ABSTRACT
Osteosarcoma is a common bone malignancy in children and adolescents. Chemotherapeutic drug resistance is the major factor impacting the surgical outcome and prognosis of patients with osteosarcoma. This investigation assessed the role and mechanism of circular RNA_ANKIB1 in the development of osteosarcoma. The circular RNA (circ) _ANKIB1, microRNA (miR)-26b-5p, enhancer of zeste homolog 2 (EZH2) expression in OS samples was investigated through RT-qPCR. The EZH2, multidrug resistance protein 1 (MRP1), P-gp, and lipoprotein receptor-related protein (LRP) protein expressions were analyzed through western blot. The association between circ_ANKIB1 and the occurrence of clinic-pathological features in OS patients was assessed; the circular features of circ_ANKIB1 were analyzed. The hFOB1.19, KHOS, U2-OS OS cells were used to study the semi-inhibitory concentration IC50 of Doxorubicin (DXR)-resistant cells, clone formation, invasion, and apoptosis. The luciferase assay was used to study the binding of circ-ANKIB1 with miR-26b-5p and the targeting of miR-26b-5p with EZH2. In vivo experiments were performed via subcutaneous tumorigenic experiments. MiR-26b-5p in OS tissues and cells and DXR-resistant OS tissues and cells was silenced while circ_ANKIB1 and EZH2 were elevated. Circ_ANKIB1 silencing elevated miR-26b-5p, repressed EZH2, MRP1, P-gp, LRP, IC50, and elevated OS advancement. Circ_ANKIB1 bind miR-26b-5p. Reduced miR-26b-5p revered the influence of silencing circ_ANKIB1 on DXR resistant OS cells. MiR-26b-5p targeted EZH2, and EZH2 elevation reversed the impact of increasing miR-26b-5p on DXR resistant cells. Circ_ANKIB1 silencing suppressed DXR-resistant OS cells in vivo. In conclusion, Circ_ANKIB1 binds miR-26b-5p and modulates EZH2 to accelerate the chemo-resistance of osteosarcoma.
Graphical abstract
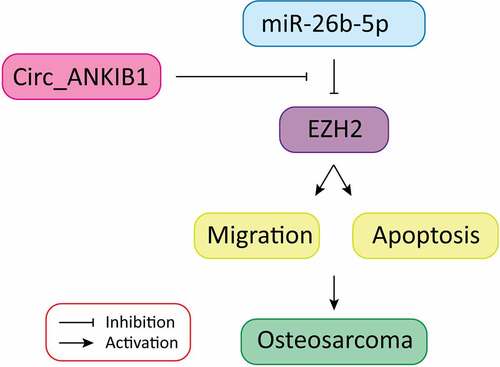
1 Introduction
Osteosarcoma (OS) is a prevalent bone malignancy in children and adolescents [Citation1]. As a mesenchymal tumor, OS tends to emerge in the metaphysis of long bones with abundant blood supply and fast growth, covering the proximal humerus, distal femur, or proximal tibia [Citation2]. The OS has a rapid progression leading to high morbidity and mortality [Citation3]. The overall survival rate of OS has been significantly increased in the past few decades due to advances in surgery and chemotherapy [Citation4]. Chemotherapeutic drugs have been adopted for the treatment of OS since the 1970s, with doxorubicin (DXR) being the primary chemotherapy regimen [Citation5]. Nevertheless, chemoresistance is the major adverse occurrence influencing the efficacy of chemotherapy drugs and reducing the survival rate of patients with OS [Citation6]. Consequently, there is a need to understand the molecular mechanisms leading to chemotherapy resistance, to improve the prognosis of OS.
Circular RNA (circRNA) is a form of non-coding RNA discovered a few decades ago. According to reports, circ RNA has a crucial role in the occurrence and progression of tumors [Citation7]. According to reports, the competitive endogenous RNA (ceRNA) mechanism, including gene transcription products like long non-coding RNA (lncRNA), competitively combine with microRNA (miRNA) via sponge adsorption [Citation8]. After that, circRNA is provided with miRNA adsorption to modulate OS [Citation9]. Circ RNA has also been reported to have a role in tumor chemo-resistance via the ceRNA mechanism. According to Wang et al, hsa_circ_0092276 facilitates the resistance of DXR to breast cancer (BC) via modulating the miR-348/ATG7 axis [Citation10]. In addition, circ RNA has been confirmed to modulate the resistance of OS cells to cisplatin or DOX via the ceRNA mechanism. CircRNA ankyrin repeat and IBR domain containing 1 (ANKIB1) (circANKIB1; circRNA ID: has_circ_0009112, position: chr7:91,972,337–91,981,956) is a product of ANKIB1 mRNA splicing (http://www.circbank.cn/infoCirc.html?id=hsa_circANKIB1_024). Zhu et al. [Citation11,Citation12] state that circ_ANKIB1 is elevated in OS, and Yi-xin Du et al. maintain that circ_ANKIB1 accelerates the development and invasion of OS cells via modulating the miR-19b/SOCS3/STAT pathway [Citation13]. However, the action and mechanism of circ_ANKIB1 in OS chemo-resistance were not reported.
MiRNAs are another non-coding RNA that modulates various tumor cells’ biological functions, involving cell differentiation and advancement [Citation14]. MiRNAs mediate the chemotherapy resistance of OS, offering a promising therapeutic target [Citation15–17]. MiR-26b has been reported as a tumor suppressor gene against OS [Citation11,Citation18], but its function in chemotherapy resistance has not yet been elucidated. Enhancer of zeste homolog 2 (EZH2) is a member of the histone methyltransferase family, which has a role in the carcinogenesis and chemotherapy resistance of OS [Citation11,Citation19].
The present investigation hypothesized that circular RNA_ANKIB1 accelerates osteosarcoma chemo-resistance via binding microRNA-26b-5p and modulating EZH2. This study aimed to determine the expressions of Circ_ANKIB1, EZH2, and miRNA-26b-5p in OS, determine the effects of circ_ANKIB1 knockdown on OS and investigate whether circ_ANKIB1 is a target of miR-26b-5p. Finally, the effects of miRNA-26b-5p inhibition on circ_ANKIB1 and circ_ANKIB1 knockdown on DXR-resistant OS cells in vivo were investigated
2 Methods
2.1 Source and grouping of tissue specimens
Osteosarcoma biopsy samples were collected in Huai’an Second People’s Hospital from August 2017 to May 2020 and prepared through histopathology and surgical resection. In total, 61 OS specimens and 61 para-cancerous tissue specimens were collected. The samples were taken from patients who had not done radiotherapy or chemotherapy prior to enrollment; All the enrolled patients received DXR-based chemotherapy, and the patients were divided into the DXR-sensitive (n = 33) and the DXR-resistant (n = 28) groups, in line with the Huvos scoring system. The study was reviewed and authorized by the ethics committee of Huai’an Second People’s Hospital (Approval number: H1703211c). Informed written consent was obtained from all the participants prior to inclusion into the study.
2.2 Cell culture
Normal osteoblast cell hFOB1.19 and human KHOS, U2-OS OS cell lines were purchased from American Type Culture Collection (ATCC, Manassas, VA, USA) and used for the study. The hFOB1.19 cells were grown in a culture medium of a 1:1 mixture of Dulbecco’s Modified Eagle’s Medium without phenol red and Ham’s F12 medium, augmented with 2.5 mM of l-glutamine, 0.3 mg/mL of G418, (all from Sigma) penicillin(100 U/ml) and streptomycin (100 U/ml). (Thermo Fisher Scientific, Waltham, MA), and 10% of fetal bovine serum, FBS (Gibco, Waltham, MA, USA) in a humidified 5% CO2 atmosphere at a temperature of 37°C.
The U2-OS and KHOS cells were cultured in Dulbecco’s modified Eagle medium (DMEM) supplemented with 10% fetal bovine serum (FBS)(Gibco, Uruguay), penicillin(100 U/ml), and streptomycin (100 U/ml) (Thermo Fisher Scientific, Waltham, MA). The cells were cultured at 37°C and 5% CO2. DXR resistant cell line KHOS/DXR and U2OS/DXR were generated via exposing the parental cell lines KHOS and U2OS to increasing doses of DXR (Sigma, San Francisco, CA, USA). The DXR resistant cell lines culture was done in the same DMEM culture medium with 1 µg/mL DXR to sustain their resistant phenotype.
2.3 Cell transfection
Cells were seeded in a 6-well plate, and the relevant sequences were transfected separately. The cells were divided into 6-well plates one day before transfection to obtain a cell density of about 70% during transfection. Transfection was done using lipofectamine 2000 reagent Kit (11,668–027, Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s guidelines. Furthermore, the si-circ_ANKIB1#1/-circ_ANKIB1#2, si/mimic/inhibitor-NC, miR-26b-5p inhibitor/mimic, and Oe-EZH2 were prepared, synthesized, and transfected into the cells. All vector construction, plasmids, identification of sequencing, virus packaging, and detection of titers were provided by the Shanghai Genechem Co., Ltd. (Shanghai, China).
2.4 Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
Total RNA in tissues and cells was extracted and measured using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was reverse transcribed into cDNA using the reverse transcription kit (K1621, Fermentas, Maryland, NY, USA). Each gene was tested with a fluorescence quantitative polymerase chain reaction (PCR) kit (Takara, Dalian, China) and examined with a real-time fluorescent quantitative PCR instrument (Thermo Fisher Scientific, Massachusetts, USA). U6 was used as the internal reference for miR-26b-5p, while glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was utilized as the internal reference for circ_ANKIB1 and EZH2 [Citation20]. Calculation of the target gene was done with the 2−ΔΔCt method, N = 3, and the average was taken. Primer sequences of circ_ANKIB1, miR-26b-5p, and EZH2 were designed as shown in .
Table 1. Primer sequences
2.5 RNase R treatment
The RNAs (1 µg) were treated from cells using RNase R (2 U/µg, GeneSeed, Guangzhou, China); Incubation and inactivation were performed. Reverse transcription of the treated RNAs was done with divergent or convergent primers, which were tested. N = 3, and the average was taken.
2.6 RNA immunoprecipitation (RIP)
RNA immunoprecipitation (RIP) was done as described elsewhere [Citation21]. Determination of the RIP was done with a Magna RIP Kit (Millipore, Billerica, MA, USA). To this end, collection and resuspension of 1 × 107 OS cells were performed in 100 μL of RIP lysis buffer plus protease inhibitor cocktail and RNase inhibitors. Approximately 200 μL of the cell lysates were then incubated with 5 μg magnetic beads conjugated to anti-Argonaute 2 (AGO2) antibody (Millipore) or control rabbit IgG (Millipore). After digestion with proteinase K, extraction of the immunoprecipitated RNAs was performed. Test of enrichment of circ_ANKIB1and miR-26b-5p was finally conducted. The experiment was done in triplicate.
2.7 RNA-pull down
RNA-pull down was done as described elsewhere [Citation21]. Transfection of the cells was done using 50 nM biotin-labeled WT-bio-miR-26b-5p and MUT-bio-miR-26b-5p (Wuhan GeneCreate Biological Engineering Co., Ltd., Wuhan, China). The cells were then collected and incubated for 10 min in a specific lysis buffer (Ambion, Austin, Texas, USA). The lysates were incubated with M-280 streptavidin magnetic beads (S3762, Sigma, USA) pre-coated with RNase-free and yeast tRNA (TRNABAK-RO, Sigma, USA). The samples were then rinsed with pre-cooled lysis buffer, low/high-salt buffer. The combined RNA was purified and examined for the enrichment of the circ_ANKIB1. N = 3, and the average was taken.
2.8 The luciferase activity assay
Prediction of the targeting sites of miR-26b-5p and circ_ANKIB1/EZH2 was performed as previously described [Citation22]. Circ_ANKIB1/EZH2-3’-untranslated region (3ʹUTR) sequence primer was designed, and mutation of the target sites of its 3ʹUTR sequence primer miR-26b-5p was used to gain the 3ʹUTR MUT fragment. To attain the same sticky ends, the pMIR-Report vector plasmid and the target gene were simultaneously digested via Hind III and Mlu I restriction enzyme. The T4 ligase was used to transform the target gene and the pMIR-Report vector plasmid into the Escherichia coli JM109 strain. After the advancement of a single colony, colony PCR and recombinant plasmids were digested and tested, and sequencing was performed. The miR-26b-5p mimic and its negative control (all designed and synthesized via GenePharma Co. Ltd. Company, Shanghai, China) were separately transfected into the cells with circ_ANKIB1/EZH2-3ʹUTR WT and circ_ANKIB1/EZH2-3ʹUTR MUT. After the development of the cells with a specific density, collection and lysis of the cells were conducted. Examination of the luciferase activity was done with a luciferase detection kit (Promega, USA) and an ultra-micro ultraviolet photometer (Bio-Rad, USA). N = 3, and the average was taken.
2.9 Western blot
The cells were transferred in a centrifuge tube, 100 μL radio-immunoprecipitation assay lysis solution (R0020, Beijing solarbio science & technology co. ltd., Beijing, China) was added and homogenized at 3000 rpm. The protein concentration was determined using bicinchoninic acid (BCA) kit (AR0146, Wuhan Boster Company, Wuhan, China), and the concentration of each sample was adjusted to 3 μg/μL.
The extracted protein was loaded into the loading buffer, boiled for 10 min, and a 30 μg sample was loaded per well, and the protein was separated using 10% polyacrylamide gel electrophoresis. The protein was transferred to polyvinylidene fluoride (PVDF) membrane (P2438, Sigma, USA) via semi-dry electroporation transfer method, blocked with 5% bovine serum albumin (10-L16, Beijing Zhongsheng Likang Technology Co., Ltd., China), and the primary antibody was then added at; EZH2 (1: 1000, Abcam, Cambridge, MA, USA), P-gp (1: 500, Abcam, Cambridge, MA, USA), multidrug resistance protein 1 (MRP1) (1: 1000, Abcam, Cambridge, MA, USA) and lipoprotein receptor-related protein (LRP) (1: 1000, Abcam, Cambridge, MA, USA).
Later, corresponding horseradish peroxidase-labeled IgG (1: 1000, Wuhan Boster Company, Wuhan, China) was added and incubated. The membrane was then rinsed, and a band was developed using a chemiluminescence reagent. The GADPH (ab181602, Abcam, USA, 1: 10,000) was used as the internal reference. The band’s development was analyzed with Bio-rad Gel Dol EZ imager (GEL DOC EZ IMAGER, Bio-rad, California, USA). A gray value analysis of the target band was done via Image J software (National Institutes of Health, Bethesda, MD, USA). The experiment was done in triplicates.
2.10 Cell-counting kit-8 (CCK-8)
The cells were harvested at the logarithmic growth phase, and the pellets were collected via conventional digestion and centrifugation. Cells were then resuspended, counted, diluted, and adjusted to a 1 × 104/m concentration. Approximately 200 μL of the diluted cell suspension was added into 96-well plates and incubated. After adherence, the cells were treated with gradient concentrations of DXR. The cells were then cultured for 48 h, and the old medium was then discarded and replaced with 10% CCK-8 fresh medium as per the manufacturer’s instructions of Cell Counting Kit-8 (Dojindo, Tokyo, Japan). The cells were then incubated for 3 h, and the absorbance value was examined at 450 nm. The repression rate of the cell proliferation = (1-the experiment A /the control A) × 100%, and IC50 of the cells lines was tested which was defined as a chemotherapeutic drug concentration proving the successful construction of drug-resistant strains, and repressed the cell proliferation with 50%. N = 3, and the average was taken.
2.11 Colony formation experiment
A colony formation assay was done as previously described [Citation23]. The cells at the logarithmic growth phase were harvested, and the cell pellets were collected through conventional digestion and centrifugation. The cell pellets were resuspended and counted, and the concentration was adjusted to 1 × 105/mL and diluted with 4 ml of culture medium in 6-well plates. The cells were incubated for 3 weeks to the point of the development of clones. The cells were fixed in methanol for 15 minutes, and the number of visible clones was counted after crystal violet staining. The experiment was done in triplicates.
2.12 Flow cytometry
The flow cytometry was done as previously described [Citation24]. Cells in the logarithmic phase were detached and collected in a flow tube with 0.25% pancreatin (without ethylene diamine tetraacetic acid (EDTA)) (PYG0107, Boster, Wuhan, China). The cells were then centrifuged, and supernatants were collected and washed twice. The cells were then stained with Annexin-V-fluorescein isothiocyanate (FITC), propidium iodide (PI), and 4-(2-hydroxyethyl)-1-piperazineëthanesulfonic acid (HEPES) buffer solutions according to the instructions of Annexin-V-FITC Cell Apoptosis Detection Kit (K201-100, Biovision, USA). Approximately 1 × 106 cells were resuspended in 100 µL staining solution, incubated, and 1 mL of HEPES buffer solution was finally added. FITC and PI fluorescence was analyzed via exciting 515 nm 620 nm bandpass filters at 488 nm. The cell apoptosis was assessed via flow cytometry (BD Biosciences, Franklin Lakes, NJ, USA). The assay was repeated three times.
2.13 Scratch test
Scratch assay was done as previously described [Citation25]. The cells were seeded in a 6-well plate at 4 × 105 cells/well. After transfection, at a confluence of around 80%, a straight line was made along the central axis at an angle perpendicular to the bottom of the plate surface using a sterile 10 μL pipette tip. The serum-free medium in the culture was replaced and cultured further for 0.5 h. Cell migration was observed, and the images were captured using an inverted optical microscope at 0 and 24 h. the assay was done in triplicate.
2.14 Transwell
Transwell assay was done as previously described [Citation26]. The cell invasion in each group was examined using a Matrigel-coated Transwell chamber (Becton Dickinson). The cells at the logarithmic growth phase were harvested, transfected, resuspended, and counted. Approximately 1 × 105 of cells (200 μL) was added in the upper chamber of Transwell, and 500 μLof 1 Roswell Park Memorial Institute 1640 medium supplemented with 20% FBS was added in the lower chamber of Transwell. Cells in the Transwell chamber were collected, and the cells adjacent to the lateral membrane of the upper chamber were discarded. The cells were fixed with 4% paraformaldehyde solution, stained with crystal violet. The cells were observed, photographed under an optical microscope, and analyzed using ImageJ software (National Institutes of Health, Bethesda, MD, USA). The experiment was done in triplicates.
2.15 Subcutaneous tumorigenesis in nude mice
BALB/c nude mice (4–6 weeks) were purchased from Shanghai Slack Company, Shanghai, China. The mice were bred under specific pathogen-free (SPF) environmental conditions. The animal experiment was authorized by the committee on Anima Care and Use of Huai’an Second People’s Hospital.
Transfection of KHOS/DXR and U2OS/DXR cells was done after si-NC/circ_ANKIB1#1. The single-cell suspension (2 × 106 pcs/mL) was subcutaneously injected into the back of nude mice with a disposable sterile syringe to study the effect of circ_ANKIB1 on OS drug-resistant cells in vivo. In addition, 5 mice were used in each group. Behind a week, the mice were injected with 2 mg/kg DXR (Solarbio) 7 days after the introduction of the tumor. The volume of the tumor was monitored once a week. The mice were sacrificed after 4 weeks, and the tumor was harvested. The tumor weight and dimensions were then measured. The tumor volume was calculated using the formula; Tumor volume = 0.5 × (long axis × short axis) 2.
2.16 Statistical analysis
Data analysis was done using GraphPad Prism 8. The data were presented as mean ± standard deviation (SD). The Two-group comparison was done via t-test. The multiple groups’ comparison was made using one-way analysis of variance (ANOVA), and pairwise comparison was determined using Tukey’s multiple comparisons test. Analysis of the relevance of circ-ANKIB1 in OS patients’ clinic-pathological features was also done and. Pearson test was used to carry out the correlation analysis. P < 0.05 was regarded as significant.
3 Results
3.1 Circ_ANKIB1 and EZH2 are upregulated in OS tissues
Osteosarcoma is a common bone malignancy in children and adolescents. Chemotherapeutic drug resistance is the primary factor impacting the surgical outcome and prognosis of patients with osteosarcoma. The present investigation hypothesized that circular RNA_ANKIB1 accelerates osteosarcoma chemo-resistance via binding microRNA-26b-5p and modulating EZH2. The study aimed to determine the expressions of circ_ANKIB1, EZH2, and miRNA-26b-5p in OS, determine the effects of circ_ANKIB1 knockdown on OS and investigate whether circ_ANKIB1 is a target of miR-26b-5p. Finally, the effects of miRNA-26b-5p inhibition on circ_ANKIB1 and circ_ANKIB1 knockdown on DXR-resistant OS cells in vivo were investigated.
The circ_ANKIB1, miR-26b-5p, and EZH2 expression in OS tissues and corresponding para-cancerous tissues were determined, and the results confirmed an increased circ_ANKIB1 expression in the tumor compared to the normal tissues. However, miR-26b-5p expression was reduced in OS tissues compared to the para-cancerous tissues. EZH2 expression was also elevated in the OS tissues compared to the normal tissues ((). The analysis of the correlation of circ_ANKIB1, miR-26b-5p with EZH2 in patients with OS showed that miR-26b-5p is negatively correlated with circ_ANKIB1 () while circ_ANKIB1 and EZH2 were positively associated (). EZH2 and MiR-26b-5p demonstrated a negative correlation (). Additionally, analysis of circ_ANKIB1 and clinic-pathological features of OS patients clarified that: Tumor node metastasis (TNM) grade, differentiation, and tumor size of patients with OS were positively correlated with circ_ANKIB1, as proved in .
Table 2. Association of relative circ_ANKIB1 and OS patients’ clinic-pathological features
Figure 1. Circ_ANKIB1 and EZH2 are upregulated in OS tissues.
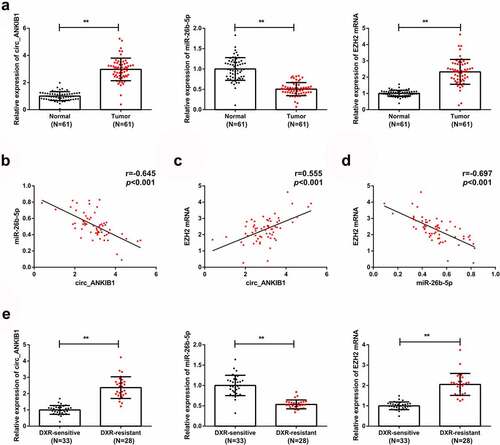
RT-qPCR was then used to determine the circ_ANKIB1, miR-26b-5p, and EZH2 mRNA expressions in DXR-resistant and DXR-sensitive OS tissues. The observations demonstrated increased circ_ANKIB1 and EZH2 mRNA expressions but reduced miR-26b-5p mRNA expressions in DXR-resistant OS tissues compared to the DXR-sensitive OS tissues ().
3.2 Circ_ANKIB1, EZH2, and miR-26b-5p expressions in DXR-resistant OS tissue
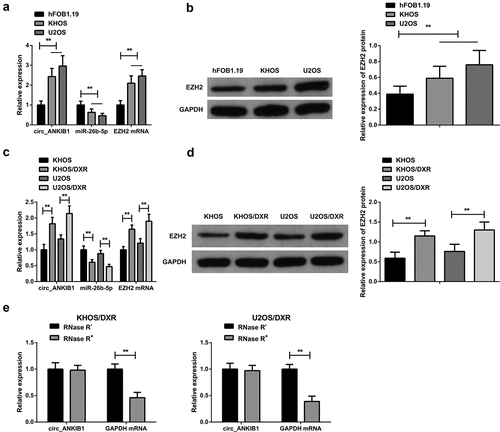
The hFOB1.19, KHOS, and U2OS cells were cultured, and DXR resistant cell lines KHOS/DXR and U2OS/DXR were constructed. RT-qPCR and western blotting were used to determine the mRNA expressions of MiR-26b-5p, circ_ANKIB1, and EZH2. According to the results, MiR-26b-5p expression was significantly reduced while circ_ANKIB1 and EZH2 expressions were elevated in KHOS and U2OS tumor cell lines compared to the hFOB1.19 normal cell line (). Additionally, miR-26b-5p expression in the drug-resistant cell lines KHOS/DXR and U2OS/DXR was significantly reduced compared to the parental lines, but circ_ANKIB1 and EZH2 expressions were significantly increased (). Additionally, circ_ANKIB1 was resistant to RNase R treatment, while digestion of GAPDH was via RNase R, clarifying that circ_ANKIB1 has a ring structure ().
Figure 2. Increased circ_ANKIB1, EZH2, but suppressed miR-26b-5p expressions in DXR-resistant OS tissue.
3.3 circ_ANKIB1 knockdown suppresses DXR-resistant OS cells
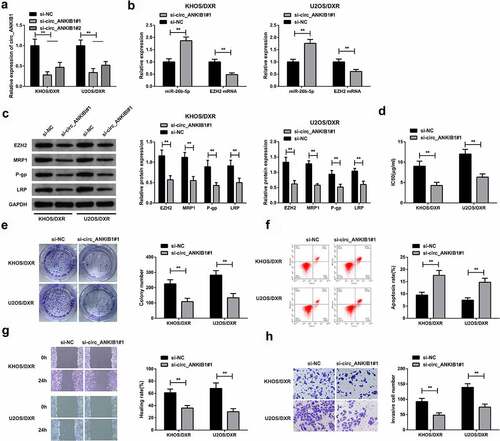
The KHOS/DXR and U2OS/DXR cells were transfected with si-circ_ANKIB1#1 and si-circ_ANKIB1#2 or the si-NC, and then si-circ_ANKIB1#1 mRNA expression was determined using RT-qPCR. The results demonstrated a significantly reduced si-circ_ANKIB1#1 expression (). Further, the expression of miR-26b-5p and EZH2 were assessed in the KHOS/DXR and U2OS/DXR cells transfected with circ_ANKIB1#1 or si-NC. The results confirmed an increased miR-26b-5p mRNA expression but a reduced EZHZ mRNA expression in KHOS/DXR and U2OS/DXR cells transfected with circ_ANKIB1#1 compared to the controls (). The EZHZ, MRP-1, P-gp, LRP expressions were determined in the si-circ_ANKIB1 or the si-NC cells. The results confirmed significantly reduced EZHZMRP1, P-gp, and LRP protein expressions in the si-circ ANKIB1-transfected KHOS/DXR and U2OS/DXR cells compared to the si-NC transfected cells (). Knockdown of circ_ANKIB1 significantly reduced the IC50 for the KHOS/DXR and U2OS/DXR cells. In addition, the clone formation was significantly reduced in the cells transfected with si- circ_ANKIB1 compared to the si-NC-transfected cells (). Knockdown of circ_ANKIB1 also significantly increased the apoptosis of KHOS/DXR and U2OS/DXR cells compared to the si-NC transfected cells (). Finally, the wound healing and cell invasion capacity were significantly reduced following transfection of the cells with si-circ ANKIB1 compared to transfection with si-NC accelerated the cell advancement. (). These observations confirmed that circ_ANKIB1 silencing effectively suppressed DXR-resistant OS cells.
Figure 3. Circ_ANKIB1 knockdown suppresses DXR-resistant OS cells.
3.4 circ_ANKIB1 is a target of miR-26b-5p in OS
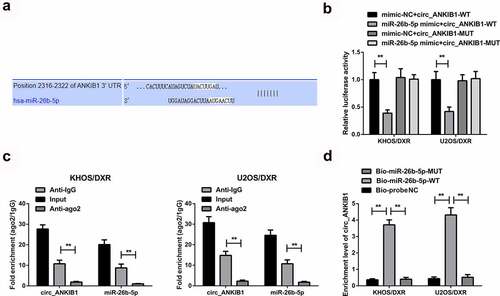
We then investigated whether circ_ANKIB1 inhibits DXR-resistant OS cells by targeting miR-26b-5p. Analysis of miR-26b-5p target gene using Pictar, miRanda, and Targetscan prediction tools showed that circ_ANKIB1 binds miR-26b-5p in the 3ʹUTR region, as shown in . In addition, the luciferase activity was significantly reduced in cells co-transfected with miR-26 mimic and wild-type circ_ANKIB1compared to the mimic-NC+ wild-type circ_ANKIB1. However, no significant differences were observed in the luciferase activity of mutant 3ʹUTR versus the mimic-NC, confirming that Circ_ANKIB1 specifically binds with miR-26b-5p, as shown in . Circ_ANKIB1 and miR-26b-5p enrichment in Anti-AGO2 pellets were significantly increased compared to the Anti-IgG, as presented in . The enrichment of Circ_ANKIB1 in the Bio-miR-5-WT was remarkably elevated compared to the abundance of Circ_ANKIB1 in the Bio-miR-5-MUT or the Bio-probe NC, as presented in .
Figure 4. Circ_ANKIB1 is a target of miR-26b-5p in OS.
3.5 MiR-26b-5p inhibition reverses the suppressive effect of circ_ANKIB1 in OS
We then aimed at assessing the role of miR-26b-5p inhibitors in the control of OS. To this effect, the KHOS/DXR and U2OS/DXT cells were con-transfected with si-circ_ANKIB1#1+ miR-26b-5p inhibitor or si-circ_ANKIB1#1 and inhibitor-NC. The expression of miR-26b-5p and EZH2 mRNA were determined through RT-qPCR. According to the observations, miR-26b-5p mRNA expression was significantly reduced in the si-circ_ANKIB1#1+ inhibitor-NC compared to the si-circ_ANKIB1#1+ miR-26b-5p inhibitor. However, EZH2 mRNA expression was significantly increased in the circ_ANKIB1#1+ miR-26b-5p inhibitor compared to the si-circ_ANKIB1#1+ inhibitor-NC, as shown in . Determination of proteins expression using western blotting confirmed a significant elevation of EZH2, MRP1, P-gp, and LRP proteins in the cells co-transfected with si-circ_ANKIB1#1 miR-26b-5p inhibitor compared to the cells co-transfected with si-circ_ANKIB1#1 and inhibitor-NC, as shown in .
Figure 5. MiR-26b-5p inhibition reverses the suppressive effect of circ_ANKIB1 against DXR-resistant.
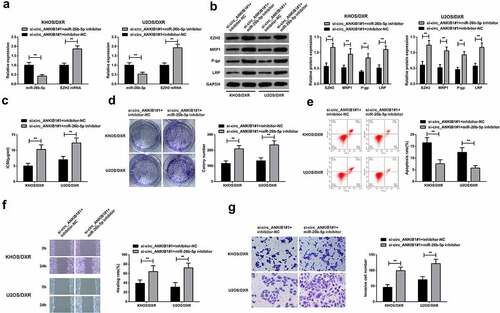
.The IC50 and colony numbers were also significantly increased in the cells co-transfected with si-circ_ANKIB1#1 and miR-26b-5p inhibitor compared to those co-transfected with si-circ_ANKIB1#1 and inhibitor-NC, as shown in and 5D. However, apoptosis was significantly reduced in the cells co-transfected with si-circ_ANKIB1#1 and miR-26b-5p inhibitor compared to the cells co-transfected with si-circ_ANKIB1#1 and inhibitor-NC, as shown in . Finally, the wound healing and cell invasion ability were also significantly increased in the cells co-transfected with si-circ_ANKIB1#1 and miR-26b-5p inhibitor compared to the cells co-transfected with si-circ_ANKIB1#1 and inhibitor-NC, as shown in . These observations confirmed that inhibition of miR-26b-5p reverses the protective role of circ_ANKIB1#1 silencing against OS.
3.6 MiR-26b-5p targets EZH2 to repress DXR-resistant OS cells
The downstream mechanism of miR-26B-5p was then investigated. According to the gene target prediction result, EZH2 was the target gene of miR-26b-5p (). The luciferase binding activity assay confirmed a significantly reduced activity in the cells co-transfected with miR-26b-5p mimic and wild-type EZH2 compared to cells co-transfected with mutant 3ʹUTR or the mimic-NC, clarifying that miR-26b-5p targeted EZH2, as shown in . The cells were then transfected with MiR-26b-5p-mimic, MiR-26b-5p mimic+ Oe-EZHZ or mimic-NC, miR-26b-5p, and EZH2 mRNA expression was then determined through RT-qPCR.
Figure 6. MiR-26b-5p targets EZH2 to repress DXR-resistant OS cells.
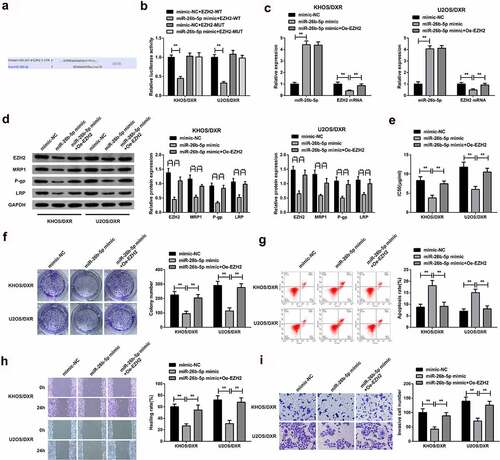
The results confirmed a significant elevation of miR-26b-5p mRNA expression but a significant decrease of EZH2 mRNA in the miR-26b-5p-mimic and MiR-26b-5p mimic+ Oe-EZHZ group compared to the mimic-NC, as shown in . Furthermore, miR-26b-5p, EZH2 and MRP1, P-gp, and LRP protein were remarkably inhibited in the miR-26b-5p-mimic and MiR-26b-5p mimic+ Oe-EZHZ group compared to the mimic-NC, as proved in . The IC50 and colony numbers were significantly reduced in the miR-26b-5p-mimic and miR-26b-5p mimic+ Oe-EZHZ group compared to the mimic-NC (). The apoptosis was significantly increased in the miR-26b-5p mimic compared to the miR-26b-5p mimic+ Oe-EZHZ or the mimic-NC group (). Finally, the wound healing and cell invasion capacity was significantly increased in the miR-26b-5p mimic+ Oe-EZHZ compared to the miR-26b-5p mimic group (). Taken together, these observations confirmed that miR-26b-5p targets EZH2 to repress DXR-resistant OS cells.
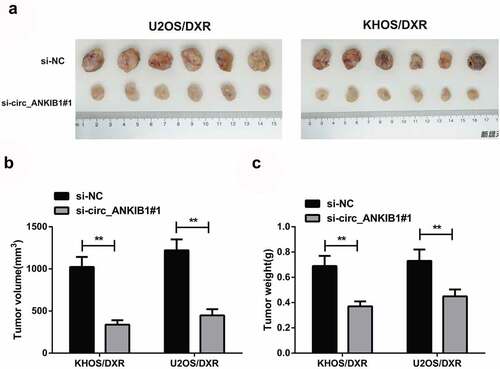
3.7 Knockdown of circ_ANKIB1 suppresses DXR-resistant OS cells in vivo
We finally investigated the effects of circ_ANKIB1 knockdown in the progression of OS in vivo. The xenograft tumor mouse model was constructed to determine the function of circ_ANKIB1 on DXR drug-resistance in vivo. The results confirmed that knockdown of circ_ANKIB1 significantly suppressed the tumor volume and weight of the tumor mice model (). These results together showed that the knockdown of circ_ANKIB1 repressed DXR-resistant OS cells.
Figure 7. Knockdown of circ_ANKIB1 suppresses DXR-resistant OS cells in vivo.
4 Discussion
Resistance to chemotherapy is always the major challenge in OS treatment [Citation11,Citation27]. DXR is among the most effective drugs in standard chemotherapy for OS, but 40–45% of patients with advanced OS do not respond or only partially respond to DXR [Citation28,Citation29] . Drug resistance in the tumor is a complex, multi-step process. Recently, with the advancement of bioinformatics analysis and elevated-throughput sequencing technology, the biological functions of circular RNA (circRNA) have been gradually identified [Citation30]. Some studies have reported that circRNA has a role in developing resistance to chemotherapy in human cancers such as OS [Citation31].
Consequently, targeting circRNA to arrest DXR resistance might be a promising treatment strategy. In this study, circ_ANKIB1was significantly elevated in OS tissues and cells. Circ_ANKIB1 in DXR-resistant OS tissues and cells was increased compared to the DXR-sensitive tissues and cells. Silencing of circ_ANKIB1 repressed the progression of DXR-resistant OS cells via miR-26b-5p /EZH2 axis.
According to recent studies, circRNA has a vital role in controlling OS chemo-resistance. For instance, Ji Y et al. maintain circ_0002060 strengthens Adriamycin resistance in OS via modulating miR-198/ABCB1 axis [Citation32]. Additionally, circ_0001721 has been reported to heighten Adriamycin resistance via miR-758/TCF4 axis and facilitate tumorigenesis of OS [Citation33]. Increased circ-LARP4 is also associated with reduced Enneking phase and prolonged survival and improved chemo-sensitivity of OS to cisplatin and Adriamycin via sponging miR-424 [Citation34]. In the present study, circ_ANKIB1 is elevated in OS and linked with TNM grade, tumor size, and lymph node metastasis, which agrees with the previous findings [Citation13]. The modulation of circ_ANKIB1 in OS chemo-resistance was further explored, which clarified thatcirc_ANKIB1 silencing repressed the progression of advancement DXR-resistant OS cell lines. Furthermore, this trend was also confirmed in the in vivo mouse experiments. As confirmed, circ_ANKIB1 has a crucial biological function in the chemo-resistance of OS. Furthermore, circ_ANKIB1 binds with miR-26b-5p. Circ_ANKIB1 had a role in the chemical resistance of OS via the competitive endogenous RNA mechanism (ceRNA mechanism)”.
MicroRNAs (miRNAs), small non-coding RNAs, control the gene, and the critical function of miRNAs within cancer pathogenesis and response to treatment has been confirmed in different cancers [Citation35]. Previous studies have reported the repression of miR-26b-5p in different tumors such as lung adenocarcinoma LAC [Citation36], Human Papillary Thyroid Cancer [Citation37], and Burkitt lymphoma [Citation38]. The results of the present study observed that miR-26b-5p was silenced in OS and DXR-resistant OS tissues and cells. Circ_ANKIB1 targeted adsorption of miR-26b-5p and miR-26b-5p inhibition reversed the effect of circ_ANKIB1 silencing on DXR resistance OS cells, which proved that circ_ANKIB1 modulated DXR resistant OS cells via miR-26b-5p binding. Elevated miR-26b-5p represses the DXR resistant OS cells, and miRNAs have a role in establishing and advancing cancer via interacting with the targeted genes [Citation39]. In this study, EZH2 was elevated in DXR-resistant OS cells, consistent with the previous findings. Yuan J et al. [Citation27]elucidate that EZH2 is elevated in DXR-resistant OS cells and targets miR-760 in arresting the progression of DXR-resistant OS cells. The results clarified that EZH2 was elevated in OS and DXR-resistant OS. Furthermore, miR-26b-5p inhibited DXR-resistant OS via targeting EZH2.
5 Conclusion
In conclusion, circ_ANKIB1 accelerates OS resistance to DXR via modulating miR-26b-5p and EZH2. This study provides a novel understanding of the biological mechanism of DXR resistance in OS, highlighting a possible target approach for developing an alternative therapeutic intervention against osteosarcoma.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Zhao X, Wu Q, Gong X, et al. Osteosarcoma: a review of current and future therapeutic approaches [J]. Biomed Eng Online. 2021;20(1):24.
- Belayneh R, Fourman MS, Bhogal S, et al. Update on Osteosarcoma [J]. Curr Oncol Rep. 2021;23(6):71.
- Kager L, Tamamyan G, Bielack S. Novel insights and therapeutic interventions for pediatric osteosarcoma [J]. Future Oncol. 2017;13(4):357–368.
- Jo VY, Doyle LA. Refinements in sarcoma classification in the current 2013 world health organization classification of tumours of soft tissue and bone [J]. Surg Oncol Clin N Am. 2016;25(4):621–643.
- König M, Osnes TA, Lobmaier I, et al. Multimodal treatment of craniofacial osteosarcoma with high-grade histology. A single-center experience over 35 years [J]. Neurosurg Rev. 2017;40(3):449–460.
- Casali PG, Bielack S, Abecassis N, et al. Bone sarcomas: ESMO-PaedCan-EURACAN clinical practice guidelines for diagnosis, treatment and follow-up [J]. Ann Oncol. 2018;29(Suppl 4):iv79–iv95.
- Tang Q, Hann SS. Biological roles and mechanisms of circular RNA in human cancers [J]. Onco Targets Ther. 2020;13:2067–2092.
- Yu D, Zhang S, Feng A, et al. Methotrexate, doxorubicin, and cisplatinum regimen is still the preferred option for osteosarcoma chemotherapy: a meta-analysis and clinical observation [J]. Medicine (Baltimore). 2019;98(19):e15582–e15582.
- Wang SY, Hu HZ, Qing XC, et al. Recent advances of drug delivery nanocarriers in osteosarcoma treatment [J]. J Cancer. 2020;11(1):69–82.
- Barrett SP, Salzman J. Circular RNAs: analysis, expression and potential functions [J]. Development. 2016;143(11):1838–1847.
- Wang S, Claret FX, and Wu W. MicroRNAs as therapeutic targets in nasopharyngeal carcinoma [J]. Front Oncol. 2019;13(9):756.
- Zhu X, Liu C, Shi J, et al. Circular RNA circANKIB1 promotes the progression of osteosarcoma by regulating miR-217/PAX3 axis [J]. J Bone Oncol. 2021;27(100347):100347.
- Du YX, Guo LX, Pan HS, et al. Circ_ANKIB1 stabilizes the regulation of miR-19b on SOCS3/STAT3 pathway to promote osteosarcoma cell growth and invasion [J]. Hum Cell. 2020;33(1):252–260.
- Mirahmadi Y, Nabavi R, Taheri F, et al. MicroRNAs as biomarkers for early diagnosis, prognosis, and therapeutic targeting of ovarian cancer [J]. J Oncol. 2021;2021(3408937):1–25.
- Lin Z, Xie X, Lu S, et al. Noncoding RNAs in osteosarcoma: implications for drug resistance [J]. Cancer Lett. 2021;504:91–103.
- Li G, Li Y, Wang DY. Overexpression of miR-329-3p sensitizes osteosarcoma cells to cisplatin through suppression of glucose metabolism by targeting LDHA [J]. Cell Biol Int. 2021;45(4):766–774.
- Yang D, Xu T, Fan L, et al. microRNA-216b enhances cisplatin-induced apoptosis in osteosarcoma MG63 and SaOS-2 cells by binding to JMJD2C and regulating the HIF1α/HES1 signaling axis [J]. J Exp Clin Cancer Res. 2020;39(1):201.
- Wu Q, Feng Q, Xiong Y, et al. RAB31 is targeted by miR-26b and serves a role in the promotion of osteosarcoma [J]. Oncol Lett. 2020;20(5):244.
- Zhu Z, Tang J, Wang J, et al. MiR-138 acts as a tumor suppressor by targeting EZH2 and enhances cisplatin-induced apoptosis in osteosarcoma cells [J]. PLoS One. 2016;11(3):e0150026.
- Ayuk SM, Abrahamse H, Houreld NN. The role of photobiomodulation on gene expression of cell adhesion molecules in diabetic wounded fibroblasts in vitro [J]. J Photochem Photobiol B. 2016;161:368–374.
- Wu Y, Xie Z, Chen J, et al. Circular RNA circTADA2A promotes osteosarcoma progression and metastasis by sponging miR-203a-3p and regulating CREB3 expression [J]. Mol Cancer. 2019;18(1):73.
- Hu S, Li J, Tong M, et al. MicroRNA‑144‑3p may participate in the pathogenesis of preeclampsia by targeting Cox‑2 [J]. Mol Med Rep. 2019;19(6):4655–4662.
- Li W, Zheng G, Xia J, et al. Cell cycle‐related and expression‐elevated protein in tumor overexpression is associated with proliferation behaviors and poor prognosis in non‐small‐cell lung cancer [J]. Cancer Sci. 2018;109(4):1012–1023.
- Li Z, Qin X, Bian W, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis [J]. J Exp Clin Cancer Res. 2019;38(1):1–13.
- Xu M, Cao F-L, Li N, et al. Leptin induces epithelial-to-mesenchymal transition via activation of the ERK signaling pathway in lung cancer cells [J]. Oncol Lett. 2018;16(4):4782–4788.
- Xing Z, Zhang Z, Gao Y, et al. The lncRNA LINC01194/miR-486-5p axis facilitates malignancy in non-small cell lung cancer via regulating CDK4 [J]. Onco Targets Ther. 2020 3151;13:3151–3163.
- Yuan J, Liu Y, Zhang Q, et al. CircPRDM2 contributes to doxorubicin resistance of osteosarcoma by elevating EZH2 via sponging miR-760 [J]. Cancer Manag Res. 2021;13:4433–4445.
- Zhang B, Zhang Y, Li R, et al. The efficacy and safety comparison of first-line chemotherapeutic agents (high-dose methotrexate, doxorubicin, cisplatin, and ifosfamide) for osteosarcoma: a network meta-analysis [J]. J Orthop Surg Res. 2020;15(1):51.
- Buondonno I, Gazzano E, Jean SR, et al. Mitochondria-targeted doxorubicin: a new therapeutic strategy against doxorubicin-resistant osteosarcoma [J]. Mol Cancer Ther. 2016;15(11):2640–2652.
- Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer [J]. J Cancer. 2019;10(21):5015–5021.
- Kun-Peng Z, Xiao-Long M, Lei Z, et al. Screening circular RNA related to chemotherapeutic resistance in osteosarcoma by RNA sequencing [J]. Epigenomics. 2018;10(10):1327–1346.
- Ji Y, Liu J, Zhu W, et al. circ_0002060 enhances doxorubicin resistance in osteosarcoma by regulating the miR-198/ABCB1 axis [J]. Cancer Biother Radiopharm. 2020.DOI:10.1089/cbr.2020.4240.
- Guan H, Xu H, Chen J, et al. Circ_0001721 enhances doxorubicin resistance and promotes tumorigenesis in osteosarcoma through miR-758/TCF4 axis [J]. Cancer Cell Int. 2021;21(1):336.
- Hu Y, Gu J, Shen H, et al. Circular RNA LARP4 correlates with decreased enneking stage, better histological response, and prolonged survival profiles, and it elevates chemosensitivity to cisplatin and doxorubicin via sponging microRNA-424 in osteosarcoma [J]. J Clin Lab Anal. 2020;34(2):e23045.
- Bracken CP, Scott HS, Goodall GJ. A network-biology perspective of microRNA function and dysfunction in cancer [J]. Nat Rev Genet. 2016;17(12):719–732.
- Han F, Huang D, Huang X, et al. Exosomal microRNA-26b-5p down-regulates ATF2 to enhance radiosensitivity of lung adenocarcinoma cells [J]. J Cell Mol Med. 2020;24(14):7730–7742.
- Zhou A, Pan H, Sun D, et al. miR-26b-5p inhibits the proliferation, migration and invasion of human papillary thyroid cancer in a β-catenin-dependent manner [J]. Onco Targets Ther. 2020;13:1593–1603.
- Niu F, Kazimierska M, Nolte IM, et al. The mir-26b-5p/kpna2 axis is an important regulator of burkitt lymphoma cell growth [J]. Cancers (Basel). 2020;12(6):1464.
- Di Leva G, Garofalo M, Croce CM. MicroRNAs in cancer [J]. Annu Rev Pathol. 2014;9:287–314.
