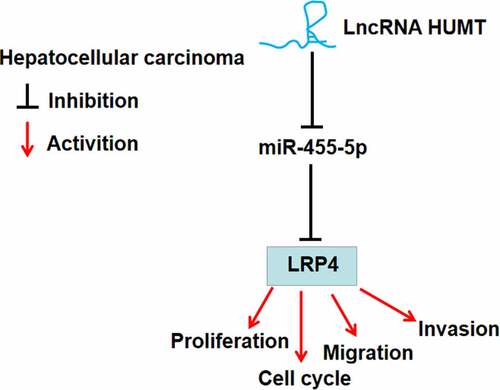
ABSTRACT
The present study aimed at investigating the effects and mechanism of long noncoding RNA highly upregulated in metastatic triple-negative breast cancer lymph node (lncRNA HUMT) in hepatocellular carcinoma (HCC). Quantitative real-time polymerase chain reaction was used to assess the expression of HUMT, microRNA (miR)-455-5p, and low-density lipoprotein receptor-related protein 4 (LRP4) in HCC tissues. Colony forming and 5-ethynyl-2′-deoxyuridine assays were performed to assess cell proliferation. Transwell assay was performed to measure cell migration and invasion. Cell cycle distribution was assessed using flow cytometry. The protein expression of LRP4, proliferating cell nuclear antigen (PCNA), matrix metallopeptidase 2 (MMP-2), and MMP-9 was detected using western blot. Luciferase reporter assay and RNA immunoprecipitation assay was used to confirm the target association between miR-455-5p and HUMT or LRP4. In our study, the level of HUMT was enhanced in HCC tissues and cells. Cell proliferation, invasion, and migration in HCC cells were repressed by knockdown of HUMT, and knockdown of HUMT arrested cells in G1 phase and decreased the levels of PCNA, MMP-2, and MMP-9. MiR-455-5p was a target of HUMT. Lowexpression of miR-455-5p reversed the inhibitive influence on HCC cells induced by of HUMT silencing. LRP4 was a target of miR-455-5p and was negatively regulated by miR-455-5p. In addition, LRP4 expression was positively modified by HUMT, and LRP4 inhibited the inhibitory effects on HCC cells induced by HUMT silencing. In conclusion, HCC cell proliferation, invasion, and migration were restrained by knockdown of HUMT, which was related to the miR-455-5p/LRP4 axis.
Graphical Abstract
KEYWORDS:
Introduction
Hepatocellular carcinoma (HCC) has been listed as the second leading cause of death and the fifth leading cause of all cancer cases, seriously endangering human life and health [Citation1]. In the past 20 years, the incidence rate of HCC has rapidly increased year by year because of the impacts of economic development and other factors, and it occurs in all age groups [Citation2,Citation3]. At present, the treatment of HCC is limited, and the commonly used methods are tumor ablation, liver resection and hepatic transplantation [Citation4]. In addition, transarterial chemical embolization and sorafenib are regarded as palliative treatments [Citation5]. Despite the rapid development of treatment methods in the past few decades, the overall long-term survival rate is still very low in HCC [Citation6]. Moreover, due to the complex pathogenesis and the high recurrence rate after resection, there is still no clear and effective treatment method of HCC [Citation7]. Thus there is an urgent need for better solutions.
Long noncoding RNAs (lncRNAs) consist of more than 200 nucleotides and act as crucial effects in a variety of cancers [Citation8–11]. It is reported that lncRNAs take part in various biological processes, including proliferation, metabolism, immunity, differentiation, and migration [Citation12,Citation13]. Imbalance of lncRNAs was found in HCC [Citation14,Citation15]. Liu et al. found that the expression of lncRNA cytoskeleton regulator RNA (CYTOR) was upregulated in HCC tumor tissues, and knockdown of CYTOR inhibited HCC progression [Citation16]. Wu et al. [Citation17] reported that high-expression of CCAAT/enhancer binding protein-alpha antisense 1 facilitated HCC progression. It has revealed that lncRNA focally amplified lncRNA on chromosome 1 facilitated HCC cell proliferation and migration by regulating miR-1236 [Citation18]. LncRNA highly upregulated in metastatic triple-negative breast cancer lymph node (HUMT), as one of the member of lncRNAs, is reported to accelerate metastasis in triple-negative breast cancer [Citation19]. Nevertheless, the mechanism of HUMT in the development of HCC remains indistinct.
MicroRNAs (miRNAs) consist of 18–25 nucleotides and can regulate target genes by complementary binding to their 3′-UTR [Citation20]. Studies have shown that miRNAs are related to cell proliferation, differentiation, apoptosis, lipid metabolism, inflammation, immune responses, and oncogenesis [Citation21–23]. Recently, miR-455-5p has been reported to participate in multiple disorders. For example, miR-455-5p had the neuroprotective effects on spinal cord ischemia reperfusion injury by inhibiting apoptosis of neurons [Citation24]. MiR-455-5p inhibits vascular smooth muscle cell proliferation and migration [Citation25]. Previous studies have revealed that miRNAs help to seek diagnostic biomarker, and offering new targets for HCC [Citation26–28]. Through targeting insulin-like growth factor-1 receptor, miR-455-5p restrained the proliferation and invasion of HCC cells [Citation29]. Low-density lipoprotein receptor-related protein 4 (LRP4), a member of the LDL receptor family, contains a large extracellular domain, a few EGF-like domains, a transmembrane domain, and a small intracellular domain [Citation30]. It is reported that LRP4 plays an important role in formation of the neuromuscular junction [Citation31]. LRP4 induces extracellular matrix productions and promotes chondrocyte differentiation [Citation32]. Mao et al. found that LRP4 promoted gastric cancer cell migration and invasion, and high expression of LRP4 was correlated with an unfavorable prognosis in GC patients [Citation33]. However, the mechanism of miR-455-5p and LRP4 in HCC is unclear.
We hypothesized that lncRNA HUMT played an important role in HCC progression. This study aimed to investigate the function of HUMT in HCC and the specific mechanism of regulating miR-455-5p/ LRP4 axis.
Materials and methods
Bioinformatics analysis
To select the candidate lncRNAs involved in HCC, the lncRNAs in The Cancer Genome Atlas (TCGA) database were mapped on heatmap. Based on TCGA database, the levels of HUMT, miR-455-5p, and LRP4 was obtained using the DESeq2 R package [Citation34]. False-discovery rate (FDR) method was used to calculate the adjusted P-values, absolute log2-fold change of >1 and FDR of < 0.05 were set as the cutoff. The overall survival of HUMT, miR-455-5p, and LRP4 in HCC patients was analyzed using the Kaplan–Meier curve method. Log rank test was performed to compare survival times between two groups (high and low).
Patient samples
The study obtained the approval of the Ethics Committee of Yantai Qishan Hospital (approval no. 20200510001). Every patient or every patient’s guardian signed the ‘informed written consent’. The levels of HUMT, miR-455-5p, and LRP4 were determined in 30 pairs HCC tissues and matched to non-cancer tissues.
Cell cultivation
From the Nanjing Cobioer Biosciences Co., LTD (Nanjing, China), human HCC cell lines (HCCLM3, MHCC97L, MHCC97H, and Huh7) were acquired. Dulbecco’s modification of Eagle’s medium (Sigma-Aldrich, USA) supplemented with 10% fetal bovine serum (FBS; HyClone, USA) and 1% penicillin/ streptomycin (Gibco, USA) was used to maintain these HCC cells. From American Type Culture Collection (Manassas, VA, USA), the human liver cell line (THLE-2) was acquired. THLE-2 cells were cultured in bronchial epithelial cell basal medium supplemented with 5 ng/mL epidermal growth factor, 70 ng/mL phosphoethanolamine, and 10% FBS at 37°C in a humidified incubator containing 5% CO2.
Cell transfection
GeneCopoeia Co., Ltd. (Guangzhou, China) synthetized small interferring (si) RNA-HUMT#1, siRNA-HUMT#2, siRNA-negative control (si-NC), miR-455-5p mimic, mimic NC, inhibitor NC, miR-455-5p inhibitor, pcDNA3.1-LRP4 and pcDNA3.1-NC. HCCLM3 and MHCC97H cells were transfected with si-HUMT#1, si-HUMT#2, si-NC using lipofectamine 2000 (Invitrogen, USA). Meanwhile, HCCLM3 and MHCC97H cells transfected with si-HUMT#1 were cotransfected with inhibitor NC, miR-455-5p inhibitor, pcDNA3.1-LRP4, and pcDNA3.1. After 48 h, quantitative real-time polymerase chain reaction (qRT-PCR) was used to assess the transfection efficiency.
qRT-PCR
TRIzol (Invitrogen, USA) was applied to extract total RNA. PrimeScript RT Reagent Kit (Takara, Japan) was applied to perform reverse transcription. SYBR Premix Ex Taq II (Takara) was performed to detect the levels of miR-455-5p, HUMT, and LRP4 in an ABI7300 real-time PCR machine (Thermo Fisher Scientific, USA). U6 and GAPDH served as internal control. The levels of HUMT, miR-455-5p, and LRP4 was analyzed using 2−ΔΔCt method [Citation35]. displays the primers.
Table 1. The primers used in qRT-PCR
Fluorescence in situ hybridization (FISH) assay
HCCLM3 and MHCC97H cells (6 × 104 cells/well) were seeded on a cover glass on the 24‐well culture. When the cell confluence reached about 80%, cells were fixed in 4% paraformaldehyde for 15 min. After being treated with protease K, glycine, and acylation reagent, cells were incubated with 250 µl prehybridization solution for 1 h at 42°C, and then incubated with hybridization solution containing lncRNA HUMT probes (RiboBio, China) overnight at 42°C. Next, the nucleus was stained with 4ʹ,6-diamidino-2-phenylindole 2hci (ab104139, 1:100, Abcam) for 5 min. Finally, cells were photographed with a fluorescence microscope (Olympus, Japan) [Citation36].
Colony formation assay
The transfected HCCLM3 and MHCC97H cells were plated into a 6-well plate. After incubation for 14 days, methanol (Sigma-Aldrich) was used to fix cells for 15 min and crystal violet (Sigma-Aldrich) was used to stain cells. Finally, the number of colony was assessed under a light microscope (Thermo Fisher Scientific) [Citation37].
5-ethynyl-2′-deoxyuridine (EdU) assay
The transfected HCCLM3 and MHCC97H cells (1.5 × 105 cells/well) were seeded into a 24-well plate. Next day, the cells were incubated with 50 µM EdU (RiboBio, China) for 2 h. Subsequently, 4% paraformaldehyde was used to fix cells for half an hour. Then cells were incubated with glycine (2 mg/ml), 0.5% Trion X-100, and stained with apollo® fluorescent dye and Hoeschst 33342. The inverted microscopy (Nikon, Japan) was applied to take the images [Citation38].
Cell cycle assessment
After transfection for 48 h, HCCLM3 and MHCC97H cells were fixed in ice-cold 70% ethanol for 4 h at 4°C. Next, cells were incubated with Propidium Iodide and RNase A (Beyotime Biotechnology) for 30 min at 37°C. Finally, cell cycle was evaluated using a flow cytometer (BD Biosciences, USA) [Citation39].
Cell invasion and migration assays
After transfection for 48 h, cell suspension was prepared. The upper chamber covered with (invasion assay) or without (migration assay) matrigel was inoculated with HCCLM3 and MHCC97H cells (1 × 105), and the lower chamber was filled with 600 μl completely medium. The cells were cultivated at 37°C overnight, and then stained by 0.1% crystal violet. Finally, the microscope was applied to take the images of invaded or migrated cells [Citation36].
Western blot
Cells were lyzed with Radio-Immunoprecipitation Assay buffer (Aspen Biotechnology, China) to extract total proteins. The concentration of proteins was measured using Bicinchoninic acid Protein Assay kit (Thermo Fisher Scientific). Next, protein samples (30 μg) were loaded to 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis and transferred onto polyvinylidene difluoride membranes (Millipore, USA). After being blocked in 5% skim milk for 1 h, the membranes were incubated with primary antibodies [LRP4, ab230188, Abcam, UK; proliferating cell nuclear antigen (PCNA), ab92552, Abcam; matrix metallopeptidase 2 (MMP-2), ab181286, Abcam; matrix metallopeptidase 9 (MMP-9), ab283575, Abcam; GAPDH, ab181602, Abcam] overnight at 4°C. After that, the membranes were incubated with horseradish peroxidase‐labeled goat anti‐rabbit secondary antibody (ab205718, Abcam) for 1 h at room temperature. Finally, the protein bands were visualized using an efficient chemiluminescenc (ECL) kit (Millipore), and the gray density of protein bands were evaluate using Image J software (National Institute Health, USA).
Dual-luciferase reporter assay
When cells confluence reached 80%, miR-455-5p mimic, mimic NC, and the pmirGLO luciferase reporter gene vector with either HUMT-wild type (WT), LPR4-WT or HUMT-mutant type (MUT), LPR4-MUT was transfected into HCCLM3 and MHCC97H cells. The relative luciferase activities were detected after transfection for 48 h [Citation40].
RNA immunoprecipitation (RIP) assay
The Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore) was used to conduct RIP assay. Briefly, HCCLM3 and MHCC97H cells were lysed in a RIP-lysis buffer. Then, the RIP buffer containing magnetic beads conjugated with anti-Ago-2 (Millipore) or anti-IgG (Millipore) were added to the cell lysates and incubated for overnight at 4°C. Next, the magnetic beads were incubated with proteinase K buffer for 30 min. Finally, the enrichment of HUMT, miR-455-5p, and LRP4 in purified RNAs was examined by qRT-PCR [Citation41].
Statistical analyses
GraphPad Prism 7 (USA) was used to perform statistical analyses. Student’s t-test was used to analyze changes in experimental results between two groups. ANOVA with Dunnett t-test was performed for multiple comparisons. The relationship among miR-455-5p, HUMT, and LRP4 was analyzed using Pearson’s correlation analysis. Differences with P < 0.05 were considered as significant.
Results
In the present study, we investigated the expression, function, and molecular mechanism of HUMT in HCC progression using bioinformatics, function-loss experiments. The findings showed that HUMT expression was upregulated in HCC tissues and cells, knockdown of HUMT inhibited HCC cell proliferation, migration, and invasion via regulating the miR-455-5p/LRP4 axis.
LncRNA HUMT is upregulated in HCC tissues
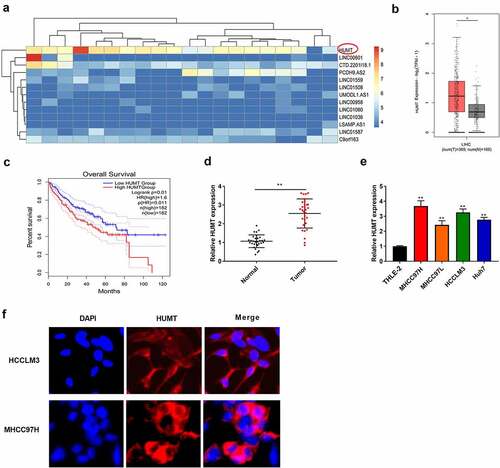
Among the top differentially expressed lncRNAs between normal and HCC tissues in TCGA database, 2 aberrantly up-regulated and 11 down-regulated candidate lncRNAs were identified (). We chose HUMT as the candidate gene for HCC in the subsequent assays. Furthermore, to probe the expression level of HUMT in HCC patients, TCGA database was used. As presented in , an obvious HUMT high expression was discovered in HCC patients relative to that in normal group. As presented in , HUMT high expression was related to worse prognosis in HCC patients (P = 0.01). In addition, HUMT was higher in HCC tissues (). illustrates that HUMT expression was higher in HCC cell lines (MHCC97H, Huh7, HCCLM3, and MHCC97L). HCCLM3 and MHCC97L cell lines with higher HUMT expression were used in the subsequent assays. FISH assay was used to verify the subcellular localization of HUMT. The results showed that HUMT was mainly localized in the cytoplasm in HCCLM3 and MHCC97L cells ().
Figure 1. LncRNA HUMT expression was higher in HCC tissues. (a) The heatmap indicated the LncRNA expression in HCC. (b) TCGA database was used to analyze HUMT expression in HCC tissues. (c) According to TCGA database, the overall survival of HUMT in HCC patients was analyzed. (d) HUMT expression in normal and tumor tissues was tested by qRT-PCR (n = 30). (e) QRT-PCR was performed to assess HUMT expression in human HCC cell lines (HCCLM3, MHCC97L, MHCC97H, and Huh7) and human liver cell line (THLE-2). (f) The subcellular localization of HUMT was assessed using FISH assay **P < 0.01 vs. Normal group or THLE-2 cells.
The proliferation and metastasis of HCC cells is inhibited by knockdown of HUMT
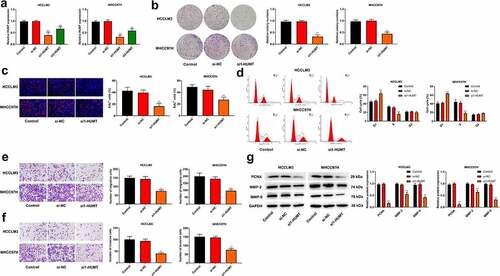
Si-HUMT #1 and si-HUMT #2 were transfected into HCCLM3 and MHCC97L cells. ) illustrates that si1-HUMT and si2-HUMT reduced HUMT expression level in HCCLM3 and MHCC97L cells. Besides, the knockdown efficiency of si1-HUMT was higher than that in si2-HUMT, thus si1-HUMT was utilized in the subsequence assays. As shown in ()), the ability of proliferation was significantly repressed by HUMT knockdown in HCCLM3 and MHCC97L cells. We then measured cell cycle distribution using flow cytometry. The results showed that knockdown of HUMT triggered G1 phase accumulation and decreased cell number of S phase in HCCLM3 and MHCC97L cells ()). In addition, as presented in (, knockdown of HUMT decreased the number of migration and invasion in HCCLM3 and MHCC97L cells. The protein expression of proliferatic and metastatic protein markers (PCNA, MMP-2, and MMP-9) was detected using western blot. As expected, the results showed that knockdown of HUMT decreased the protein expression of PCNA, MMP-2, MMP-9 in HCCLM3 and MHCC97L cells ()).
Figure 2. Knockdown of HUMT inhibited HCC cell proliferation and metastasis. (a) HUMT expression was assessed in HCCLM3 and MHCC97L cells using qRT-PCR. (b-c) Proliferation of HCCLM3 and MHCC97H cells was assessed using colony formation and EDU assays. (d) Cell cycle distribution in HCCLM3 and MHCC97H cells was assessed using flow cytometry. (e-f) HCCLM3 and MHCC97H cell migration and invasion were determined by transwell assay. (g) The protein expression of proliferatic and metastatic protein markers (PCNA, MMP-2, and MMP-9) in HCCLM3 and MHCC97H cells was detected using western blot.**P < 0.01 vs. Control or si-NC group.
MiR-455-5p is a target of HUMT in HCC
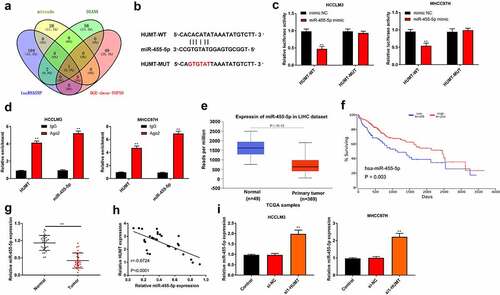
To probe the mechanisms of HUMT in HCC, we first predicted its potential target genes. One common miRNA (miR-455-5p) was obtained from the intersection of mirode, DIANA, lncRNASNP, and DGE-down top50 ()). ) displays the region of miR-455-5p targeted by HUMT. ) elucidates that the luciferase activity of HCCLM3 and MHCC97L cells containing HUMT 3’-UTR-WT instead of HUMT 3’-UTR-MUT was reduced by miR-455-5p mimic. Moreover, RIP assay showed that the enrichment of HUMT and miR-455-5p immunoprecipitated with Ago2 was increased compared with IgG precipitates in HCCLM3 and MHCC97L cells ()). According to TCGA database, miR-455-5p was lowly expressed in HCC tissues, and low expression of miR-455-5p was related to worse survival ()), manifesting that miR-455-5p was involved in HCC progression. As presented in ), miR-455-5p was downregulated in HCC tissues. Besides, there was a negative correlation between HUMT expression and miR-455-5p expression (r = −0.6724, P < 0.001, )). Afterthat, we found that miR-455-5p expression was highly expressed in si1-HUMT group of HCCLM3 and MHCC97L cells ()).
Figure 3. HUMT targets miR-455-5p in HCC. (a) One common miRNA (miR-455-5p) was gained from the intersection of venn diagram (mirode, DIANA, lncRNASNP, and DGE-down top50). (b) The binding sites of HUMT and miR-455-5p. (c) The targeting relationship between HUMT and miR-455-5p was verified by luciferase reporter assay in HCCLM3 and MHCC97H cells. (d) After anti-Ago2-mediated RIP assay, the expression of HUMT and miR-455-5p in HCCLM3 and MHCC97H cells was detected using qRT-PCR. (e)According to TCGA database, miR-455-5p expression in HCC was analyzed. (f) The overall survival of miR-455-5p in HCC patients from TCGA database. (g) QRT-PCR was performed to test miR-455-5p expression in normal and tumor tissues (n = 30). (h) The relationship between miR-455-5p and HUMT was analyzed by Pearson correlation analysis. (i) MiR-455-5p expression was examined in HCCLM3 and MHCC97H cells by qRT-PCR. **P < 0.01 vs. Normal, control, mimic NC, IgG or si-NC group.
HUMT regulates proliferation and metastasis through miR-455-5p in HCC
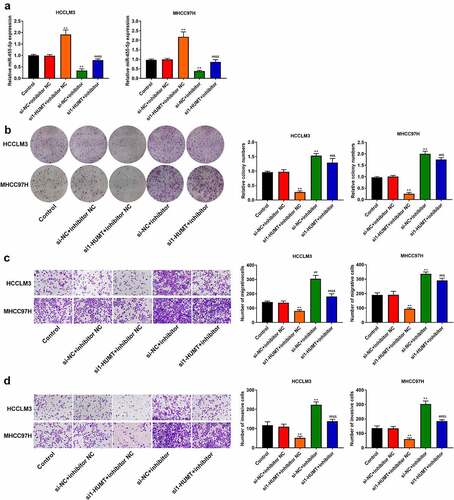
To further probe the effects of miR-455-5p in the regulation of HUMT on HCC cell proliferation and motility, rescue assays were performed. shows that lowexpression of miR-455-5p weakened the promotive effect of HUMT ablation on miR-455-5p expression in HCCLM3 and MHCC97L cells. In addition, the down-regulation of miR-455-5p partly reversed the repressive effects of HUMT depletion on cell proliferation, migration, and invasion by in HCCLM3 and MHCC97L cells ().
Figure 4. MiR-455-5p was related to the regulation of HUMT on HCC cell proliferation, migration, and invasion. (a) MiR-455-5p expression in HCCLM3 and MHCC97H cells was detected using qRT-PCR. (b) HCCLM3 and MHCC97H cell proliferation was detected using colony formation assay. (c-d) HCCLM3 and MHCC97H cell migration and invasion were determined by transwell assay. **P < 0.01 vs. Control or si-NC+inhibitor NC group. ##P < 0.01 vs. si1-HUMT+inhibitor NC group. &P < 0.05 and &&P < 0.01 vs. si-NC +inhibitor group.
LRP4 is a target of miR-455-5p in HCC
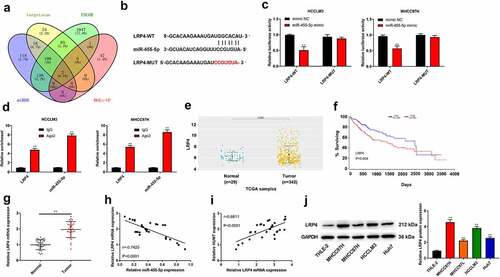
The potential target genes of miR-455-5p were predicted by bioinformatics analysis. The intersection of targetscan, ENCOR, miRDB, and DGE-up showed 2 common genes, and LRP4 was chosen for further research. The region of miR-455-5p targeted by LRP4 was displayed in ). Besides, ) elucidates that the luciferase activity of HCCLM3 and MHCC97L cells containing LRP4 3’-UTR-WT was weakened by miR-455-5p mimic. Moreover, RIP assay showed that the enrichment of LRP4 and miR-455-5p immunoprecipitated with Ago2 was increased compared with IgG precipitates in HCCLM3 and MHCC97L cells ()). On the basis of the data acquired from TCGA database, LRP4 was highly expressed in HCC tissues (P = 0.0055), and high expression of LRP4 was related to worse survival ()). Moreover, we found that LRP4 was higher in HCC tissues, and there was a negative correlation between LRP4 expression and miR-455-5p expression ()). Nevertheless, as illustrated in ), there was a positive correlation between LRP4 expression and HUMT expression. In addition, the protein expression of LRP4 in the human liver cell line (THLE-2) and HCC cell lines (HCCLM3, MHCC97L, MHCC97H, and Huh7) was detected using western blot. The results showed that the protein expression of LRP4 in HCC cell lines was higher than that in THLE-2 line ().
Figure 5. MiR-455-5p targets LRP4 in HCC. (a) The intersection of veen diagram (targetscan, ENCOR, miRDB, and DGE-up) showed 2 common genes. (b) The binding sites of miR-455-5p and LRP4. (c) The targeting relationship between LRP4 and miR-455-5p was confirmed by luciferase reporter assay in HCCLM3 and MHCC97H cells. (d) After anti-Ago2-mediated RIP assay, the expression of LRP4 and miR-455-5p in HCCLM3 and MHCC97H cells was detected using qRT-PCR. (e) TCGA database was used to analyze the level of LRP4 in HCCs. (f) The overall survival of LRP4 in HCC patients from TCGA database. (g) QRT-PCR assay was applied to test LRP4 expression in normal and tumor tissues (n = 30). (h, i) Pearson correlation analysis was performed to assess the relationship among HUMT, miR-455-5p and LRP4. (j) The protein expression of LRP4 in the human liver cell line (THLE-2) and HCC cell lines (HCCLM3, MHCC97L, MHCC97H, and Huh7) was detected using western blot. **P < 0.01 vs. Normal, mimic NC or THLE-2 group.
LRP4 overexpression reversed the inhibitory role of silencing HUMT on HCC cell proliferation and metastasis
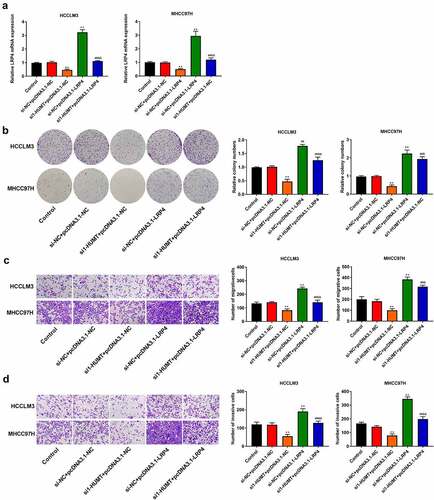
We used qRT-PCR to test LRP4 expression in HCCLM3 and MHCC97H cells. ) shows that overexpression of LRP4 inhibited the repressive effect of HUMT knockdown on LRP4 expression. In addition, through colony formation and Transwell assays, we found that the LRP4 over-expression reversed the inhibitory effects of HUMT knockdown on cell proliferation, invasion, and migration in HCCLM3 and MHCC97H cells ()).
Figure 6. LRP4 overexpression inhibited the inhibitory role of silencing HUMT on HCC proliferation, migration, and invasion. (a) QRT-PCR assay was applied to detect LRP4 expression in HCCLM3 and MHCC97H cells. (b) Proliferation of HCCLM3 and MHCC97H cells was assessed using colony formation asssay. (c, d) The transwell assay was used to assess HCCLM3 and MHCC97H cell migration (c) and invasion (d). **P < 0.01 vs. Control and si-NC+pcDNA3.1-NC. ##P < 0.01 vs. si-HUMT+pcDNA3.1-NC. &P < 0.05 and &&P < 0.01 vs. si-NC + pcDNA3.1-LRP4.
Discussion
LncRNAs are involved in growth, invasion, and migration in HCC [Citation42,Citation43]. As a highly dysregulated lncRNA, HUMT has been demonstrated to play a role in triple-negative breast cancer [Citation19]. However, the mechanism related to HUMT in HCC progression remains unclear. We surveyed the influences of HUMT on HCC progression in this study. We found that HUMT was related to miR-455-5p/LRP4 axis in HCC.
We identified that HUMT expression was increased in HCC tissues and cells, and then explored the function and underling mechanisms. The findings indicated that knockdown of HUMT inhibited HCC cell proliferation and metastasis and arrested cells in G1 phase. Knockdown of HUMT decreased the protein expression of PCNA, MMP-2, MMP-9 in HCCLM3 and MHCC97L cells. Moreover, HUMT exerted its function through the miR-455-5p/LRP5 axis. Through protein kinase B/mammalian target of rapamycin/vascular endothelial growth factor signaling, HUMT exerted its function of proliferation and metastasis by recruiting Y-box binding protein 1 protein in triple-negative breast cancer [Citation19]. Agreed with the above findings, our study confirmed that HUMT participated in cell proliferation and metastasis in HCC.
To figure out whether HUMT acts as a competing endogenous (ce) RNA in HCC, we performed bioinformatics analysis, RIP assay, and luciferase reporter assay. The results showed that miR-455-5p was a target of HUMT in HCC cells. Furthermore, miR-455-5p lowexpression reversed the repressive impact of HUMT knockdown on HCC cell proliferation and metastasis, indicating the roles of HUMT was involved in miR-455-5p. In addition, we searched and confirmed the target relationship between miR-455-5p and LRP4 by bioinformatics analysis and luciferase reporter assay. Functional analysis showed that cell proliferation, invasion, and migration were promoted by lowexpression of miR-455-5p in HCC cells, which is also consistent with former researches [Citation29,Citation44]. Under the regulation of miR-140-5p, LRP4 accelerated cell migration and invasion in gastric cancer [Citation33]. In this study, LRP4 and HUMT have the binding sites of miR-455-5p, which supported that HUMT acted as a ceRNA. Our results also displayed the promoting effects of LRP4, revealed by abrogating the facilitative role of HUMT knockdown in HCC progression. Besides, LRP4 expression was regulated by HUMT and miR-455-5p, which indicated that HUMT could regulate LRP4 by sponging miR-455-5p in HCC.
Conclusion
Taken together, HUMT expression was upregulated in HCC tissues and cells. HCC cell proliferation, invasion, and migration were inhibited by knockdown of HUMT, which was involved in the miR-455-5p/LRP4. These findings may provide novel clinical implications for HCC treatment.
Highlight
LncRNA HUMT expression is upregulated in hepatocellular carcinoma tissues.
Knockdown of HUMT inhibits hepatocellular carcinoma cell growth and metastasis.
HUMT acts as a competing endogenous RNA of miR-455-5p to regulate LRP4 expression.
Author’s contributions
Conception and design: Xianzhi Zou and Peng Sun; Perform research: Hui Xie and Lu Fan; Data analysis and interpretation: Kun Ding, Jiyang Wang and Yang Li; Manuscript writing: Xianzhi Zou; Final approval of manuscript: All authors
Availability of data and material
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
The protocol of this research has been approved by the Ethics Committee of Yantai Qishan Hospital (approval no. 20200510001). All patients have signed written informed consent.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Wang H, Liu Y, Shen K, et al. A comparison between radiofrequency ablation combined with transarterial chemoembolization and surgical resection in hepatic carcinoma: a meta-analysis. J Cancer Res Ther. 2019;15(7):1617–1623.
- Hartke J, Johnson M, Ghabril M. The diagnosis and treatment of hepatocellular carcinoma. Semin Diagn Pathol. 2017;34(2):153–159.
- Sim HW, Knox J. Hepatocellular carcinoma in the era of immunotherapy. Curr Probl Cancer. 2018;42(1):40–48.
- Fang L, Meng X, Luo W, et al. Treatment of primary hepatic carcinoma through ultrasound-guided microwave ablation. Niger J Clin Pract. 2019;22(10):1408–1411.
- Grandhi MS, Kim AK, Ronnekleiv-Kelly SM, et al. Hepatocellular carcinoma: from diagnosis to treatment. Surg Oncol. 2016;25(2):74–85.
- Chaudhari VA, Khobragade K, Bhandare M, et al. Management of fibrolamellar hepatocellular carcinoma. Chin Clin Oncol. 2018;7(5):51.
- Zhu ZX, Huang JW, Liao MH, et al. Treatment strategy for hepatocellular carcinoma in China: radiofrequency ablation versus liver resection. Jpn J Clin Oncol. 2016;46(12):1075–1080.
- Wawrzyniak O, Ż Z, Rolle K, et al. Circular and long non-coding RNAs and their role in ophthalmologic diseases. Acta Biochim Pol. 2018;65(4):497–508.
- Wei JW, Huang K, Yang C, et al. Non-coding RNAs as regulators in epigenetics (Review). Oncol Rep. 2017;37(1):3–9.
- Huang L, Dai G. Long non-coding RNA DCST1-AS1/hsa-miR-582-5p/HMGB1 axis regulates colorectal cancer progression. Bioengineered. 2022;13(1):12–26.
- Yan H, Zhou Y, Chen Z, et al. Long non-coding RNA HCG11 enhances osteosarcoma phenotypes by sponging miR-1245b-5p that directly inhibits plakophilin 2. Bioengineered. 2022;13(1):140–154.
- Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. 2016;17(1):47–62.
- Cao L, Wang Y, Wang Q, et al. LncRNA FOXD2-AS1 regulates chondrocyte proliferation in osteoarthritis by acting as a sponge of miR-206 to modulate CCND1 expression. Biomed Pharmacothe. 2018;106:1220–1226.
- Ji Y, Sun H, Liang H, et al. Evaluation of LncRNA ANRIL potential in hepatic cancer progression. Journal of Environmental Pathology, Toxicology and Oncology: Official Organ of the International Society for Environmental Toxicology and Cancer. 2019;38(2):119–131.
- Zhang Z, Wang S, Liu Y, et al. Low lncRNA ZNF385D‑AS2 expression and its prognostic significance in liver cancer. Oncol Rep. 2019;42(3):1110–1124.
- Liu Y, Geng X. Long non-coding RNA (lncRNA) CYTOR promotes hepatocellular carcinoma proliferation by targeting the microRNA-125a-5p/LASP1 axis. Bioengineered. 2022;13(2):3666–3679.
- Wu C, Tang ZY, Chen HY, et al. High-expression of lncRNA CEBPA-AS1 promotes liver cancer progression. Eur Rev Med Pharmacol Sci. 2019;23(19):8295–8302.
- Li B, Mao R, Liu C, et al. LncRNA FAL1 promotes cell proliferation and migration by acting as a CeRNA of miR-1236 in hepatocellular carcinoma cells. Life Sci. 2018;197:122–129.
- Zheng S, Yang L, Zou Y, et al. Long non-coding RNA HUMT hypomethylation promotes lymphangiogenesis and metastasis via activating FOXK1 transcription in triple-negative breast cancer. J Hematol Oncol. 2020;13(1):17.
- Lu TX, Rothenberg ME. MicroRNA. J Allergy Clin Immunol. 2018;141(4):1202–1207.
- Mellis D, Caporali A. MicroRNA-based therapeutics in cardiovascular disease: screening and delivery to the target. Biochem Soc Trans. 2018;46(1):11–21.
- Tiwari A, Mukherjee B, Dixit M. MicroRNA key to angiogenesis regulation: miRNA biology and therapy. Curr Cancer Drug Targets. 2018;18(3):266–277.
- Liu Y, Liu J, Cui J, et al. Role of lncRNA LINC01194 in hepatocellular carcinoma via the miR-655-3p/SMAD family member 5 axis. Bioengineered. 2022;13(1):1115–1125.
- Liu B, Zheng W, Dai L, et al. Bone marrow mesenchymal stem cell derived exosomal miR-455-5p protects against spinal cord ischemia reperfusion injury. Tissue Cell. 2022;74:22.
- Zhang X, Liu Y, Zhao J, et al. MiR-455-5p serves as a biomarker of atherosclerosis and inhibits vascular smooth muscle cell proliferation and migration. Per Med. 2021;18(3):213–221.
- Gailhouste L, Liew LC, Yasukawa K, et al. MEG3-derived miR-493-5p overcomes the oncogenic feature of IGF2-miR-483 loss of imprinting in hepatic cancer cells. Cell Death Dis. 2019;10(8):553.
- Zhang X, Xu X, Ge G, et al. miR‑498 inhibits the growth and metastasis of liver cancer by targeting ZEB2. Oncol Rep. 2019;41(3):1638–1648.
- Hu S, Zhang J, Fang X, et al. Identification of microRNA hsa-miR-30c-5p as an inhibitory factor in the progression of hepatocellular carcinoma and investigation of its regulatory network via comprehensive analysis. Bioengineered. 2021;12(1):7154–7166.
- Hu Y, Yang Z, Bao D, et al. miR-455-5p suppresses hepatocellular carcinoma cell growth and invasion via IGF-1R/AKT/GLUT1 pathway by targeting IGF-1R. Pathol Res Pract. 2019;215(12):152674.
- Jing H, Chen P, Hui T, et al. Synapse-specific Lrp4 mRNA enrichment requires Lrp4/MuSK signaling, muscle activity and Wnt non-canonical pathway. Cell Biosci. 2021;11(1):021–00619.
- Weatherbee SD, Anderson KV, Niswander LA. LDL-receptor-related protein 4 is crucial for formation of the neuromuscular junction. Development. 2006;133(24):4993–5000.
- Asai N, Ohkawara B, Ito M, et al. LRP4 induces extracellular matrix productions and facilitates chondrocyte differentiation. Biochem Biophys Res Commun. 2014;451(2):119–131.
- Mao Z, Wang Z, Zhang S, et al. LRP4 promotes migration and invasion of gastric cancer under the regulation of microRNA-140-5p. Cancer Biomarkers. 2020;29(2):3666–3679.
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15(12):014–0550.
- Chen H, Jin G. Downregulation of Salusin-β protects renal tubular epithelial cells against high glucose-induced inflammation, oxidative stress, apoptosis and lipid accumulation via suppressing miR-155-5p. Bioengineered. 2021;12(1):6155–6165.
- Yin Z, Zhou Y, Ma T, et al. Down-regulated lncRNA SBF2-AS1 in M2 macrophage-derived exosomes elevates miR-122-5p to restrict XIAP, thereby limiting pancreatic cancer development. J Cell Mol Med. 2020;24(9):5028–5038.
- Ye Y, Huang S, Wu Y. UNBS5162 and amonafide inhibits tumor progression in human melanoma by the AKT/mTOR pathway. Cancer Manag Res. 2019;11:2339–2348.
- Yao Y, Chen X, Yang H, et al. Hsa_circ_0058124 promotes papillary thyroid cancer tumorigenesis and invasiveness through the NOTCH3/GATAD2A axis. J Exp Clin Cancer Res. 2019;38(1):019–1321.
- Ma Y, Chen Y, Lin C, et al. Biological functions and clinical significance of the newly identified long non‑coding RNA RP1‑85F18.6 in colorectal cancer. Oncol Rep. 2018;40(5):2648–2658.
- Li D, Tang X, Li M, et al. Long noncoding RNA DLX6-AS1 promotes liver cancer by increasing the expression of WEE1 via targeting miR-424-5p. J Cell Biochem. 2019;120(8):12290–12299.
- Zhang R, Zhu W, Ma C, et al. Silencing of circRNA circ_0001666 represses EMT in Pancreatic cancer through upregulating miR-1251 and downregulating SOX4. Front Mol Biosci. 2021;8(684866).
- Zhang Z, Wang S, Yang F, et al. LncRNA ROR1‑AS1 high expression and its prognostic significance in liver cancer. Oncol Rep. 2020;43(1):55–74.
- Jiao Y, Li Y, Ji B, et al. Clinical value of lncRNA LUCAT1 expression in liver cancer and its potential pathways. J Gastrointestin Liver Dis. 2019;28(4):2648–2658.
- Sheng LQ, Li JR, Qin H, et al. Blood exosomal micro ribonucleic acid profiling reveals the complexity of hepatocellular carcinoma and identifies potential biomarkers for differential diagnosis. World J Gastrointest Oncol. 2020;12(10):1195–1208.
