBS. The beads were cleaned twice using a buffer composed of 20 mM HEPES (pH 7.5), 350 mM NaCl, 1% Nonidet (N)P-40, 0.5% deoxycholate, 0.25% SDS, and 5 mM dithiothreitol for 3 min, washed twice in a buffer composed of 20 mM Tris (pH 7.4), 150 mM NaCl, and 0.1% NP-40 for 5 min. Following be processed using proteinase K and Turbo DNase, qRT-PCR was conducted via purified RNA [Citation34].
2.14. Statistical analysis
All data collected were imported into SPSS v23.0 software (SPSS Inc, USA) and analyzed. Measurement data were expressed as mean ± standard deviation. Paired t-tests were used for before vs after treatment comparisons in the same group. The statistic analysis tool ANOVA was used for group comparison. P values less than 0.05 were considered the results were significant.
3. Results
m6A methylation links to the pathogenesis of several diseases. We hypothesized that METTL14 may affect RPE cell phagocytosis, tight junction, apoptosis and cell cycle by regulating m6A methylation. To clarify the significance of the m6A RNA modification in RP, we tested phagocytosis, apoptosis, and cell cycle distribution in a human RPE cells following lentivirus-mediated knockdown of METTL14. Differentially expressed genes and changes in m6A level were evaluated by RNA-seq and MeRIP-seq.
3.1. Downregulation of METTL14 in RP is associated with cytopathic changes in RPE cells
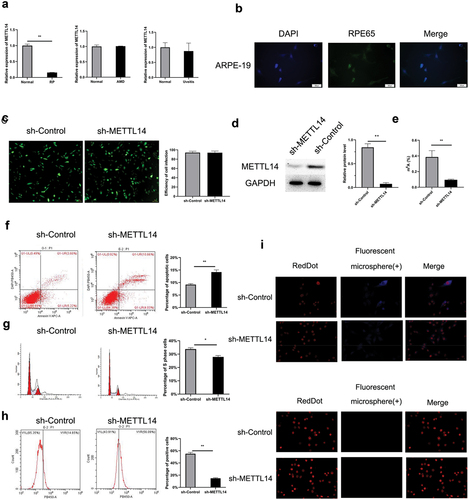
To elucidate whether METTL14 affected RPE-associated ocular diseases, the research investigated the expression of the METTL14 gene in the Gene Expression Omnibus (GEO) database (GSE12086, GSE115828, and GSE66936). The findings indicated that METTL14 expression was downregulated in patients with RP but not in those with AMD or uveitis (). Cultured ARPE-19 cells were infected with a lentivirus expressing shRNA against METTL14, and a decrease in METTL14 level was confirmed (). METTL14 knockdown decreased m6A level in ARPE-19 cells (); this was accompanied by a decrease in phagocytosis, increase in apoptosis, and cell cycle arrest ().
Figure 1. METTL14 silencing induces cytopathic changes in ARPE-19 cells. (a) Expression level of METTL14 in RP patients (GSE12086, lymphoblast samples), AMD patients (GSE115828, retinal samples), and uveitis patients (GSE66936, peripheral blood samples) in the GEO database. (b) Detection of RPE65 protein expression in ARPE-19 cells by immunocytochemistry. (c) Efficiency of METTL14 shRNA lentivirus infection in ARPE-19 cells. (d) METTL14 protein expression detected by Western blotting. (e) m6A abundance. (f–h) Flow cytometry analysis of cell apoptosis (f), cell cycle distribution (g), and cytophagocytosis (h) following METTL14 knockdown. (i) Cytophagocytosis of ARPE-19 cells. *P < 0.05, **P < 0.01.
3.2. Detection of METTL14 mRNA expression and methylation via RNA-seq and MeRIP-seq approaches
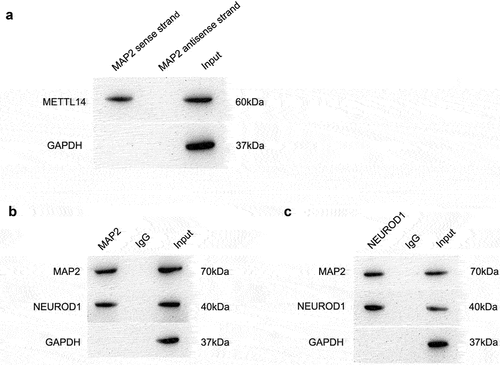
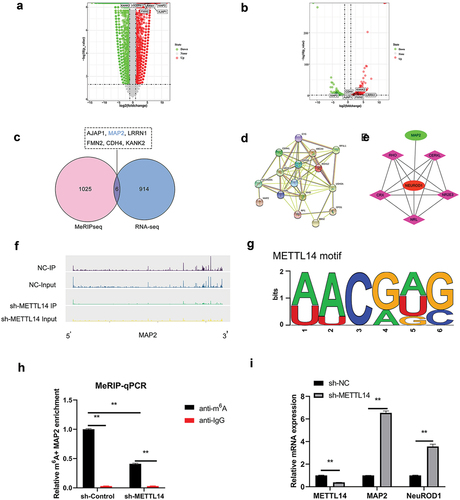
To clarify METTL14 action in RP, METTL14-depleted ARPE-19 cells were analyzed on RNA-seq and MeRIP-seq approaches. Transcripts with 3 250 upregulated and 4 262 downregulated were detected by RNA-seq (). Meanwhile, the MeRIP-seq results revealed that peak m6A abundance was decreased in 346 transcripts and increased in 685 (). Differentially expressed genes (DEGs) were assigned thresholds (the genes with a copy number in both the control and shRNA-mediated METTL14 knockdown groups <100 and |log2(fold change)| ≥2), 920 DEGs and 1031 genes with altered peak m6A abundance were identified (). Six transcripts (AJAP1, MAP2, LRRN1, FMN2, CDH4, and KANK2) showed overlap between the RNA-seq and MeRIP-seq gene sets (). Protein–protein interaction (PPI) analysis was carried out between these 6 genes and over 110 genes related to RPE-associated ocular diseases (). Only MAP2 was found to interact with RP-related pathogenic genes such as NEUROD1 (). The m6A peaks of MAP2 were markedly reduced in RPE cells following METTL14 knockdown (), which was accompanied by increased expression of MAP2 and NEUROD1 as detected by qPCR (). MAP2 was therefore chosen as a candidate target of METTL14 for subsequent research.
Figure 2. RNA expression and methylation following METTL14 knockdown detected by RNA-seq and MeRIP-seq. (a,b) Results of RNA-seq and MeRIP-seq. The x axes show the log fold change in DEGs expression; the y axes show negative log of the statistical significance of expression changes. (c) Venn diagram of RNA-seq and MeRIP-seq results with 6 overlapped genes. (d) PPI network of overlapped genes and >110 pathogenic genes in RP and AMD. (e) Interaction network between MAP2 and NEUROD1. (f) m6A peak in MAP2. (g) Motif of METTL14. (h) MAP2 m6A abundance in RPE cells following METTL14 knockdown detected by MeRIP-qPCR. (i) Detection of METTL14, MAP2, and NEUROD1 expression by qPCR following METTL14 knockdown. **P < 0.01.
3.3. Analysis of MAP2 function in RPE cells
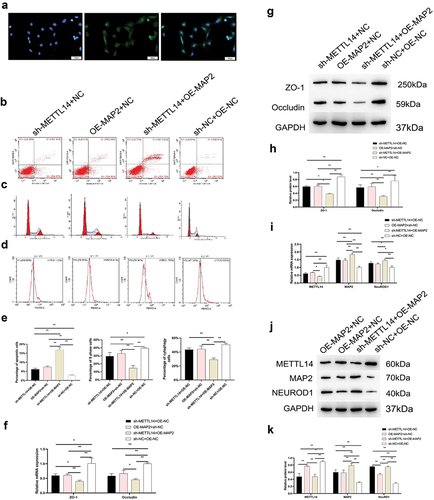
As with METTL14 knockdown, MAP2 overexpression impaired phagocytosis and undermined the integrity of tight junctions in RPE cells while increasing apoptosis and inducing cell cycle arrest (); these effects were accompanied by upregulation of NEUROD1 ().
Figure 3. MAP2 function in RPE cells. (a) RPE65 protein expression in RPE cells detected by immunocytochemistry. (a–d) Apoptosis, phagocytosis, and cell cycle distribution in the sh-METTL14+ OE-NC, sh-METTL14+ OE-MAP2, OE-MAP2+ NC, and sh-NC+OE-NC groups of RPE cells. (e) Quantitative analysis of results shown in panels B–D. (f–h) Zona occludens (ZO)-1 and occludin expression in the 4 groups detected by qPCR (f) and Western blotting (g, h) in the 4 groups. (i–k) METTL14, MAP2, and NEUROD1 expression detected by qPCR (i) and Western blotting (j, k) in the 4 groups. NC, negative control; OE, overexpression; sh, short hairpin. *P < 0.05, **P < 0.01.
3.4. Mechanism of METTL14–MAP2–NEUROD1 interactions
RNA pull-down assay revealed that METTL14 could directly bind to MAP2 mRNA (). To further clarify the pathogenic mechanism of MAP2, we examined the PPI network and found that the METTL14–MAP2 interaction was linked to more than 110 genes implicated in RPE-associated ocular diseases. A co-IP assay was performed to evaluate binding between MAP2 and NEUROD1. A complex containing both proteins was pulled down with anti-MAP2 antibody (), and MAP2 was also identified in pull-down complex using anti-NEUROD1 antibody (). These results suggest that MAP2 directly binds to NEUROD1.
3.5. METTL14 inhibits MAP2 expression and translation via the YTHDF2 signaling pathway
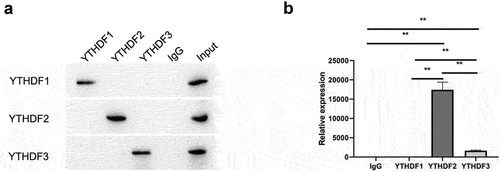
As a methyltransferase, METTL14 acts in conjunction with an RNA m6A reader to regulate target gene expression. Common readers include YTHDF1, YTHDF2, and YTHDF3. We predicted that YTHDF2 is the main reader associated with mediating MAP2 expression by METTL14. To test this hypothesis, this study examined the interaction between YTHDF1, YTHDF2, and YTHDF3 and MAP2 by RIP-qPCR, and found that MAP2 mRNA was pulled down using antibodies against YTHDF2 and YTHDF3 ().
4. Discussion
The RPE is an essential supporting tissue for the normal physiologic activities of photoreceptors. Damage to the RPE can lead to the irreversible impairment or death of photoreceptors and RPE-associated ocular diseases such as RP and AMD. The pathogenesis of RPE-associated ocular diseases is poorly understood. The results of this study demonstrate that METTL14 may be of vital significance in RP development through regulation of the MAP2 expression.
RNA m6A modification is intimately correlated with pre-mRNA editing, translation, stability, nuclear export, and maturation of microRNA [Citation35]. It was reported that YTHDF1 inhibits melanoma by regulating histidine triad nucleotide binding protein (HINT)2 mRNA translation, while METTL3 was shown to target intercellular adhesion molecule (ICAM)-1 to regulate replication and apoptosis of epithelial cells in lens with diabetic cataract [Citation36]. However, the relationship between METTL14 and ocular diseases has not been elucidated. METTL14 is an essential component of the m6A methyltransferase complex and it is implicated in various disorders and the development of malignant tumors [Citation37]. Our analysis of data in the GEO database revealed downregulation of METTL14 expression in RP patients. Our in vitro experiments using ARPE-19 cells showed that METTL14 knockdown increased apoptosis resulting from cell cycle arrest of RPE cells, suggesting that METTL14 is involved in the development of RPE-associated ocular diseases.
Our study provides the first evidence that METTL14 can directly regulate MAP2 expression through m6A modification. MAPs are closely related to the maintenance of cell morphology and migration in eukaryotic cells and the maintenance of nerve fiber networks. MAP2 participates in microtubule assembly and synaptic growth and mediates communication between microtubules and other components of nerve cells; abnormal expression of MAP2 links to neurodegenerative diseases [Citation38,Citation39]. The highly expressed MAP2 gene in the retina of AMD patients indicates a close relationship with RPE-associated ocular diseases [Citation10]. We found that MAP2 is involved in various aspects of RPE cell function; upregulation of MAP2 decreased phagocytosis, impaired tight junctions, inhibited proliferation, and increased apoptosis in RPE cells. We also demonstrated that MAP2 directly interacted with NEUROD1, a pathogenic gene in RP. Collectively, these data suggest that METTL14, MAP2, and NEUROD1 contribute to the pathogenesis of RPE-associated ocular diseases, although the detailed mechanism remains to be determined.
m6A readers (YTHDF1/2/3, eukaryotic initiation factor [eIF]3, and insulin like growth factor 2 mRNA-binding protein [IGF2BP]1/2/3) bind to m6A-modified motifs either directly or indirectly, thereby affecting RNA functions [Citation39]. In this study, we found that YTHDF2 and YTHDF3 antibodies could bind to MAP2 mRNA. YTHDF2 is known to promote RNA degradation [Citation40]. In our study, MAP2 expression was increased in METTL14-depleted RPE cells. The findings in the present research demonstrate that MAP2 serves as a target of YTHDF2, which is by reports that YTHDF2 is the reader protein of METTL14 [Citation28]. This study is the first to demonstrate that METTL14 regulates the expression of MAP2 via the modification of m6A. And YTHDF2 was recognized as an m6A reader of MAP2 mRNA. This study is the first to propose METTL14/YTHDF2/MAP2/NEUROD1 axis, which provides new ideas for the treatment of RP.
5. Conclusion
The results of our study demonstrate that METTL14 in conjunction with YTHDF2 negatively regulates the expression of MAP2 via m6A modification. We also found that MAP2 interacts with NEUROD1 to induce pathologic changes in RPE cells. These findings provide a basis for investigations on the role of m6A modification in RPE-associated ocular disorder, indicating that therapeutic strategies targeting the METTL14/YTHDF2/MAP2/NEUROD1 axis may be an effective treatment for RP.
Highlights
METTL14 regulates the expression of MAP2 via the modification of m6A.
MAP2 could bind to NEUROD1.
YTHDF2 was recognized as an m6A reader of MAP2 mRNA.
Author contributions
LY performed the bioinformatics analysis and experiments and wrote the manuscript. CM participated in data collection and performed statistical analyses. SH and XM are the corresponding authors and supervised the study and made significant revisions to the manuscript. All authors reviewed and approved the final manuscript.
Ethics statement
This study gained the approval of the Ethics Committee of The First Affiliated Hospital of Chongqing Medical University (approval no. 2019-099).
Acknowledgements
The authors express appreciation to the members of the Chongqing Key Laboratory of Ophthalmology for their help, support, and suggestions.
Disclosure statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Data availability statement
All data created or analyzed during this study are presented in the published article.
Additional information
Funding
References
- Kwon W, Freeman SA. Phagocytosis by the retinal pigment epithelium: recognition, resolution, recycling. Front Immunol. 2020;11:604205.
- Zhang Y, Li M, Wang W, et al. Carvedilol activates nuclear factor E2-related factor 2/ antioxidant response element pathway to inhibit oxidative stress and apoptosis of retinal pigment epithelial cells induced by high glucose. Bioengineered. 2022;13:735–745.
- Zhu Z, Duan P, Song H, et al. Downregulation of circular RNA PSEN1 ameliorates ferroptosis of the high glucose treated retinal pigment epithelial cells via miR-200b-3p/cofilin-2 axis. Bioengineered. 2021;12:12555–12567.
- Wang Z, Huang Y, Chu F, et al. Integrated Analysis of DNA methylation and transcriptome profile to identify key features of age-related macular degeneration. Bioengineered. 2021;12:7061–7078.
- Wang S, Tang YJ. Sulforaphane ameliorates amyloid-beta-induced inflammatory injury by suppressing the PARP1/SIRT1 pathway in retinal pigment epithelial cells. Bioengineered. 2021;12:7079–7089.
- Pan J, Zhao L. Long non-coding RNA histone deacetylase 4 antisense RNA 1 (HDAC4-AS1) inhibits HDAC4 expression in human ARPE-19 cells with hypoxic stress. Bioengineered. 2021;12:2228–2237.
- Dong H, Wang M, Li Q. Exosomal miR-4488 and miR-1273g-5p inhibit the epithelial-mesenchymal transition of transforming growth factor beta2-mediated retinal pigment epithelial cells by targeting ATP-binding cassette A4. Bioengineered. 2021;12:9693–9706.
- Wang F, Li H, Xu M, et al. A homozygous missense mutation in NEUROD1 is associated with nonsyndromic autosomal recessive retinitis pigmentosa. Invest Ophthalmol Vis Sci. 2014;56:150–155.
- Yamashita T, Liu J, Gao J, et al. Essential and synergistic roles of RP1 and RP1L1 in rod photoreceptor axoneme and retinitis pigmentosa. J Neurosci. 2009;29:9748–9760.
- Pow DV, Sullivan RK. Nuclear kinesis, neurite sprouting and abnormal axonal projections of cone photoreceptors in the aged and AMD-afflicted human retina. Exp Eye Res. 2007;84:850–857.
- Pu Y, Chen X, Chen Y, et al. Transcriptome and differential methylation integration analysis identified important differential methylation annotation genes and functional epigenetic modules related to vitiligo. Front Immunol. 2021;12:587440.
- Liu Y, Luo S, Zhan Y, et al. Increased expression of PPAR-gamma modulates monocytes into a M2-like phenotype in SLE patients: an implicative protective mechanism and potential therapeutic strategy of systemic lupus erythematosus. Front Immunol. 2020;11:579372.
- Berner AK, Kleinman ME. Therapeutic approaches to histone reprogramming in retinal degeneration. Adv Exp Med Biol. 2016;854:39–44.
- Donato L, Scimone C, Alibrandi S, et al. Transcriptome analyses of lncRNAs in A2E-stressed retinal epithelial cells unveil advanced links between metabolic impairments related to oxidative stress and retinitis pigmentosa. Antioxidants (Basel). 2020;9.
- Zhong H, Tang HF, Kai Y. N6-methyladenine RNA modification (m(6)A): an emerging regulator of metabolic diseases. Curr Drug Targets. 2020;21:1056–1067.
- Jiang Y, Wan Y, Gong M, et al. RNA demethylase ALKBH5 promotes ovarian carcinogenesis in a simulated tumour microenvironment through stimulating NF-kappaB pathway. J Cell Mol Med. 2020;24:6137–6148.
- Li Z, Li N, Guo C, et al. The global DNA and RNA methylation and their reversal in lung under different concentration exposure of ambient air particulate matter in mice. Ecotoxicol Environ Saf. 2019;172:396–402.
- Li B, Wang X, Li Z, et al. Transcriptome-wide analysis of N6-methyladenosine uncovers its regulatory role in gene expression in the lepidopteran Bombyx mori. Insect Mol Biol. 2019;28:703–715.
- Li Q, Li X, Tang H, et al. NSUN2-Mediated m5C methylation and METTL3/METTL14-mediated m6A methylation cooperatively enhance p21 translation. J Cell Biochem. 2017;118:2587–2598.
- Zhao W, Qi X, Liu L, et al. Epigenetic regulation of m(6)A modifications in human cancer. Mol Ther Nucleic Acids. 2020;19:405–412.
- Chen XY, Zhang J, Zhu JS. The role of m(6)A RNA methylation in human cancer. Mol Cancer. 2019;18:103.
- Shi H, Wang X, Lu Z, et al. YTHDF3 facilitates translation and decay of N(6)-methyladenosine-modified RNA. Cell Res. 2017;27:315–328.
- Du H, Zhao Y, He J, et al. YTHDF2 destabilizes m(6)A-containing RNA through direct recruitment of the CCR4-NOT deadenylase complex. Nat Commun. 2016;7:12626.
- Wu J, Frazier K, Zhang J, et al. Emerging role of m(6) A RNA methylation in nutritional physiology and metabolism. Obes Rev. 2020;21:e12942.
- Guo Y, Wang P, Ma JH, et al. Modeling retinitis pigmentosa: retinal organoids generated from the iPSCs of a patient with the USH2A mutation show early developmental abnormalities. Front Cell Neurosci. 2019;13:361.
- Muratori C, Bona R, Federico M. Lentivirus-based virus-like particles as a new protein delivery tool. Methods Mol Biol. 2010;614:111–124.
- He J, Zhou M, Yin J, et al. METTL3 restrains papillary thyroid cancer progression via m(6)A/c-Rel/IL-8-mediated neutrophil infiltration. Mol Ther. 2021;29:1821–1837.
- Chen X, Xu M, Xu X, et al. METTL14-mediated N6-methyladenosine modification of SOX4 mRNA inhibits tumor metastasis in colorectal cancer. Mol Cancer. 2020;19:106.
- Arumugam S, Mary B, Kumar M, et al. Analysis of hepatic and retinal cell microRNAome during AAV infection reveals their diverse impact on viral transduction and cellular physiology. Gene. 2020;724:144157.
- Liu J, Li Y, Pu Q, et al. A polysaccharide from Lycium barbarum L.: structure and protective effects against oxidative stress and high-glucose-induced apoptosis in ARPE-19 cells. Int J Biol Macromol. 2021;201:111–120.
- Dong X, Wu W, Ma L, et al. Collectin-11 is an important modulator of retinal pigment epithelial cell phagocytosis and cytokine production. J Innate Immun. 2017;9:529–545.
- Bian ZM, Field MG, Elner SG, et al. Distinct CD40L receptors mediate inflammasome activation and secretion of IL-1beta and MCP-1 in cultured human retinal pigment epithelial cells. Exp Eye Res. 2018;170:29–39.
- Pan J, Liu F, Xiao X, et al. METTL3 promotes colorectal carcinoma progression by regulating the m6A-CRB3-Hippo axis. J Exp Clin Cancer Res. 2022;41:19.
- Hu X, Li F, He J, et al. LncRNA NEAT1 recruits SFPQ to regulate MITF splicing and control RPE cell proliferation. Invest Ophthalmol Vis Sci. 2021;62:18.
- Bian J, Zhuo Z, Zhu J, et al. Association between METTL3 gene polymorphisms and neuroblastoma susceptibility: a nine-centre case-control study. J Cell Mol Med. 2020;24(16):9280–9286.
- Jia R, Chai P, Wang S, et al. m(6)A modification suppresses ocular melanoma through modulating HINT2 mRNA translation. Mol Cancer. 2019;18:161.
- Gu C, Wang Z, Zhou N, et al. Mettl14 inhibits bladder TIC self-renewal and bladder tumorigenesis through N(6)-methyladenosine of Notch1. Mol Cancer. 2019;18:168.
- Lang F, Singh RK, Pei Y, et al. EBV epitranscriptome reprogramming by METTL14 is critical for viral-associated tumorigenesis. PLoS Pathog. 2019;15:e1007796.
- Visvanathan A, Patil V, Abdulla S, et al. N(6)-methyladenosine landscape of glioma stem-like cells: METTL3 is essential for the expression of actively transcribed genes and sustenance of the oncogenic signaling. Genes (Basel). 2019;10(2):141.
- Williams GD, Gokhale NS, Horner SM. Regulation of viral infection by the RNA modification N6-methyladenosine. Annu Rev Virol. 2019;6:235–253.