ABSTRACT
This study aimed to investigate the roles of the lysine (K)-specific demethylase 5C (KDM5C)–bone morphogenetic protein-7 (BMP-7) signaling pathway in the pathogenesis of severe preeclampsia (sPE). A total of 180 pregnant patients were enrolled in the study and classified into three groups: an early-onset sPE group (EOsPE) (n = 60), a late-onset sPE group (LOsPE) (n = 60), and a control group (normal pregnancy; n = 60). The messenger RNA (mRNA) and protein expression levels of bone morphogenetic protein receptor II (BMPRII), BMP-7, and KDM5C were detected in placenta samples from the two sPE groups, and their sites were evaluated using immunohistochemistry (IHC). The sPE groups showed an increased KDM5C mRNA expression, and the EOsPE group showed a decreased BMP-7 and BMPRII mRNA expression compared with the LOsPE group. However, contradictory results were discovered in terms of protein expression. Immunostaining of KDM5C, BMP-7, and BMPRII was observed in villous trophoblast and extravillous trophoblast cells. Compared with the control group, the staining intensity of KDM5C in the placental tissue trophoblast cell nucleus and vascular endothelial cells of the sPE groups was weaker, while that of BMP-7 and BMPRII was stronger, and the staining intensity was more subjective in the LOsPE group. Consistent findings were obtained by IHC and Western blot analysis. KDM5C nuclear-cytoplasmic translocation may regulate sPE through BMP-7 and its receptors. The KDM5C–BMP-7 signaling pathway may also lead to less invasion and increased apoptosis of the trophoblast cells, which is involved in the pathogenesis of sPE.
Graphical Abstract
Introduction
Preeclampsia (PE) is a rare disorder experienced during pregnancy, affecting 3–5% of pregnancies [Citation1]. It has a high incidence of fetal and maternal mortality and morbidity [Citation2]. In general, it is classified by clinical symptoms into two stages: early-onset PE (before 34 weeks of gestation) and late-onset PE (at or after 34 weeks of gestation) [Citation3]. Its etiology and pathogenesis are not entirely understood, but many scholars believe it is caused by shallow placenta implantation, affecting parts of the placenta’s decidual vascular dysplasia, which leads to placental ischemia and hypoxia and instigates the blood clotting system. This subsequently causes vascular endothelial cell injury and finally results in high blood pressure and proteinuria, with many clinical manifestations [Citation4]. There is some resemblance in biological behavior between tumors and PE, including the lack of trophoblastic infiltration, which leads to placenta ischemia and hypoxia and, eventually, the occurrence of PE.
Belonging to the SMCY homolog family, lysine (K)-specific demethylase 5C (KDM5C) reverses, maintains, and plays a crucial role in the functional discrimination between core promoters and enhancers by maintaining the dynamic balance of histone 3 lysine 4 (H3K4) methylation states and reversing the tri- and di-methylation of H3K4 [Citation4]. The expression of KDM5C is high in various cancers, including hepatocellular carcinoma, kidney cancer, colon cancer, breast cancer, and prostate cancer, making it a possible oncogene. The proliferation of cancer cells can also be promoted by KDM5C, and a significant delay in the G1/S transition may be largely delayed by a knockdown of KDM5C [Citation5].
Belonging to the transforming growth factor beta (TGF-β) superfamily, bone morphogenetic proteins (BMPs) play a role in the growth stage and osteogenesis regulation. Recently, these proteins have been found to have important functions in tumor biology. BMP conducts the signal through bone morphogenetic protein receptors (BMPRs) on the cell surface (serine/threonine kinase receptors) [Citation6]. Signal transduction requires not only type-I but also type-II BMPRs. As type-I and type-II receptors, BMPRIA and BMPRIB preferentially bind ligands of the Dpp class (such as BMP2 and BMP4) and the 60A class (such as BMP-7), respectively [Citation7]. The complex of these two receptor types then regulates the target gene expression by phosphorylating and translocating Smad1, 5, and 8 proteins to the nucleus [Citation8]. BMP-7 has also been found to display anti-invasive properties because of its role in impeding the expression of integrin induced by TGF-β [Citation9].
The invasion and migration of trophoblasts to the maternal endometrium and, in some cases, the shallow myometrium, occurs during normal pregnancy; this process is similar to the infiltration of a malignant tumor into the surrounding tissues as well as to distant metastases. One possible explanation for the pathogenesis of PE is a failure in the remodeling of the maternal spiral arteries and the abnormal invasion of trophoblasts [Citation10]. High expression levels of KDM5C in invasive human hepatocellular carcinoma cells have also been reported. Such ectopic expression promotes cell migration and invasion as well as epithelial-mesenchymal transition through the inactivation of BMP-7 [Citation11].
It can be suggested that the abnormal expression of the KDM5C–BMP-7 signaling pathway in the placenta plays a role in the pathogenesis of PE. However, information on this in the context of human pregnancies is currently insufficient. Zhu et al. used this method to verify that the high mobility group box 1–receptor for advanced glycation endproducts signaling pathway is involved in the occurrence of PE [Citation11]. In the present study therefore, the expression patterns of KDM5C, BMP-7, and BMPRII in early-onset severe PE (EOsPE) placenta, late-onset sPE (LOsPE) placenta, and normal controls were examined by Western blot, immunohistochemistry (IHC), and quantitative real-time polymerase chain reaction (qRT-PCR).
Methods
Case selection
A total of 180 pregnant patients from the Third Affiliated Hospital of Zhengzhou University were enrolled in this study between September 2011 and April 2016. They were classified into three groups: an EOsPE group (n = 60), an LOsPE group (n = 60), and a control group (n = 60). SPE was diagnosed based on the guidelines of the International Society for the Study of Hypertension in Pregnancy [Citation12]. Patients that had multiple gestations, fetal malformations, polycystic ovary syndrome, renal disease, diabetes mellitus, or chronic hypertension were excluded.
The study was approved by the Partners Human Research Committee, and all patients provided informed consent and written approval for the storing of their information in the hospital database and its use for research purposes.
Sample collection
Within 15 minutes after delivery, placental biopsies were collected. To avoid sampling bias, five small individual biopsies were taken: one from the placental center and one from each quadrant. Any areas of necrosis, infarction, or calcification were excluded, and full-thickness placental blocks were included. Sterile filter paper was used to remove the blood on the tissue, after which 0.9% sodium chloride was used to wash one part of the sample, which was then fixed in 10% formalin to generate a tissue microarray. The other part was frozen immediately in liquid nitrogen and stored it at −80°C.
qRT-PCR
In accordance with the manufacturer’s instructions, TRIzol reagent (Invitrogen, Carlsbad, California, USA) was applied to the extraction of total RNA from the placental tissues of all three groups. Prior to agarose gel electrophoresis, nucleic acids were used for the evaluation of RNA integrity. The preparation of complementary DNA (cDNA) was completed using a ReverTraAce qRT-PCR kit (Toyobo, Shanghai, Japan). On an Applied Biosystems 7500 (ABI, Foster City, California, USA) with the SYBR Green Real-Time PCR Master Mix (Toyobo, Shanghai, Japan), qRT-PCR was performed to examine the expression levels of KDM5C, BMPRII, and BMP-7 in all three groups. lists the utilized primers that were designed by Sangon Biotech (Shanghai, China).
Table 1. Primers used for qRT-PCR quantifications
During cycling, a denaturation of 10 minutes was performed at 95°C, followed by 15 seconds of subsequent cycling (35 times) at 95°C and 1 minute at 60°C. During melt curve analysis, the temperature was increased from 60°C to 95°C at an interval of 0.2°C to ensure the desired product was amplified. This procedure was repeated three times. The expression of glyceraldehyde 3-phosphate dehydrogenase was adopted for standardized results. The mRNA expression of all the target genes was measured using the ΔΔCt method.
Western blot analysis
A radioimmunoprecipitation assay buffer with a protease inhibitor cocktail (Solarbio, Beijing, China) and 1 mmol/L phenylmethylsulphonyl fluoride was used for the extraction of the total cellular protein from the snap-frozen tissues. A bicinchoninic acid assay (CWBIO, Beijing, China) was applied to the determination of the total protein concentration. Afterward, 10% sodium dodecyl sulfate sulphate polyacrylamide gel electrophoresis was used to separate samples with 100 μg of protein, followed by electrophoretic transfer to nitrocellulose membranes. A blocking buffer (5% skim milk, 0.1% Tween 20®, and 0.01 M tris-buffered saline) was then used to block the membranes at room temperature for 2 hours. Anti-KDM5C (1:100 dilution, ab34718, Abcam, USA), anti-BMP-7 (1:150 dilution, ab56023, Abcam, USA), and anti-BMPRII (1:150 dilution, ab78422, Abcam, USA) were used to incubate the membranes overnight at 4°C. β-actin (1:120,000 dilution, ab6276, Abcam, USA) was used to express all results. Secondary fluorescent antibodies (1:5000 dilution, Odyssey CIX, Lincoln, USA) were then used to incubate the membranes for 2 hours at room temperature, after which an infrared laser scanning imaging system (Odyssey CIX, Lincoln, USA) was used to measure the fluorescence intensity. Based on the ratio of the internal target protein, analysis of protein expression for all target genes was conducted.
IHC in the placental tissues
At room temperature, 10% buffered formalin was used to fix the tissues, which were then placed in paraffin. IHC analysis was performed on consecutive serial sections (4 mm thick) of each block. All tissue sections underwent microwave treatment for 20 minutes in 10 mmol/L sodium citrate buffer under pH 6.0 for antigen recapture after deparaffinization and hydration in distilled water. To block endogenous peroxide activity, 3% hydrogen peroxide was used to incubate these sections at 37°C for 15 minutes. The sections were then incubated at 4°C overnight with anti-KDM5C (ab34718, Abcam, USA), anti-BMP-7 (ab56023, Abcam, USA), and BMPRII (ab78422, Abcam, USA) at a dilution of 1:100. Negative control sections were incubated at 4°C overnight with a phosphate-buffered solution (PBS), after which a two-step IHC detection reagent (ZSGB-BIO, Beijing, China) was applied in a second round of incubation. Slides were immersed in a 3,3’-diaminobenzidine tetrahydrochloride substrate kit for the visualization of reaction products (ZSGB-BIO, Beijing, China), before being counterstained in hematoxylin for 5 minutes, dehydrated, cleared, and covered with a slip. Image-pro Express software and an inversion fluorescence microscope (OLYMPUS IX-71, Tokyo, Japan) were used for the assessment of the slides.
Immunostaining of KDM5C, BMP-7, and BMPRII in the placenta were graded on a semi-quantitative scale, with 3 denoting strong staining/a brownish-black color, 2 denoting moderate staining/a dark brown color, 1 denoting weak staining/a pale brown color, and 0 denoting no staining/no color. Two pathologists separately evaluated the intensity of each slide’s immunostaining.
Statistical analysis
Mean ± SD was used to express quantitative data. Under the normal distribution of the three groups’ data, a least significant difference t-test was performed for assessment of statistical differences. A nonparametric Kruskal–Wallis test was used. When the data from any two groups were normally distributed, a t-test was performed for assessment of statistical differences. If the data distribution of any group was skewed, a nonparametric Mann–Whitney U test was conducted. For the IHC of the tissue microarrays, a χ2 test was carried out to compare the three groups in terms of the intensity of staining in the placental tissues [Citation11]. Statistical significance was identified by probability level with a two-tailed P < 0.05 value. SPSS 19.0 software was used for all statistical analyses.
Results
Clinical data of the three groups
In order to understand the difference between the experimental groups and the control group, statistical analysis of the clinical data was conducted. No significant differences were identified between the three groups in weight gain during pregnancy, body mass index at delivery, and maternal age of the patients. However, patients in the two sPE groups had significantly higher systolic and diastolic blood pressure values than those in the control group, with those in the EOsPE group having higher values than those in the LOsPE group. Compared with the control group, the sPE groups showed a downward trend in number of children, the Apgar score of newborns, neonatal weight, and gestational age at delivery, although the values of the LOsPE group were higher than those of the EOsPE group (see ).
Table 2. Demographic characteristics for early-onset sPE, late-onset sPE and normal pregnancies
mRNA expressions of BMP-7, BMPRII, and KDM5C in the placenta
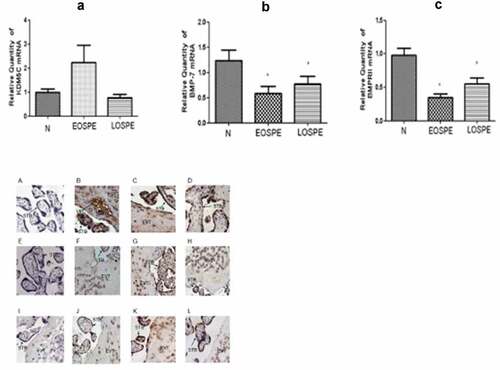
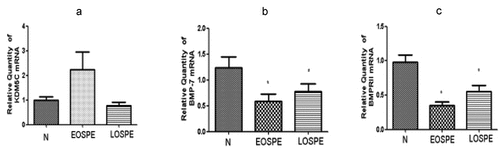
To investigate the mRNA changes of BMP-7, BMPRII, and KDM5C in the placental tissues of all three groups, qRT-PCR assays were performed. Compared with the control group, the mRNA expression of KDM5C was higher in the EOsPE group (P > 0.05). The mRNA expressions of BMP-7 and BMPRII were lower in both sPE groups than in the control group, with the EOsPE group displaying lower expression levels than the LOsPE group (P < 0.05; see ).
Figure 1. The quantitative real-time PCR of KDM5C, BMP-7, and BMPRII mRNA expressions in the placenta in the three groups.
Protein expressions of BMP-7, BMPRII, and KDM5C in the placenta
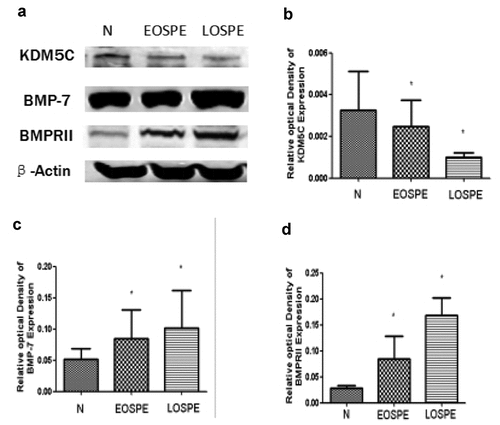
Western blot analysis was performed to investigate the changes in BMP-7, BMPRII, and KDM5C proteins in the placental tissues of all three groups, with β-actin as the internal control for each sample. Compared with the control group, the relative expression of KDM5C was induced in both sPE groups, with that of the EOsPE group having a smaller decrease than that of the LOsPE group (P < 0.05). Both sPE groups showed an increase in the expressions of BMP-7 and BMPRII, with a smaller increase in the EOsPE group than in the LOsPE group (P < 0.05; see ).
Figure 2. The protein levels of KDM5C, BMP-7, BMPRII, and β-actin in the placenta by Western blot.
Protein expression localization of BMP-7, BMPRII, and KDM5C in the placenta
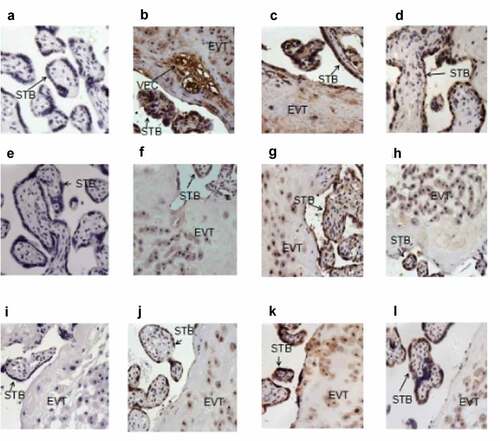
To understand the location and expression of BMP-7, BMPRII, and KDM5C in the placental tissues of all three groups, IHC was performed. shows the expression of nuclear KDM5C in most syncytia trophoblasts, extravillous cytotrophoblasts, and vascular endothelial cells, with a statistical difference being identified between the three groups in syncytia trophoblast KDM5C expressions. BMP-7 was mostly detected in the membrane of extravillous cytotrophoblasts and syncytia trophoblasts, as well as in the cytoplasm. Compared with the control group, the sPE groups had a higher immunostaining intensity of BMPRII, which is a receptor of BMP-7. Compared with the control group, the sPE groups had weaker staining intensities of nuclear KDM5C in the placental tissues (χ2 = 5.19; P < 0.05) and higher staining intensities of cytoplasmic BMP-7 and membrane BMPRII in the placental tissues (χ2 = 7.79, 12.4; P < 0.05). gives a summary of the results, categorized by the intensity of immunostaining.
Table 3. Immunostaining of nuclear KDM5C, cytoplasmic BMP-7 and membrane BMPRII in early-onset sPE, late-onset sPE and control group
Figure 3. KDM5C, BMP-7 and BMPRII staining in the placental tissue section.
Discussion
PE is currently one of the leading causes of high maternal and neonatal morbidity and mortality. It not only increases the risk of premature birth and intrauterine growth restriction but also significantly increases the risk of long-term cardiovascular disease for both the mother and the fetus. At present, the only effective treatment is childbirth, but this can lead to iatrogenic premature birth. EOsPE in particular presents clinicians with difficult choices and severe challenges due to the early stage of pregnancy at which it occurs, the immaturity of the fetus, the rapid progress of the disease, and the risk of more worrying outcomes for the mother and fetus. At present, the etiology and pathogenesis of PE are unclear, and there is a lack of effective diagnostic markers and therapeutic targets. Therefore, it is of great clinical significance to discover and study the pathogenic genes of PE.
Despite current knowledge on the etiopathology of PE being insufficient, its correlation with poor placentation and the involvement of endothelial dysfunction and excessive maternal inflammation has been well reported. Furthermore, higher placental impairment in EOsPE and chronic inflammatory conditions in LOsPE have been found [Citation13]. Some researchers have identified that KDM5C inhibits the expression of BMP-7 to promote hepatocellular carcinoma cell invasion [Citation11]. However, little knowledge has been acquired regarding KDM5C’s role and underlying mechanisms in severe PE. Therefore, the present study investigated the relationship between the KDM5C–BMP-7 signaling pathway and sPE.
In the present study, a decrease in KDM5C was found in EOsPE and LOsPE placental tissues, with a higher expression level in the latter. This suggests that reduced KDM5C results in shallow placenta implantation, i.e., the infiltration of trophoblast cells causes parts of the placenta decidual vascular dysplasia, leading to placental ischemia, hypoxia, and, eventually, to PE. As epigenetic enzymes, histone lysine demethylases (KDMs) are able to achieve the removal of not only activating but also repressive histone marks [Citation11]. KDM5A, B, C, and D constitute the KDM5 family of HDMs and trimethylated histone H3K9 [Citation14]. By promoting transcriptional activation, the proteins in this family play a crucial role in major processes like cellular proliferation and differentiation, germ cell development, stem cell renewal, and hormone response [Citation15]. Some researchers have also found that KDM5C impedes p53 expression to promote gastric cancer cell invasion and proliferation [Citation16].
Ji et al. found that BMP-7 is a target of KDM5C [Citation11], and it has been previously reported that BMP-7 impedes the TGF-β-induced expression of integrin β to prevent invasion [Citation9]. Based on a microarray analysis, Shon et al. found that EOsPE and LOsPE arere responsible for different expressions of fetal genes in placental tissue and that the TGF-β pathway may be involved in placental disorders [Citation9]. Lu et al. found that the downregulation of cathepsin C alleviates endothelial cell dysfunction by suppressing the p38 MAPK–NF-κB pathway in PE [Citation17]. The present study found that BMP-7 and BMPRII expression increased in the two sPE groups compared with the control group, and this increase was more apparent in the LOsPE group. Human and animal studies on the osteo-inductive properties of BMP-7 have also identified its potential role in facilitating osseous healing [Citation18]. High content of calcium in bone tissue and BMP-7 can promote bone calcification, indicating that BMP-7 can promote calcium deposits. Since calcification is apparent in the pathophysiology of PE, it can be suggested that increased BMP-7 causes increased calcification, i.e., the invasive basal layer trophoblast cells in the uterus is blocked, resulting in an interruption in blood exchange and causing a series of maternal and neonatal fetal complications, such as intrauterine fetal anoxia, intrauterine fetal growth restriction, placental abruption, and cardiovascular disease.
The present study identified differences between the three groups at the mRNA and protein levels: there was a higher KDM5C mRNA expression in the EOsPE group than in the control group, although no obvious changes were found in the LOsPE group. Compared with the control group, both sPE groups showed reduced mRNA expressions of BMP-7 and BMPRII, with a higher expression in the LOsPE group than in the EOsPE group.
There are many complicated pathophysiological changes in PE and many signaling pathways involved in its occurrence; these mutually affect folding diseases, so inconsistencies in mRNA and protein levels are possible. Furthermore, the results of the present study do not exclude regional and ethnic differences. Further research should aim to identify useful serum markers to predict the incidence of PE as soon as possible and at the cellular level, as well as investigate changes in the three factors in order to gain a better understanding of the pathogenesis of PE.
Conclusion
The expression of KDM5C is induced in sPE, with this decrease being more noticeable in LOsPE. In contrast, BMP-7 and BMPRII expression levels are increased in sPE, more noticeably in LOsPE. A possible explanation is KDM5C nuclear-cytoplasmic translocation within the inhibited BMP-7 expression involved in PE. The results of the present study indicate that the ectopic expression of KDM5C in PE placenta promotes trophoblast cell calcification and placenta decidual vascular dysplasia, which then leads to placental ischemia. Finally, the findings of the present study suggest that the KDM5C–BMP-7 signaling pathway plays a role in the pathogenesis of PE.
Abbreviations
KDM5C: (K)-specific demethylase 5CBMP-7: bone morphogenetic protein-7sPE: severe preeclampsiamRNA: messenger RNABMPRII: bone morphogenetic protein receptor IIIHC: immunohistochemistryPE: preeclampsiaH3K4: histone 3 lysine 4TGF-β: transforming growth factor betaBMPs: bone morphogenetic proteinsBMPRs: bone morphogenetic protein receptorsEOsPE: early-onset severe PELOsPE: late-onset sPEqRT-PCR: quantitative real-time polymerase chain reactioncDNA: complementary DNAPBS: phosphate-buffered solution
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Lindheimer MD, Taler SJ, Cunningham FG. Hypertension in pregnancy. J Am Soc Hypertens. 2010;4:68–78.
- Sibai BM. Diagnosis and management of gestational hypertension and preeclampsia. Obstet Gynecol. 2003;102:181–192.
- Valensise H, Vasapollo B, Gagliardi G, et al. Early and late preeclampsia: two different maternal hemodynamic states in the latent phase of the disease. Hypertension. 2008;52:873–880.
- Escudero CA, Herlitz K, Troncoso F, et al. Role of extracellular vesicles and microRNAs on dysfunctional angiogenesis during preeclamptic pregnancies. Front Physiol. 2016;7:98.
- Brookes E, Laurent B, Õunap K, et al. Mutations in the intellectual disability gene KDM5C reduce protein stability and demethylase activity. Hum Mol Genet. 2015;24:2861–2872.
- Alarmo EL, Rauta J, Kauraniemi P, et al. Bone morphogenetic protein 7 is widely overexpressed in primary breast cancer. Genes Chromosomes Cancer. 2006;45:411–419.
- Buijs JT, Henriquez NV, van Overveld PG, et al. Bone morphogenetic protein 7 in the development and treatment of bone metastases from breast cancer. Cancer Res. 2007;67:8742–8751.
- Montesano R. Bone morphogenetic protein-4 abrogates lumen formation by mammary epithelial cells and promotes invasive growth. Biochem Biophys Res Commun. 2007;353:817–822.
- Shon SK, Kim A, Kim JY, et al. Bone morphogenetic protein-4 induced by NDRG2 expression inhibits MMP-9 activity in breast cancer cells. Biochem Biophys Res Commun. 2009;385:198–203.
- Zhu L, Zhang Z, Zhang L, et al. HMGB1-RAGE signaling pathway in severe preeclampsia. Placenta. 2015;36:1148–1152.
- Ji X, Jin S, Qu X, et al. Lysine-specific demethylase 5C promotes hepatocellular carcinoma cell invasion through inhibition BMP7 expression. BMC Cancer. 2015;15:801.
- von Dadelszen P, Magee LA, Roberts JM. Subclassification of preeclampsia. Hypertens Pregnancy. 2003;22:143–148.
- Cheng SB, Sharma S. Preeclampsia and health risks later in life: an immunological link. Semin Immunopathol. 2016;38:699–708.
- Whetstine JR, Nottke A, Lan F, et al. Reversal of histone lysine trimethylation by the JMJD2 family of histone demethylases. Cell. 2006;125:467–481.
- Kawazu M, Saso K, Tong KI, et al. Histone demethylase JMJD2B functions as a co-factor of estrogen receptor in breast cancer proliferation and mammary gland development. PLoS One. 2011;6:e17830.
- Xu L, Wu W, Cheng G, et al. Enhancement of proliferation and invasion of gastric cancer cell by KDM5C via decrease in p53 expression. Technol Cancer Res Treat. 2017;16:141–149.
- Lu F, Gong H, Lei H, et al. Downregulation of cathepsin C alleviates endothelial cell dysfunction by suppressing p38 MAPK/NF-κB pathway in preeclampsia. Bioengineered. 2022;13:3019–3028.
- Grauer JN, Patel TC, Erulkar JS, et al. Young investigator research award winner. Evaluation of OP-1 as a graft substitute for intertransverse process lumbar fusion. Spine (Phila Pa 1976). 2000;26:127–133. (Phila Pa 1976). 2001
