ABSTRACT
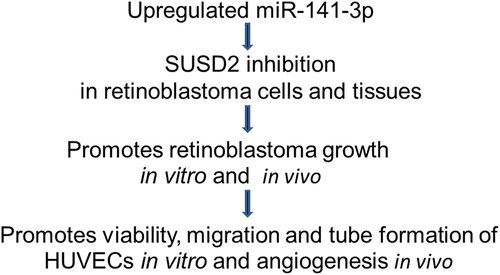
Retinoblastoma, often referred to as eye cancer, is a common primary pediatric intraocular malignancy. In this framework, micro ribose nucleic acids (miRNAs) play essential roles in retinoblastoma oncogenesis and development. However, the function and mechanism of the miR-141-3p/sushi domain-containing protein 2 (SUSD2) axis in retinoblastoma are unclear. To address these issues, miR-141-3p and SUSD2 expressions between the retinoblastoma patients and the normal control are identified by analyzing the Gene Expression Omnibus (GEO) datasets. Moreover, bioinformatics analysis, a dual-luciferase reporter assay, functional loss, and gain together with rescue experiments are employed to explore the biological function and molecular mechanisms of the miR-141-3p/SUSD2 axis in retinoblastoma oncogenesis and development. Our data showed that SUSD2 levels are considerably decreased in retinoblastoma cells and tissues. SUSD2 overexpression inhibited viability, promoting apoptosis of retinoblastoma cells and inhibiting tube formation of primary human umbilical vein endothelial cells (HUVECs) in vitro. The bioinformatics analysis and dual-luciferase reporter tests showed that SUSD2 is directly regulated by miR-141-3p. The miR-141-3p inhibition suppressed retinoblastoma growth and angiogenesis, while miR-141-3p overexpression increased retinoblastoma growth and angiogenesis, which is partially reversed when SUSD2 is over-expressed both in vivo and in vitro. In conclusion, SUSD2 is a tumor-suppressor in retinoblastoma. miR-141-3p/SUSD2 axis played an essential role in regulating angiogenesis and retinoblastoma progression, serving as a new biomarker for management of retinoblastoma.
Graphical abstract
KEYWORDS:
1. Background
Retinoblastoma is the most common pediatric intraocular malignancy. In this context, several comprehensive therapeutic strategies have been in practice, including chemoreduction, enucleation, local treatments, such as freezing-, laser-, and radiation-therapies, and systemic chemotherapy. Despite applying various combinations of treatment strategies, the clinical management of retinoblastoma is still unsatisfying and highly challenging due to the poor prognosis of patients, especially in developing countries [Citation1,Citation2]. Therefore, it is required to discover novel therapeutic targets and diagnostic biomarkers toward exploring molecular mechanisms underlying retinoblastoma oncogenesis and progression. Furthermore, angiogenesis plays a critical role in the oncogenesis and metastasis of various cancers, including retinoblastoma. Moreover, it is evident that angiogenesis is one of the most central factors of malignant biological behaviors [Citation1,Citation3].
UHRF1 (ubiquitin-like with PHD and RING finger domains 1), a protein-coding gene, is often significantly increased in various cancer cells and positively associated with aggressive phenotypes, such as the increased proliferation ability and decreased apoptosis rates. Inhibition of UHRF1 expression improves the therapeutic efficacy of histone deacetylase inhibitors in retinoblastoma [Citation4,Citation5], indicating the importance of UHRF1 in retinoblastoma oncogenesis and progression. After identification of the differentially expressed messenger ribose nucleic acids (DEMs) in the Gene Expression Omnibus (GEO) dataset GSE135424, between the UHRF1 control-knockdown (YT) and UHRF1-knockdown (YS) Y79 retinoblastoma cells based on |logFC|≥ 2 and P-value < 0.05, we observed that UHRF1 knockout in Y79 retinoblastoma cells significantly upregulated the expression of sushi domain-containing protein 2 (SUSD2) gene. However, the function and mechanism of SUSD2 in retinoblastoma oncogenesis and progression have yet remained to be reported.
Previous reports identified that SUSD2 could serve as a tumor suppressor in various cancers. Moreover, it was evident that the SUSD2 could be negatively correlated to the metastasis and positively correlated to the survival of high-grade serous ovarian cancer [Citation6]. Moreover, the reduced SUSD2 expression could be associated with shortened survival time and an independent prognostic factor for lung adenocarcinoma patients [Citation7]. In addition, the decreased SUSD2 expression acted as a tumor suppressor in lung cancer and RCC [Citation8]. Accordingly, we speculated that SUSD2 might function as a tumor suppressor in retinoblastoma.
MicroRNAs (miRNAs), a type of short non-coding RNAs, usually control target gene expression via binding with the 3ʹ-untranslated region (3ʹ-UTR), thereby contributing to the regulation of proliferation, differentiation, and apoptosis of cells [Citation9]. In recent times, several reports showed that miRNAs were significant in the development, progression, and treatment of malignancies, such as retinoblastoma [Citation10,Citation11]. Based on the TargetScan bioinformatics assay, miR-141-3p has been evidenced to have a prospective binding site with 3ʹ-UTR of SUSD2. Nevertheless, the function and mechanism of miR-141-3p in retinoblastoma oncogenesis and progression remain unclear. By identifying the overlapping differentially expressed miRNAs in the retinoblastoma versus the normal tissues (GSE7072) or serum (GSE41321), miR-141-3p is identified to be considerably upregulated in retinoblastoma patients. Therefore, we further speculated that miR-141-3p might promote retinoblastoma development by targeting SUSD2.
To address the above hypothesis, we explored the potential function and mechanism of the miR-141-3p/SUSD2 axis in retinoblastoma development both in vitro and in vivo, as well as the involvement of angiogenesis. After analyzing miR-141-3p and SUSD2 expressions between the retinoblastoma patients and the normal control from the GEO datasets, we speculated that miR-141-3p might promote retinoblastoma development by targeting SUSD2. To explore these aspects, bioinformatics analysis, a dual-luciferase reporter assay, functional loss, and gain together with rescue experiments were used, correlating to retinoblastoma oncogenesis and development.
2. Experimental section
2.1. Microarray data information and data processing of differentially expressed RNAs
GEO, a well-known open database of gene profile/microarray/high throughput sequencing, was employed to extract the gene expression profile of GSE135424 (GPL16791Illumina HiSeq 2500, Homo sapiens) between the UHRF1 control-knockdown (YT) and UHRF1-knockdown (YS) Y79 retinoblastoma cell line. Further, the differentially expressed mRNAs (DEMs) between UHRF1 control-knockdown (YT) and UHRF1-knockdown (YS) Y79 retinoblastoma cells were identified based on P-value < 0.05 and |logFC|≥ 2 and visualized by Heat map and Volcano plots. The gene ontology (GO) and KEGG pathway enrichment of DEMs were further evaluated using the online DAVID tool.
The miRNA expression profiles of both GSE7072 (GPL4879 Human miRNA 2k custom array) between the retinoblastoma and normal retina tissues and GSE41321 (GPL14767, Agilent-021827 Human miRNA Microarray G4470C, Feature Number version) between the retinoblastoma and non-retinoblastoma serum were extracted. Differentially expressed miRNAs between retinoblastoma and non-retinoblastoma patients were identified based on |logFC|≥ 2 and P-value < 0.05, and in GSE7072 and GSE41321 datasets. Then, the overlapping differentially expressed miRNAs in GSE7072 and GSE41321 datasets were identified by the online Venn software. The differentially expressed miRNA with log FC < 0 was referred to as a down-regulated miRNA, while the differentially expressed miRNA with log FC > 0 was indicated as an up-regulated miRNA.
2.2. Patient specimens and informed consents
The current study was approved by the Ethical Committee on Scientific Research of Shanghai Eighth People’s Hospital. Retinoblastoma and self-matched paracancerous tissues were collected from retinoblastoma patients during surgery treatment in our hospital and approved with written informed consents for the use in this study.
2.3. Reagents
Endothelial Cell Growth Supplement (ECGS) and Matrigel were procured from BD technologies (New Jersey, USA). Fetal bovine serum (FBS), DMEM, penicillin, and streptomycin were obtained from HyClone Laboratories Inc. (Logan, USA). Lipofectamine®3000, pmirGLO vector, pcDNA3.1 vector, and Trizol reagent were purchased from Invitrogen (Waltham, USA). The cell counting kit (CCK)-8 test kit was acquired from Dojindo Corp. Ltd. (Kyushu, Japan). Dual-Luciferase Reporter assay system was obtained from Promega Corp. Ltd. (Madison, USA). The protein extraction kit was acquired from Beyotime Technologies (Shanghai, China). PVDF membranes from Millipore (Billerica, US), electrochemical luminescence (ECL) kit from Tanon (Shanghai, China), antibodies against SUSD2, ACTIN, VEGF and CD31 from Cell Signaling Technology (Danvers, US), and Annexin V‑propidium iodide (PI) dual-staining apoptosis test kit from Nanjing Keygen Biotech Co., Ltd., (Nanjing, China) were obtained.
2.4. Cells, culture conditions, and transfection
Human umbilical vein endothelial cells (HUVECs) were purchased from PromoCell GmbH (Heidelberg, Germany) and cultured in fresh Ham’s Kaighn’s Modification F12 (F12K) supplemented with 10% FBS, 2 mM L-Glutamine, 0.05 mg/ml Endothelial Cell Growth Supplement (ECGS), and 0.1 mg/ml Heparin sodium salt from the porcine intestinal mucosa. Human retinoblastoma cells (Y79 and WERI-RB1) and normal human retinal pigment epithelial cells (ARPE-19) were obtained from the Cell Center of the Chinese Academy of Sciences (Shanghai, China) and cultured in DMEM containing 10% FBS, 100 μg/mL streptomycin, and 100 U/mL penicillin in 5% CO2 cell culture incubator at 37°C.
miR-141-3p inhibitors, inhibitor negative control (NC inhibitor), miR-141-3p mimics, and miR-141-3p negative control (NC mimics) were acquired from GenePharma Co., Ltd. (Shanghai, China). SUSD2 3ʹ-UTR sequence covering mutant (mut) or wild-type (WT) binding sites of miR-141-3p were respectively cloned into dual-luciferase reporter plasmid (pmirGLO). Lipofectamine 3000 was used for cell transfection in a serum-free medium. Full-length SUSD2 cDNA was cloned into the pcDNA3.1 vector. Empty (NC) and SUSD2 containing pcDNA3.1 plasmid (OE-SUSD2) were respectively transfected into retinoblastoma cells using lipofectamine®3000, and stably transfected cells were screened with 2 mg/ml of G418 [Citation12,Citation13].
2.5. Tube form ation assay
Initially, HUVECs at a density of 2 × 104 in 100 µl of retinoblastoma cell-conditioned medium (supernatant) were plated in each well of a 96-well cell culture plate containing 50 µl Matrigel and incubated at 37°C for 4 h with 5% CO2. Further, the tube formation images were acquired under an inverted microscope, and the branch points in each frame were enumerated.
2.6. Dual-luciferase reporter evaluation
Based on the TargetScan bioinformatics assay, it was evident that the miR-141-3p showed a potential binding site with 3ʹ-UTR of SUSD2. The pmirGLO vector was used to confirm the direct binding between miR-141-3p with 3ʹ-UTR of SUSD2. Constructs of mutant (mut) reporter of pmirGLO/SUSD2 3ʹ-UTR-mut (Luci-SUSD2-mut) and wild-type reporter of pmirGLO/SUSD2 3ʹ-UTR (Luci-SUSD2-WT) were co-transfected with miR-141-3p mimics (miR-141-3p) or miR-141-3p negative control (NC) in Y79 cell line. Finally, the luciferase activity was determined with a dual-luciferase reporter assay system after 48 h of transfection, following the manufacturer’s instructions [Citation14].
2.7. qRT-PCR assay
Initially, the Trizol reagent was applied for total RNA extraction, followed by cDNA reverse transcription for mRNA with PrimeScrip™ RT Master Mix and for miRNAs with PrimeScript miRNA cDNA Synthesis Kit. Further, the primers provided by Sangon Co., Ltd. (Shanghai, China) were employed, and the relative RNA expression was determined by the 2−ΔΔCt method [Citation12]. The primers used are listed in .
Table 1. Primers used in this study.
Table 2. The up-regulated miRNAs in serum of retinoblastoma patients.
Table 3. The down-regulated miRNAs in serum of retinoblastoma patients.
2.8. Cell viability
Initially, cells at a density of 2 × 104 were seeded in each well of a 96-well cell culture plate. At the corresponding transfection time period, the working CCK-8 solution (10 μL/well) was added into cells. After 2 h incubation, the absorbance (OD) value of each well was acquired at 450 nm wavelength with a microplate reader [Citation12]. Further, the viability curve was built according to optional density (OD) values.
2.9. Colony formation assay
Cells to be tested were seeded in the 6-well plates (1000/well) and cultured in complete medium for 7–10 days. Colonies were fixed with 10% formalin and stained with crystal violet. [Citation12] Finally, the resultant colonies were observed.
2.10. Transwell migration assay
Cells to be tested were seeded at a density of 5 × 104/well in a 200 μl serum-free medium in the insert of the 12-well transwell plates with 300 μl of 10% FBS in the lower chamber. After 24 h, the migrated cells were fixed with 4% paraformaldehyde stained with 1% crystal violet solution for 10 min and snapped with a microscope [Citation12].
2.11. Cell apoptosis assay
Cells to be tested were analyzed by using the Annexin V/PI double-staining apoptosis test kit. Briefly, 5 μl of reagent was added into the cells, mixed, and incubated at room temperature for 15 min in the dark. After being mixed with 400 μl of 1× binding buffer, apoptosis of the cells was analyzed with flow cytometry (CytoFLEX flow cytometer; CytExpert 2.1 Beckman Coulter Inc. Brea, USA) [Citation12].
2.12. Western blotting
Total protein was isolated with a protein extraction kit, followed by measuring the protein concentration with the BCA method and separating 20 μg with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). After being transferred onto PVDF membrane, blocked in 5% skimmed milk for 1 h at RT, cultured with primary antibody at 4°C overnight, washed with TBST, cultured with the second antibody for 1 h at RT, the protein bands were developed with ECL reagent [Citation12].
2.13. Establishment and treatment of xenografts
4-week-old BALB/c-nu mice were bought from Shanghai SLAC Laboratory Animal Co., Ltd (Shanghai, China). Xenograft mouse models bearing retinoblastoma were generated by subcutaneous injection of 100 mL Matrigel with 1 × 107 Y79 retinoblastoma cells transfected with miR-141-3p inhibitor, NC inhibitor, miR-141-3p mimic, NC mimic, or miR-141-3p mimic + OE-SUSD2, into BALB/c-nu mice (n = 4). The longest and the shortest diameters of tumors were determined with Vernier Calipers every 5 days, and the tumor volumes were calculated with the formulation: V = ½(longest diameter × shortest diameter)2. The entire tumors of sacrificed mice were removed entirely and weighed. All animal experiments were approved by the Institutional Animal Care and Use Committee of Huazhong University of Science and Technology and completed following the institutional rules [Citation15].
2.14. Immunohistochemical (IHC) assay
After deparaffinization, antigen retrieval, and nonspecific binding block, the sections were incubated with a primary antibody, a secondary antibody, and then the chromogen diaminobenzidine for immunohistochemical staining. Positive staining was observed under a light microscope [Citation15].
2.15. Statistical analysis
Statistical analyses were conducted with SPSS 22.0 (SPSS Inc., Chicago, US). The measured data were shown as mean ± standard deviation (S.D.). Notably, the differences between groups were compared with the Student’s two-tailed t-test at a defined level of statistical significance of P < 0.05 [Citation15].
3. Results
The aim of this study was to investigate the function and mechanism of the miR-141-3p/SUSD2 axis in retinoblastoma. After analyzing miR-141-3p and SUSD2 expressions between the retinoblastoma patients and the normal control from the GEO datasets, we speculated that miR-141-3p might promote retinoblastoma development by targeting SUSD2. To address the above hypothesis, bioinformatics analysis, dual-luciferase reporter assay, functional loss, and gain together with rescue experiments were applied to explore the biological function and molecular mechanisms of miR-141-3p/SUSD2 axis in retinoblastoma oncogenesis and development.
3.1. Downregulated SUSD2 expression is identified in retinoblastoma patient tissues and cells
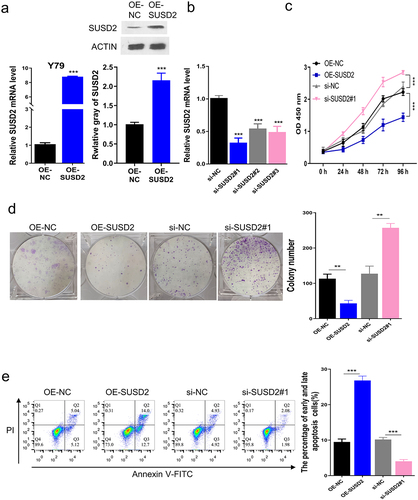
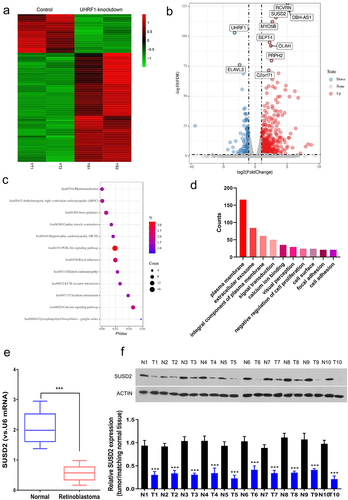
To explore the key factors involved in oncogenesis and progression of retinoblastoma, we first extracted the GSE135424 datasets from GEO and identified the differentially expressed mRNAs (DEMs) between the UHRF1 knockdown and the control retinoblastoma Y79 cells based on LogFC≥2 and P < 0.05. Among the identified 519 DEMs, 383 DEMs were significantly up-regulated while 135 were significantly down-regulated. The identified DEMs were shown as the cluster heat map () and the volcano plots (). The Gene Ontology (GO) function annotation () and the KEGG pathway enrichment () of the DEMs were performed, respectively. SUSD2 was found to be significantly up-regulated in UHRF1 silenced Y79 cells. Therefore, we hypothesized that SUSD2 might be a tumor suppressor since UHRF1 has been reported to be overexpressed in different human cancers, including retinoblastoma [Citation5]. Furthermore, the SUSD2 expression profile in retinoblastoma patient tissues was investigated, confirming significant downregulation of SUSD2 expression at mRNA () and protein () levels in retinoblastoma patient tissues.
Figure 1. SUSD2 is downregulated in retinoblastoma tissues and cells. Identification of differentially expressed mRNAs (DEMs) between the UHRF1 knockdown and the control retinoblastoma Y79 cells (GSE135424) with LogFC≥2, P < 0.05. (a) Heat map of DEMs. Green, low expression; Red, high expression. (b) Volcano plot indicated DEM distribution. (c) KEGG pathway enrichment analysis of DEMs. (d) Gene ontology (GO) enrichment analysis of DEMs; FDR = false discovery rate; mRNAs = messenger RNAs. SUSD2 expression was dramatically down-regulated in retinoblastoma (t) than in adjacent normal (n) tissues from retinoblastoma patients detected by (e) qRT-PCR (n = 25) and (f) Western blotting (n = 10). ***P < 0.001.
3.2. SUSD2 promotes apoptosis and inhibits the proliferation of retinoblastoma cells
To determine the function of SUSD2 in retinoblastoma oncogenesis, Y79 cells were transfected with pcDNA3.1 vector containing SUSD2 sequence to overexpress SUSD2, in which the empty pcDNA3.1 vector was served as the negative control (NC). The qRT-PCR and Western blotting analyses revealed that SUSD2 was successfully overexpressed in Y79 cells after transfection of SUSD2 containing pcDNA3.1 vector versus the NC (). Meanwhile, the qRT-PCR analysis showed that SUSD2 was successfully silenced in Y79 cells after transfection of si-SUSD2 vs. the NC (). The aggressive retinoblastoma phenotypes in Y79 cells were detected after SUSD2 was overexpressed. Moreover, the viabilities (0–96 h) of Y79 cells were detected with the CCK-8 assay, indicating that the time-dependent SUSD2 overexpression inhibited the cell viability compared with the OE-NC. In contrast, SUSD2 silencing time-dependently promoted cell viability compared with the si-NC (). Moreover, the colony proliferation ability was determined by colony formation assay, demonstrating that SUSD2 overexpression inhibited the colony proliferation ability compared with the OE-NC, while SUSD2 silence promoted the colony proliferation ability compared with the si-NC (). Finally, the cell apoptosis rate was determined by flow cytometric assay, which showed a significant increase of apoptosis after SUSD2 overexpression while a decrease of apoptosis after SUSD2 silence in Y79 cells ().
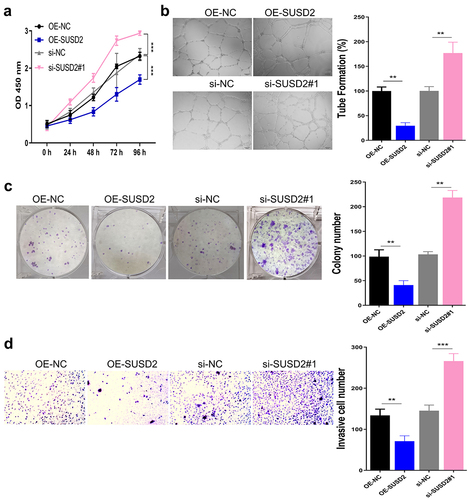
3.3. SUSD2 overexpression promotes blood vessel formation in vitro
Since solid cancer progression and distant metastasis count on the formation of cancer-related blood vessels to provide required oxygen and nutrients, more specifically, angiogenesis is essential for cancer growth, invasion, and metastasis [Citation16]. Therefore, we further examined the viability, the tube formation ability, the colony proliferation ability, and the migration ability of HUVECs after culture with a conditioned medium of Y79 cells. It was observed from the experimental results that the viability (), the tube formation ability (), the colony proliferation ability (), and the migration ability () of HUVECs were significantly inhibited with SUSD2 overexpression while significantly increased with SUSD2 silence in Y79 cells (P < 0.001)
3.4. SUSD2 acts as a specific target of miR-141-3p
Since the miRNAs play a major role in oncogenesis by regulating the cancer-related genes, we analyzed the DEMs between retinoblastoma tissues and normal retinal tissues from the GEO dataset (GSE7072). Simultaneously, we also analyzed the serum DEMs between the retinoblastoma patients and the normal control from the GEO dataset (GSE41321). The overlapping DEMs between GSE7072 and GSE41321 datasets were further screened by Venn online tool (http://bioinformatics.psb.ugent.be/webtools/Venn), in which 6 significantly up-regulated miRNAs (), ) and 7 significantly down-regulated (), ) genes were identified. To further explore the potential miRNAs harboring complementary binding sequences with SUSD2-3ʹUTR, bioinformatics analysis was performed using the online tool TargetScan, to regulate the SUSD2 in retinoblastoma oncogenesis. Moreover, miR-141-3p was identified to harbor specific binding sites with SUSD2-3ʹUTR (). Thus, we assumed that miR-141-3p might directly interact and regulate SUSD2.
Figure 4. miR-141-3p specifically targets SUSD2. The overlapping DEMs in retinoblastoma versus normal tissues (GSE7072) or serum (GSE41321). Venn maps to identify (a) the up-regulated overlapping DEMs. (b) The down-regulated overlapping DEMs with LogFC≥2, P < 0.05, in GSE7072 and GSE41321 datasets. (c) Illustration of potential miR-141-3p binding sites in SUSD2 3ʹUTR predicted by TargetScan database and the mutation sites used for specific binding assay. (d) Dual-luciferase reporter gene activity in Y79 cells transfected with wild type (WT) or mutated (Mut) reporter of SUSD2 3ʹUTR in the presence of miR-141-3p mimics (miR-141-3p) or negative control (NC). (e) miR-141-3p expression in retinoblastoma tissues was significantly higher compared with paracancerous normal tissues (n = 25). (f) miR-141-3p expression in retinoblastoma (Y79 and WERI-RB1) cells was significantly higher vs. normal human retinal pigment epithelial cells (ARPE-19) detected by qRT-PCR. SUSD2 expression detected by qRT-PCR (g) and Western blotting (h) in Y79 cells after miR-141-3p overexpression with miR-141-3p mimics or miR-141-3p science with miR-141-3p inhibitor. **P < 0.01 and,***P < 0.001.
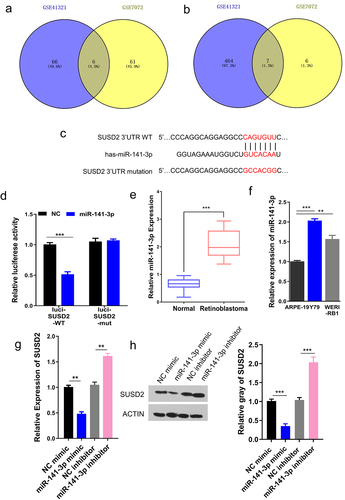
Moreover, dual-luciferase reporter activity was employed to validate specific binding between miR-141-3p with SUSD2 3ʹ-UTR by sub-cloning mutated or WT SUSD2 3ʹ-UTR reporter gene into dual-luciferase reporter vector (pmirGLO). Notably, the luciferase activity of the pmirGLO-SUSD2 3ʹ-UTR-WT reporter gene in Y79 cells was considerably reduced after transfecting miR-141-3p mimics, remaining unchanged when predicted miR-141-3p binding sites in SUSD2 3ʹ-UTR were mutated (). To further identify the involved mechanism, miR-141-3p expression in retinoblastoma was investigated, showing that miR-141-3p expression in retinoblastoma was significantly upregulated vs. paracancerous normal tissues (). Meanwhile, the qRT-PCR analysis showed that the miR-141-3p expression was found to be significantly upregulated in retinoblastoma Y79 and WERI-RB1 cells than in normal human retinal pigment epithelial ARPE-19 cells (). Moreover, SUSD2 expression was identified by qRT-PCR and Western blotting in Y79 cells after miR-141-3p overexpression with miR-141-3p mimics or silence with miR-141-3p inhibitor, revealing that the mRNA () and the protein () expressions of SUSD2 were reduced with miR-141-3p mimic, while increased with miR-141-3p inhibitor. Together, these experimental results suggested the specific binding between miR224-3p with SUSD2-3ʹUTR down-regulated SUSD2 expression in retinoblastoma cells.
3.5. miR-141-3p promotes proliferation and suppresses apoptosis of retinoblastoma cells, as well as promotes angiogenesis of endothelial cells by targeting SUSD2 in vitro
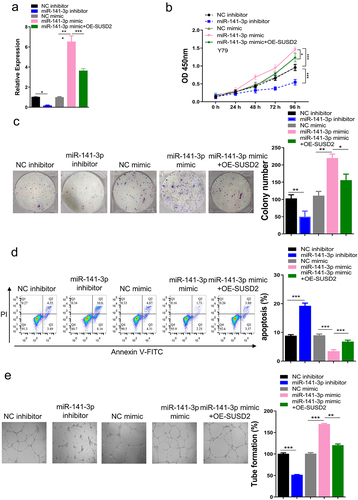
To further explore the function and mechanism of the miR-141-3p/SUSD2 axis in retinoblastoma oncogenesis, we determined the functional gain and loss, as well as the rescue experiments in Y79 cells, followed by analyzing the viability and apoptosis of Y79 cells, tube formation ability of endothelial cells after being cultured with Y79 cell-conditioned medium. The experimental results showed a significantly inhibited angiogenesis with SUSD2 overexpression in Y79 cells. As depicted in , miR-141-3p expression was considerably decreased after transfection of miR-141-3p inhibitor compared with transfection of NC inhibitor. Moreover, miR-141-3p expression was considerably upregulated after transfection of miR-141-3p mimics vs. transfection of NC mimics. Inhibition of miR-141-3p by employing the miR-141-3p inhibitor repressed the OD values of Y79 cells (0–96 h). To this end, the overexpression of miR-141-3p by miR-141-3p mimics increased OD values of Y79 cells (0–96 h). Nevertheless, the increased OD values by miR-141-3p mimics were partly reversed when SUSD2 was overexpressed (). Similarly, inhibition of miR-141-3p also presented the reduced colony proliferation ability of Y79 cells, while overexpression of miR-141-3p by miR-141-3p mimics increased colony proliferation ability of Y79 cells. In contrast, the increased colony proliferation ability by miR-141-3p mimics was partly reversed when SUSD2 was overexpressed (). In addition, the miR-141-3p inhibition by miR-141-3p inhibitor stimulated Y79 cell apoptosis, while miR-141-3p overexpression by miR-141-3p mimics decreased apoptosis of Y79 cells, and the decreased apoptosis by miR-141-3p mimics was partly reversed when SUSD2 was over-expressed (). Tube formation ability of HUVECs was repressed after being cultured with a conditioned medium of miR-141-3p inhibitor transfected Y79 cells. Moreover, the tube formation ability of HUVECs was increased after being cultured with a conditioned medium of miR-141-3p mimics transfected Y79 cells, and the increased tube formation ability by miR-141-3p mimics was partly reversed when SUSD2 was over-expressed (OE-SUSD2) ().
Figure 5. miR-141-3p promotes proliferation and suppresses apoptosis of retinoblastoma cells, as well as promotes angiogenesis of endothelial cells by targeting SUSD2 in vitro. (a) After successful knockdown of miR-141-3p with miR-141-3p inhibitor, overexpression of miR-141-3p using miR-141-3p mimic with or without SUSD2 overexpression using OE-SUSD2 in Y79 cells, (b) viability, (c) colony proliferation, and (d) apoptosis of Y79 cells. (e) Tube formation ability of HUVECs after culture with conditioned medium of Y79 cells. **P < 0.01 and ***P < 0.001.
3.6. miR-141-3p stimulates retinoblastoma growth and angiogenesis by targeting SUSD2 in vivo
To further confirm the function and mechanism of miR-141-3p/SUSD2 axis in retinoblastoma oncogenesis in vivo, xenograft mouse models bearing retinoblastoma were generated by subcutaneous injection of Y79 cells transfected with miR-141-3p inhibitor, NC inhibitor, miR-141-3p mimic, NC mimic, or miR-141-3p mimic + OE-SUSD2. Similar to the results in vitro, the mass (), volume (), and the weights () of retinoblastoma tumors were significantly inhibited by miR-141-3p inhibitor, increased by miR-141-3p mimics. In contrast, the increased mass (), volume (), and the weights () of tumors by miR-141-3p mimics were partly reversed when SUSD2 was over-expressed (OE-SUSD2), indicating that the miR-141-3p stimulated retinoblastoma growth in vivo via targeting SUSD2.
Figure 6. miR-141-3p promotes retinoblastoma growth and angiogenesis by targeting SUSD2 in vivo (n = 4). (a) Mass, (b) volume. (c) weights of the xenograft tumors. Key factor expressions by immunohistochemistry (d), Western blot assay (e), and qRT-PCR (f) analysis. **P < 0.01 and ***P < 0.001.
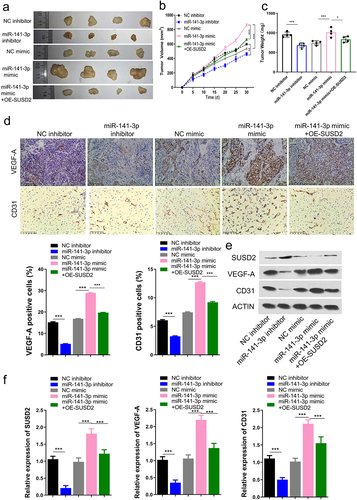
Moreover, immunohistochemistry, Western blotting, and qRT-PCR assays confirmed that the expressions of VEGF and CD31, the two key factors of blood vessel formation, were significantly inhibited by miR-141-3p inhibitor and increased by miR-141-3p mimics. At the same time, the increased expressions of VEGF and CD31 by miR-141-3p mimics were partly reversed when SUSD2 was over-expressed (OE-SUSD2) (). The SUSD2 protein expression was also determined by Western blotting assay, indicating that SUSD2 protein expression was significantly upregulated by miR-141-3p inhibitor, downregulated by miR-141-3p mimics, and reversed by SUSD2 over-expression (). These findings indicated that the miR-141-3p stimulated blood vessel formation in vivo via targeting SUSD2. SUSD2 expression was identified by qRT-PCR and Western blotting in Y79 cells after miR-141-3p overexpression with miR-141-3p mimics, revealing that the mRNA and protein expressions of SUSD2 were reduced with miR-141-3p mimic.
4. Discussion
Indeed, the prognosis of retinoblastoma is still unsatisfying even under the comprehensive clinical management strategies. Therefore, understanding the fundamental molecular mechanisms in retinoblastoma oncogenesis and progression is essential. Recent shreds of evidence reported that miRNAs were essential in retinoblastoma development toward identifying new diagnostic, therapeutic, and prognostic biomarkers in the clinical management of retinoblastoma patients [Citation10,Citation11]. In this context, UHRF1 has greatly attracted the attention of researchers in the field of cancer research, not only as a vital epigenetic regulator for histone modifications and DNA methylation but also as a critical regulator for cancer cell survival [Citation4]. Notably, the UHRF1 expression is upregulated in retinoblastoma and contributes to retinoblastoma development.
To further explore the valuable biomarkers in retinoblastoma development, bioinformatical analysis was performed in the current study on the basis of the GEO dataset GSE135424. SUSD2 was found to be significantly upregulated when UHRF1 was silenced in Y79 retinoblastoma cells, indicating the importance of SUSD2 in retinoblastoma progression. This hypothesis was further confirmed by the following findings. SUSD2 was first discovered to be significantly downregulated in retinoblastoma cells and tissues, indicating the tumor suppressor role of SUSD2.
In recent years, the regulatory effects of miRNAs in key factors associated with cancer oncogenesis and progression have attracted increased attention. Thus, we subsequently performed the bioinformatics assay based on the two GEO datasets (GSE7072 and GSE41321), followed by Venn software analysis, in which 13 commonly changed miRNAs were identified with 6 upregulated and 7 downregulated in retinoblastoma patients. Moreover, bioinformatical analysis by TargetScan showed that miR-141-3p, one of the 6 significantly upregulated miRNAs, had the potential binding sites in the 3ʹ-UTR sequences of SUSD2. These findings were further verified by dual-luciferase activity. Furthermore, the expression of SUSD2 was found to be negatively regulated by miR-141-3p expression in retinoblastoma cells. Moreover, SUSD2 overexpression reversed miR-141-3p mimics promoted aggressive phenotypes of retinoblastoma cells and retinoblastoma-related angiogenesis. Together, the current work provided new shreds of evidence supporting the importance of the miR-141-3p/SUSD2 axis in retinoblastoma development
5. Conclusion
In summary, our current study explored the discovery of a tumor suppressor, SUSD2, in retinoblastoma, which was regulated by miR-141-3p. Notably, the miR-141-3p/SUSD2 axis regulated retinoblastoma growth, as well as the angiogenesis of HUVECs both in vitro and in vivo. Therefore, targeting miR-141-3p/SUSD2 would provide novel biomarkers for early diagnosis, therapeutics, and prognosis in retinoblastoma management.
Availability of data and material
The data during the current study are available from the corresponding author on reasonable request.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Chen S, Chen X, Luo Q, et al. Retinoblastoma cell-derived exosomes promote angiogenesis of human vesicle endothelial cells through microRNA-92a-3p. Cell Death Dis. 2021;12(7):695.
- Xu Y, Fu Z, Gao X, et al. Long non-coding RNA XIST promotes retinoblastoma cell proliferation, migration, and invasion by modulating microRNA-191-5p/brain derived neurotrophic factor. Bioengineered. 2021;12(1):1587–1598.
- Choi SH, Kim AR, Nam JK, et al. Tumour-vasculature development via endothelial-to-mesenchymal transition after radiotherapy controls CD44v6(+) cancer cell and macrophage polarization. Nat Commun. 2018;9(1):5108.
- He H, Lee C, Kim JK. UHRF1 depletion sensitizes retinoblastoma cells to chemotherapeutic drugs via downregulation of XRCC4. Cell Death Dis. 2018;9(2):164.
- Kim JK, Kan G, Mao Y, et al. UHRF1 downmodulation enhances antitumor effects of histone deacetylase inhibitors in retinoblastoma by augmenting oxidative stress-mediated apoptosis. Mol Oncol. 2020;14(2):329–346.
- Sheets JN, Patrick ME, Egland KA. SUSD2 expression correlates with decreased metastasis and increased survival in a high-grade serous ovarian cancer xenograft murine model. Oncotarget. 2020;11(24):2290–2301.
- Guo W, Shao F, Sun S, et al. Loss of SUSD2 expression correlates with poor prognosis in patients with surgically resected lung adenocarcinoma. J Cancer. 2020;11(7):1648–1656.
- Cheng Y, Wang X, Wang P, et al. SUSD2 is frequently downregulated and functions as a tumor suppressor in RCC and lung cancer. Tumour Biol. 2016;37(7):9919–9930.
- Cai Y, Yu X, Hu S, et al. A brief review on the mechanisms of miRNA regulation. Genomics Proteomics Bioinformatics. 2009;7(4):147–154.
- Wang L, Wang C, Wu T, et al. Long non-coding RNA TP73-AS1 promotes TFAP2B-mediated proliferation, metastasis and invasion in retinoblastoma via decoying of miRNA-874-3p. J Cell Commun Signal. 2020;14(2):193–205.
- Castro-Magdonel BE, Orjuela M, Camacho J, et al. miRNome landscape analysis reveals a 30 miRNA core in retinoblastoma. BMC Cancer. 2017;17(1):458.
- Hu S, Chang J, Ruan H, et al. Cantharidin inhibits osteosarcoma proliferation and metastasis by directly targeting miR-214-3p/DKK3 axis to inactivate beta-catenin nuclear translocation and LEF1 translation. Int J Biol Sci. 2021;17(10):2504–2522.
- Zheng S, Guo Y, Dai L, et al. Long intergenic noncoding RNA01134 accelerates hepatocellular carcinoma progression by sponging microRNA-4784 and downregulating structure specific recognition protein 1. Bioengineered. 2020;11(1):1016–1026.
- Gu Q, Hou W, Shi L, et al. Circular RNA ZNF609 functions as a competing endogenous RNA in regulating E2F transcription factor 6 through competitively binding to microRNA-197-3p to promote the progression of cervical cancer progression. Bioengineered. 2021;12(1):927–936.
- Zhu Y, Ke KB, Xia ZK, et al. Discovery of vanoxerine dihydrochloride as a CDK2/4/6 triple-inhibitor for the treatment of human hepatocellular carcinoma. Mol Med. 2021;27(1):15.
- Viallard C, Larrivee B. Tumor angiogenesis and vascular normalization: alternative therapeutic targets. Angiogenesis. 2017;20(4):409–426.