ABSTRACT
Combating climate change and ensuring energy supply to a rapidly growing global population has highlighted the need to replace petroleum fuels with clean, and sustainable renewable fuels. Biofuels offer a solution to safeguard energy security with reduced ecological footprint and process economics. Over the past years, lignocellulosic biomass has become the most preferred raw material for the production of biofuels, such as fuel, alcohol, biodiesel, and biohydrogen. However, the cost-effective conversion of lignocellulose into biofuels remains an unsolved challenge at the industrial scale. Recently, intensive efforts have been made in lignocellulose feedstock and microbial engineering to address this problem. By improving the biological pathways leading to the polysaccharide, lignin, and lipid biosynthesis, limited success has been achieved, and still needs to improve sustainable biofuel production. Impressive success is being achieved by the retouring metabolic pathways of different microbial hosts. Several robust phenotypes, mostly from bacteria and yeast domains, have been successfully constructed with improved substrate spectrum, product yield and sturdiness against hydrolysate toxins. Cyanobacteria is also being explored for metabolic advancement in recent years, however, it also remained underdeveloped to generate commercialized biofuels. The bacterium Escherichia coli and yeast Saccharomyces cerevisiae strains are also being engineered to have cell surfaces displaying hydrolytic enzymes, which holds much promise for near-term scale-up and biorefinery use. Looking forward, future advances to achieve economically feasible production of lignocellulosic-based biofuels with special focus on designing more efficient metabolic pathways coupled with screening, and engineering of novel enzymes.
Graphical Abstract
1. Introduction
Climate change, environmental degradation, and pollution-related health problems in the population have all been linked to the increasingly excessive consumption of fossil fuels by the transportation sector, globally [Citation1]. These issues have diverted industry, decision-makers, and scientists’ attention towards generating alternative fuels from renewable sources [Citation2,Citation3]. The United Nations (UN) has also introduced affordable and clean energy as a prime concern among Sustainable Development Goals (SDG) and set different targets to make it available for everyone without competing for future generations [Citation4]. Most recently, an inclusive focus was given on greener transport vehicles to cut down the net carbon emissions and global warming in the 26th annual summit of COPs (Conference of the Parties) held in Glasgow, United Kingdom [Citation5].
Biofuels primarily produced using bio-based materials, i.e., starch, sugar, lignocellulose, animal fats, and other biopolymers, are considered cost-effective and environmentally benign alternatives to petroleum-based fuels impoverishment [Citation6,Citation7]. Lignocellulose is deemed the cheapest and most abundant biopolymer among the available bio-based materials on the earth [Citation8,Citation9]. Several studies have reported bioethanol, biodiesel, and other valuable petrochemicals, that is, butanol, isopentanol, terpenes, etc., production from lignocellulosic biomass (LCB). Its feasibility has been hampered by the inherent recalcitrance of the biomass [Citation10–12]. The increased understanding of cell wall polysaccharides and/or lignin biosynthesis and recent advances in metabolic engineering have enabled the possibility of increased digestibility of the LCB with the improved release of fermentable sugars. Lipid biosynthesis and storage engineering are also advantageous to enhance biofuel production [Citation13,Citation14].
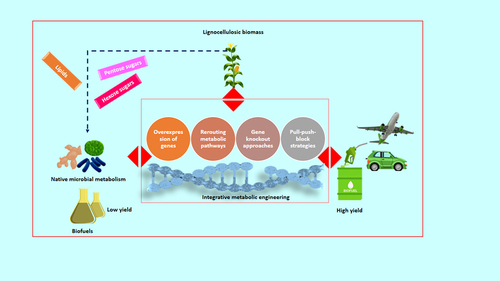
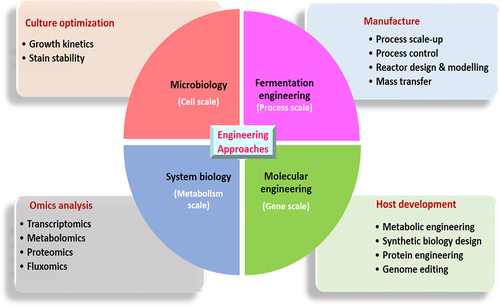
The microbial-assisted bioconversion processes greatly affect the cost-competitive biofuel production [Citation15]. It seems that the capital and operational cost associated with the microbial bioconversion process are the highest contributors to the overall cost production of biofuels [Citation16]. Several native and non-indigenous microorganisms can catalyze lignocellulose substrates (LCS) into a broad array of biofuels. Still, its feasibility directly relies on the inherent metabolic facet of that particular microorganism [Citation17,Citation18]. Nevertheless, some of these substances or their precursors can also be synthesized from distinct metabolic pathways occurring naturally in microorganisms. The ideal microorganism/strain for sustainable scale-up and biorefinery use should utilize various substrates and produce a higher yield of end products and better sturdiness against inhibitors [Citation19]. These metabolic shortcomings can be addressed by tailoring or redesigning the metabolic facets of each microbe/stain. Substantial progress has been made in this regard during the last couple of years. Numerous engineering strategies were employed to design and optimize pathways in microorganisms for advancing biofuel production have been reconnoitred in . Furthermore, these approaches would help to improve the substrate spectrum and metabolic fluxes towards biofuel pathways, as well as enhance the possibility of high-yielding phenotypes [Citation20–22]. In addition, cell surface display engineering has enabled precise modification of surface enzymes to enhance the biocatalytic capabilities of microbial cells. This contributes greatly to the expansion of biofuels production from lignocellulose substrate [Citation23,Citation24]. Through genome editing, the ability to create new robust industrial phenotypes with high volumetric productivity, improved substrate utilization, and increased inhibitor tolerance has been demonstrated [Citation25,Citation26]. Recent efforts to develop genetically modified microorganisms or recombinant strains are under intense development. A number of microbial platforms and strains with actual scale-up features have been patented or are in the process of being accepted [Citation27–29].
Figure 1. Different levels of engineering approaches at microbes for enhanced production of Biofuels.
This paper aims to review the metabolic engineering strategies used to produce robust microbial workhorses that use LCB as the primary feedstock for biofuel production. In recent years, there have been a number of breakthroughs in this holistic approach, mainly focusing on the different microbial domains and their metabolic pathways that produce biofuels. These are described in detail in this article, along with considerations of future development opportunities.
2. Recent metabolic advances in the lignocellulosic feedstock
2.1. Cell wall engineering
The natural recalcitrance of LCB is a considerable barrier to biofuel sustainability. To overcome this problem, researchers have previously focused on disrupting structural and compositional complexity through distinct strategies, such as pre-treatments and consolidating bioprocessing [Citation30,Citation31]. However, such methods effectively solubilize the lignocellulose polymers into fermentable sugars but employed several intricacies, that is, destruction of valuable sugars fraction, generation of inhibitor compounds, and irreversible salts, which can make downstream processes complex and expensive [Citation16,Citation32]. Recent advances in metabolic engineering have enabled the altered synthesis of the various wall polymers to increase polysaccharide accessibility and reduction of polymer-derived processing inhibitor, along with high hexose/pentose ratio in the biomass [Citation33].
Cellulose is a major homopolysaccharides in the plant cell wall. Additionally, it consists entirely of glucose, an indispensable substrate for producing biofuels [Citation34]. Significant efforts have been devoted to cellulose biosynthesis and remodeling in the lignocellulose feedstock for the purpose of cost-effective biofuel production. The expression regulation of genes including cellulose synthase (CesA), sucrose synthase (SuSy), glycosyltransferases (GT), and glycosyl hydrolases (GH) has resulted in drastic changes in cellulose content, crystallinity, and total biomass as well [Citation35–37]. Recent work in transgenic poplar has demonstrated that overexpression of PdDUF266A dramatically improved cellulose content (up to 37%) and total biomass (17–34%) than wild type [Citation38]. Furthermore, the cellulose depolymerization and ratio of the total sugar release after enzymatic saccharification were also increased up to 13% and 38%, respectively, as compared to control. In a different investigation, overexpression of the PsnSuSy2 gene (derived from hybrid poplar) in tobacco increased its cellulose content by 18%, and decreased cellulose crystallinity by 11%, when compared with control [Citation39].
Similarly, the genes involved in synthesizing mixed-linked glucan have been explored to improve hexose contents as β-1, 3 bonds can prevent the aggregation of cellulosic components and render the cell wall more digestible. Recent work has shown that overexpression of cellulose synthase-like F6 (from rice) gene in Arabidopsis can increase glucose accumulation in the matrix cell wall fraction by four times and saccharification by up to 42% compared to control lines [Citation40]. In a different investigation, overexpression of UDP-rhamnose/UDP-galactose Transporter1 (URGT1) together with galactan synthase (GalS1) and glucose epimerase (UGE2) increased stem galactose levels to four times than wild-type plants [Citation41].
Also, some recent efforts targeting xylan backbone (comprises a linear chain of b-1, 4-D-xylosyl residues) to alter recalcitrance and increase saccharification efficiency, but there are trades in many cases offs to be considered [Citation13,Citation42]. In planta, the expression of various enzymes involved in xylan biosynthesis (i.e., acyltransferase, ferulic acid esterases, etc.) can be the attractive targets to facilitate the biomass saccharification efficiency [Citation43,Citation44]. Recently, overexpression of the rice OsAT10 (a BAHD acyltransferase gene) in switchgrass enhances saccharification efficiency up to 40% [Citation45]. Similarly, OsAT10 overexpression increases saccharification efficiency in sorghum OsAT10 lines [Citation46].
Lignin, the end product of the phenylpropanoid pathway, is the foremost cause behind the inherent recalcitrance of the LCB [Citation47]. Recently, substantial progress has been made to manipulate the genes encoding enzymes of the phenylpropanoid pathway, such as phenylalanine ammonia-lyase, cinnamate 4-hydroxylase, hydroxycinnamoyl-CoA shikimate/quinate, coniferyl aldehyde dehydrogenase, cinnamyl alcohol dehydrogenase, laccase, and more [Citation48]. The overexpression or down-regulation of related genes significantly reduces the lignin contents and facilitates the lignocellulose saccharification efficiency [Citation49–51]. Recently, overexpression of rPvGRF9 genes significantly increased per plant sugar yield with less lignin content in switchgrass [Citation52].
Nevertheless, these recent efforts help to modulate plant cell wall components considerably. Still, its compositional disparities among the species and tissue often need to be optimized or redesigned to facilitate lignocellulose quality for conventional and future biorefinery use.
2.2. Lipid engineering
The increased understanding of lipid biosynthesis and storage has also enabled the possibility of cost-effective biofuel production, primarily biodiesel, and more recently jet fuel [Citation53]. Lipids are energy-dense biomolecules frequently stored as triacylglycerols (TAGs) in the plants’ seeds and non-seed tissues (vegetative tissues). The TAGs currently utilized for biodiesel production are mainly derived from edible seed feedstock. Several food crops, such as corn, soybean, oilseed crops, that is, Camelina sativa, Jatropha curcas, etc., have been established for high seed oil content thorough engineering of lipid biosynthetic pathways and genes [Citation54,Citation55]. At the same time, established pre-eminence of food versus fuel debates, it is unlikely the TAGs production from current oilseed feedstock will contribute substantially to the biofuels industry in the future. The vegetative tissues (i.e., leaf, stem, etc.) that constitute most of the plant biomass are also capable of synthesizing TAGs, but their amount typically accounts for only a minor portion of total lipids [Citation56,Citation57].
For the last couple of years, metabolic engineering to increase the TAGs biosynthetic capacity among vegetative tissues has attracted wider attention as a sustainable oil/lipid yielding platform for lignocellulosic biorefinery [Citation58]. The attempts to encourage the production of TAG in vegetative tissue have mainly focused on the integrated concept (pull, push, package, and protect) of single or multiple genetic interventions [Citation59–69], resulting in augmented TAG levels with better compositional facets in distinct non-food crops (). However, in a model plant, Tobacco, the highest TAG yield (30–33% DW) to date was achieved by combinatorial optimization of TAG biosynthetic pathways [Citation67], but it is not always straightforward as these pathways are highly conserved across different species [Citation70].
Table 1. Summary of some important metabolic engineering approaches to increase oil/lipid synthesis in vegetative tissues in non-food crops
Several avenues might be explored for utilizing vegetative tissues as high-yielding TAG platform for biofuels, including (a) modulation of lipid transporters (i.e., plastidial lipid transporter TGD5 (trigalactosyldiacylglycerol5) and fatty acid exporter 1 (FAX1); peroxisomal transport protein CTS (comatose), etc.) to optimize the lipid fluxes between the subcellular compartments, (b) development of more specific promoters (i.e., vegetative tissue-specific, developmental stage-specific, etc.) to maximizing the unusual fatty acids yields and minimizing the adverse phenotypic effects, (c) engineering of lipids metabolism and lipid droplets (LDs) for accumulation of high-value neutral lipids, that is, wax esters, volatile lipids, etc., and to enhance the energy density over LDs through the engineering of extracellular surface (cuticle) of a given plant without any adverse effects on plant growth performance [Citation71–73].
However, a recent techno-economic analysis proposed that engineered hemp (Cannabis sativa) with a minimum of 10% lipid content can produce up to 326 gallons of total biofuels per hectare of agricultural land than soybean with the production cost of USD 4.13/gallon [Citation74].
3. Overview on major metabolic pathways of microorganism for biofuel productions
Both pentose and hexose sugars (i.e., xylose, arabinose, glucose, etc.) derived from the LCS are the prime carbon source for the production of biofuels by industrially tractable microbes, such as yeasts, Escherichia coli, Clostridium spp., and more [Citation75]. These microorganisms have distinct catalytic enzymes and display specific metabolic paths to produce distinct biofuels, including higher alcohols and hydrocarbons [Citation76,Citation77]. In general, microbes use xylose and glucose as initiating molecules for the anaerobic synthesis of ethanol via Entner-Doudoroff (ED-P) and Embden-Mayer Hoff-Parnas (EMP-P) pathways [Citation78]. The higher alcohols include linear-chain alcohols and branched-chain alcohols can be synthesized via either fatty acid or amino acid pathways. Besides, fatty acid metabolism is imperative for producing linear-chain alcohols, while amino acid metabolism is advantageous for branched-chain alcohols [Citation79].
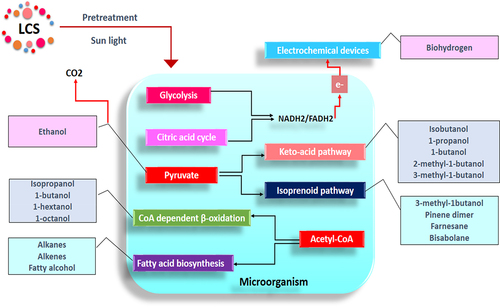
Several detailed reports have been published on molecular engineering and system biology strategies employed to design and optimize biofuel production microbial metabolic pathways [Citation80–82]. Much of the reports highlighted that the advanced biofuel production could be achieved mainly through four major metabolic pathways: (1) the keto-acid pathway, (2) the isoprenoid pathway, (3) fatty acid synthesis pathway, and (4) CoA dependent β- oxidation pathway or reverse b-oxidation pathway. Pyruvate is the precursor molecule for initiating the keto-acid pathway used to produce isobutanol, 1-propanol, 2-methyl-1-butanol, and more. It is also a precursor molecule of the isoprenoid pathway used to produce 1-isopropyl-4 methylcyclohexane, pinene dimer, farnesane, and bisabolane. Similarly, acetyl-CoA is the precursor molecule for initiating the fatty acid synthesis pathway and CoA-dependent β- oxidation pathway. The fatty acid synthesis pathway is advantageous for producing alkanes, alkenes, and fatty alcohols, while the CoA-dependent β- oxidation pathway is for isopropanol, 1-butanol, and more (). Most recently, the polyketide biosynthetic pathway mediated by polyketide synthases (PKSs) and cognate thioesterase (TE) has also been tweaked for the production of alkanes and alkenes [Citation60,Citation83].
4. Recent metabolic advances in microorganism for biofuel production
4.1. Bacteria
Bacteria are promising hosts for the generation of a variety of fuel-like compounds. These have also been given preference to metabolic pathways manipulation because of their substrate suitability, fast growth rate, and well-tractable physiology and genetics. The user-friendly hosts, that is, Zymomonas mobilis and E. coli, Clostridium, etc., have been the center of recent metabolic engineering studies to develop a model microbial chassis for sustainable biorefinery practices in the modern era [Citation84].
The Z. mobilis is a natural ethanologenic bacterium with excellent industrial features like high carbon substrate uptake rates, high ethanol tolerance (up to 16% v/v), and a low aeration cost due to anaerobic exposure fermentation [Citation85]. The main drawback of native Z. mobilis is its inability to utilize pentose sugars for ethanol production. Several recent metabolic engineering efforts have been undertaken to broaden the substrate spectrum of Z. mobilis via the introduction and efficient expression of heterologous genes coupled with utilization and assimilation of pentoses, such as xylose and arabinose. Some efficient pentose utilizing strains have been developed through combining metabolic engineering and adaptive laboratory methods. These recombinant strains reportedly harbored improved xylose utilization, co-utilization of mixed sugars and enhanced ethanol production [Citation86–88]. In addition, xylose transport has also been identified as a major barrier to the efficient utilization of xylose by Z. mobilis. The native Z. mobilis has been reported to transport xylose via Glf (Glucose facilitated diffusion protein) transporter. The Glf has a high affinity for glucose, causing competitive inhibition of xylose uptake in mixed sugar supplemented medium [Citation89]. Recently, some xylose-specific transporters, that is, XylE, and ABC type transporter, were successfully introduced in Z. mobilis to enhance the rate and extent of xylose uptake [Citation90,Citation91]. An experiment demonstrated , recently, that overexpression of ABC type transporter enhances the rate of xylose utilization by 48.9% as compared to parental stains in the presence of glucose [Citation91]. Recent efforts in Z. mobilis engineering have also enabled several strains to tolerate furfural and acetic acids, which are the predominant toxic inhibitors during lignocellulosic ethanol production [Citation92,Citation93]. Nouri et al [Citation94] developed recombinant strains with improved tolerance against the multiple inhibitors by overexpressing the genes hfq (a transcription regulator) and sigE (a transcription factor). Furthermore, strain engineered to express sigE also showed 2-fold and 4-fold higher ethanol yields than the hfq-overexpressing strain and parental strain, respectively. Recently, a dioxygenase coding gene (ZMO1721) was overexpressed to develop a recombinant strain resistant to many phenolic aldehydes, including 4-hydroxybenzaldehyde, syringaldehyde, and vanillin[Citation95].
Other than ethanol, Z. mobilis possesses endogenous abilities to produce other fuel alcohols, such as isobutanol and 2, 3-butanediol. For the 2, 3-butanediol production, recombinant strains were successfully developed by expression of acetolactate synthase, acetolactate decarboxylase, and butanediol dehydrogenase encoding genes [Citation96,Citation97]. The best-enhanced strain was able to produce more than 10 g/l of 2, 3-butanediol from glucose and xylose as well as sugar streams derived from deacetylation and mechanical refining [Citation96]. Some recombinant strains have recently been developed for isobutanol production by the regulation of alcohol dehydrogenase (adhA) and 2-ketoisovalerate decarboxylase (kivd2) [Citation98]. Recently, a maximum of 4.0 g/l of isobutanol was achieved by the overexpression of gene kivd2 combined with a synthetic heterologous operon als-ilvC-ilvD[Citation99].
E. coli is another bacterium that has been extensively investigated for biofuel production. It is naturally capable of utilizing both pentose and hexose, and is amenable to produce ethanol by an endogenous ethanologenic pathway. A disadvantage of this pathway is that only 1 mole of ethanol can be produced from 1 mole of glucose [Citation100]. This yield is relatively inefficient as compared to the homeoethalogenic pathways of Zymomonas and Saccharomyces species. Due to this, initial engineering effort has been devoted to modify existing E. coli pathways for ethanol production [Citation101]. Several recombinant strains have recently been developed by introducing relevant foreign genes, such as pyruvate decarboxylase (pdc), alcohol dehydrogenase (adh), and more. The resulting engineered strains were able to effectively direct carbon flux to higher ethanol production [Citation102]. The glycolytic pathway of E. coli KO11 was recently modified through knocking out phosphoglucose isomerase (pgi) to redirect carbon flux from glucose via ED-P and PP-P pathways [Citation103]. (Huerta‐Beristain et al. 2017). Further engineering KO11 by the adaptive evolution and deletion of pta, ack, and ldh genes provided the strain KO11 PPAL, which led to higher yield of ethanol. Recently, the intracellular NADH/NAD+ ratio in the E. coli KO11 was improved by the overexpression of formate dehydrogenase from Mycobacterium vaccae. The enhanced stains produced more than 36% ethanol as the original KO11 strain in culture for 24 h [Citation104]. Apart from this, simultaneous utilization of both xylose and glucose is also a considerable barrier towards effective ethanol production. In recent research, E. coli MG1655 was engineered through the combined genetic and evolutionary engineering to utilizing cellobiose and xylose simultaneously. The recombinant strain was capable of utilizing approximately 6 g/L of cellobiose and 2 g/L of xylose in approximately 36 h [Citation105]. A strain GX 50 was also engineered by transposon-mediated mutagenesis and metabolic evolution to enhanced xylose utilization in the presence of glucose [Citation106].
Recently, recombinant E. coli B0013-2021HPA was engineered through introducing a mutation to pyruvate–glucose phosphotransferase system (ptsG). The enhanced strain was able to utilize almost all xylose, galactose as well as arabinose from lignocellulose hydrolysates and efficiently convert complex substrate mixtures to ethanol at 42°C under oxygen-limited fermentation conditions [Citation107]. Recent effort has also been devoted to engineered E. coli strains with marked tolerance against the hydrolysate toxins, ethanol, and byproducts [Citation108–110]. In recent research, E. coli strain LY180 was constructed by introducing multidrug resistance pumps, such as SugE and MdtJI [Citation111]. The recombinant stain reportedly harbored furfural efflux and improved ethanol production in the presence of furfural or 5-hydroxymethylfurfural. More recently, a recombinant stain was developed through the combined adaptive laboratory evolution and CRISPR-enabled trackable genome engineering. The evolved strain could tolerate up to 4.7 g/L furfural and also exhibited marked cross tolerance towards end products including isobutanol, butanol, and ethanol as well as NaCl, and high temperatures [Citation112].
In addition to ethanol, recent metabolic engineering efforts have also been devoted to develop E. coli strains for the production of isobutanol, isopropanol, and other bioalcohols. Several recombinant strains have been recently constructed with a goal of enhanced bioalchohals yield and tolerance [Citation113,Citation114]. Recently, a redesigned quorum sensing system combined with a metabolic toggle switch (QT-MTS) has been employed within E. coli model to redirect metabolic flux toward the target synthetic pathway [Citation115]. The strain harboring MTS was reported to have 26-folds improvement in the intermediate pyruvate and final isobutanol production titer as compared control strain.
Clostridium sp have also been metabolically engineered for the production of butanol and higher alcohols [Citation116]. Recently, wild C. acetobutylicum strain was engineered by the disrupting hydrogenase gene hydA. The recombinant strain was able to produce 18.3% more butanol with by-product acetone decreased by 31.2%. Further exogenous supplementation of methyl viologen enhanced the butanol yield up to of 0.28 g/g from the corn stover [Citation117]. In a different investigation, recombinant Clostridium cellulovorans adhE2 strain was engineered through the introduction of thiolase (thlACA) from C. acetobutylicum and 3-hydroxybutyryl-CoA dehydrogenase (hbdCT) from C. tyrobutyricum [Citation118]. The engineered C. cellulovorans strain was able to n-butanol from cellulose at a 50% higher yield (0.34 g/g). In addition, Clostridium spp. have also been metabolically engineered to produce biohydrogen [Citation119]. Recently, recombinant C. acetobutylicum strains were developed by the overexpression of glucose-6-phosphate dehydrogenase and FeFe hydrogenase. The engineered strain was able to produce more than 1.4-fold higher ethanol yield than the wide type [Citation120].
The fastest-growing bacterium, Vibrio natriegens, is recently engineered for heterologous production of 1, 3-Propanediol from glycerol. Systematic engineering of cellular transcriptional regulators and glycerol oxidation pathway leads to the 2.36 g/L/h output production of 1, 3-Propanediol [Citation121]. The Cupriavidus necator H16 is an attractive living system, which can be metabolically engineered coupled with a gene knockdown process for directing carbon flux away from producing Poly[(R) −3 Hydroxybutyrate] and resulting in the production of biofuels like products [Citation122]. Similarly, in Clostridium ljungdahii, an artificial isopropanol producing pathway was constructed through which this bacterium can efficiently use syngas and coproduce isopropanol 3-hydroxybutyrate and ethanol [Citation123].
Methanotrophic bacteria are also an important source of bioproducts for biofuel production. Systems metabolic engineering of such bacterium has led to the advancement in methane-based bio-manufacturing of biofuels [Citation124,Citation125]. Some recent achievements by bacterial engineering are summarized in .
Table 2. Summary of major achievements in increasing biofuel production by engineering bacterial pathways
4.2. Cyanobacteria
Cyanobacteria are the photosynthetic prokaryotes with more potent photosynthetic metabolomics than land plants. This metabolomics is advantageous to make them potential cell factories, which can efficiently produce biofuels and bio-based chemicals through sequestering of carbon. Also, its well-tractable physiology and more specific growth requirements attract attention to be engineered for industrial purposes [Citation126].
Cyanobacteria are a good choice for solar-powered bioethanol production due to the presence of ethanologenic pathway [Citation127]. Consequently, sufficient efforts have been devoted to engineering the existing pathways of some model cyanobacteria species, that is, Synechocystis and Synechococcus for improved ethanol production [Citation128,Citation129]. Several recombinant strains have been developed by employing the gene dosage, induced expression as well as cassette optimization. These engineered strains were able to produce up to 0.5 g/l ethanol per day [Citation130–132].
In recent research, recombinant Synechocystis PCC 6803 strains were constructed through induced expression of four Calvin-Benson-Bassham (CBB) cycle enzymes, i.e., transketolase (TK), ribulose-1, 5-bisphosphate carboxylase/oxygenase (RuBisCO), aldolase (FBA), and fructose-1, 6/sedoheptulose-1, 7-bisphosphatase (FBP/SBPase) coupled with ethanol synthesis enzymes PDC and ADH [Citation23]. The engineered strain that overexpressed the CBB cycle enzymes was able to produce 37–69% more ethanol and 7–10% more total biomass than the strain that expressed only the ethanol biosynthesis pathway. In a different investigation, ethanol production of the Synechocystis PCC 6803 was increased by 2-9-folds by combined expression of multiple native CBB enzymes [Citation133]. More recently, a recombinant S. elongatus PCC7942 strain was developed through induced expression the ictB, ecaA, and groESL, and pdc-adhII genes [Citation134]. The engineered strain was able to improve ethanol production and cell growth under a stimulated flue gas consisted CO2 (25%), SO2 (80–90 ppm) and NO (90–100 ppm).
Recent efforts have also been directed towards engineering cyanobacteria for the production of isobutanol and 1-butanol. Numerous recombinant strains of Synechocystis and Synechococcus were recently constructed through the introduction, overexpression, and evaluation of different genes related to isobutanol and 1-butanol biosynthesis [Citation135,Citation136]. Recently, a recombinant Synechocystis PCC 6803 strain was developed through the introduction of the genes kdc (keto-acid decarboxylase) and adh under the control of the CcaS/CcaR system. The enhanced strain was able to produce 238 mg/l of isobutanol and 75 mg/l of 3-methyl-1-butanol under red and green light illumination in 5 days [Citation137]. In a different investigation, the introduction of NADH-dependent nitrate assimilation in Synechococcus PCC 7002 significantly enhanced the photosynthetic production of 2-methyl-1-butanol and isobutanol [Citation138]. Also, metabolic profiles of high salinity stress engineered S. elongatus revealed that enhanced isobutanol production was caused by lipid degradation with the increase in NADH. The engineered strain proved to be a practical and feasible system for cost-effective isobutanol production [Citation139]. Another strain of S. elongatus PCC 11801 has recently evolved in laboratory conditions through adaptive tolerance to higher concentrations of butanol [Citation140]. Notwithstanding the above-mentioned advances, cyanobacterial bioethanol and higher alcohols require further improvement to become economically viable at an industrial scale.
The Synechocystis has recently been metabolically engineered to produce 1-Octanol, one of the emerging precursor molecules for high-value product formation, including fuel. The most efficient computational tools have been proved to be useful in determining appropriate cleavage sites of thioesters in the above cyanobacteria, which further play an important role in selecting 1-octanol [Citation141]. Similarly, isobutene (a gaseous fuel) production in Synechocystis sp. PCC 6803 has been enhanced by the introduction of the α-ketoisocaproate dioxygenase gene from Rattus norvegicus (RnKICD), which resulted in optimization of the isobutene pathway [Citation142]. Also, the industrial scale-up of terpenoids through sequential heterologous expression of bottleneck enzymes of Methylerythritol 4-phosphate pathway in Synechocystis sp. PCC 6803 has been achieved by Rodrigues and Lindberg [Citation143].
In addition, large number of cyanobacteria are naturally capable of producing biohydrogen, which can be mainly synthesized by: (a) the action of bidirectional or reversible hydrogenase and (b) as a by-product of nitrogen fixation via nitrogenases [Citation144]. Many novel hydrogen-producing strains have been obtained under various culture conditions in recent years [Citation145,Citation146]. Numerous efforts have been attempted to improve H2 yield by modulating both nitrogenase and hydrogenase-based H2 production systems [Citation147,Citation148], but it requires further improvement for economic H2 production at the industrial scale. summarizes other efforts performed to improve the biofuel production potential of different cyanobacteria.
Table 3. Major engineering efforts performed to improve the biofuel production capacity of different cyanobacteria species/strains
4.3. Yeasts
Yeasts, including native, thermotolerant, halophilic, etc., have attracted much attention as cell factories convert lignocellulose to biofuels and bio-products. The large spheroidal cells, simple bioprocessing, minimal nutritional requirements, and better inhibitor tolerance relative to many bacteria and cyanobacteria can expand this scenario [Citation149]. Although Saccharomyces cerevisiae has been the promising host for ethanol production from the earliest time, it cannot efficiently utilize the sugar polymers, such as xylose, and cellobiose available in LCB [Citation81]. Also, glucose repression of xylose fermentation and toxic fermentation inhibitors in lignocellulosic hydrolysates adversely affects productivity and yield. Despite the competitive production costs and ideal utilization of substrate, rapid sugar metabolism after eliminating ethanol production is advantageous to producing advanced biofuels and biochemicals [Citation150].
Several metabolic engineering strategies to address the issues mentioned above have been undertaken and implemented to yeast in the last couple of years (). For instance, engineering the PPP and XAP pathways (either oxidoreductase or isomerase-based) has been reported to maximize xylose utilization and alleviate glucose repression on xylose fermentation [Citation151]. Hoang et al. [Citation152] constructed an efficient xylose-fermenting S. cerevisiae strain through combinatorial CRISPR–Cas9-mediated rational and evolutionary engineering. This isomerase-based xylose-fermenting strain, named XUSE, demonstrates the efficient conversion of xylose into ethanol with a high yield of 0.43 g/g and exhibited simultaneous fermentation of glucose and xylose with negligible glucose inhibition. Significantly, higher ethanol yields were also achieved by engineering the high osmolarity glycerol pathway [Citation153]. Recently, the highest ethanol yield (0.492 g/g total sugars) from lignocellulosic hydrolysates was achieved by overexpressing a mutant SFA1in S. Cerevisiae [Citation154].
Figure 3. Metabolic engineering of yeast (S. cerevisiae) for the production of biofuels from lignocellulosic sugars, PPP = pentose phosphate pathway, HLXM = heterologous xylose metabolic pathways, HLM = heterologous metabolic pathway, HLCM = heterologous cellobiose metabolic pathways.
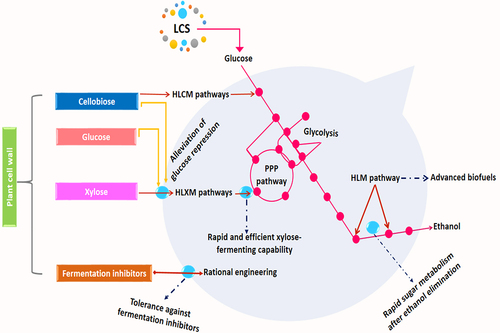
Studies also focused on transporter and transcription factor engineering to develop more powerful ethanol-producing strains as yeast lacks xylose-specific transporters. Some novel xylose transporter, such as XltA (from Aspergillus niger) and Xut3 (from Scheffersomyces stipitis), were successfully introduced into yeast to maximize xylose uptake [Citation155]. Soon after, the directed evolution of xylose-specific transporter AN25 from Neurospora crassa was found suitable for improved xylose transportation and glucose-xylose co-utilization [Citation156]. Improved xylose uptake and consumption with higher ethanol yield were obtained through the directed or adaptive evolution of Nuclear-Localized Hexokinase 2(HXK2) and endogenous hexose transporters (HXT2) and CYC8 [Citation157,Citation158]. More recently, Dzanaeva et al. [Citation159] evaluated the impact of transcription factors Znf1, Sip4, Adr1, Tup1, and Hap4 on xylose catabolism in the xylose-fermenting strain of S. cerevisiae. The increase of ethanol production from xylose compared to that of parental strain confirms their involvement in regulating xylose growth and fermentation.
Prompted by the accomplishments with ethanol producer S. cerevisiae, the nonconventional yeast Spathaspora passalidarum, and Pichia stipites have also been explored for natural and engineered xylose metabolism to improve bioethanol production [Citation160]. These yeasts have several novel alcohol dehydrogenase (ADH) encoding genes (PsADH1 to PsADH7 and SpADH1) essential for xylose assimilation and ethanol production. Interestingly, PsADH1 shows a broad spectrum for substrate-specificity and can also utilize xylitol as a substrate with moderate activity [Citation161]. The oleaginous yeast, Yarrowia lipolytica, which only has native pentose-specific transporters (TRP6 and TRP22), is also being exploited to engineer xylose metabolism. Overexpression of these native transporters with xylitol dehydrogenase improves dxylose utilization and metabolism [Citation162]. This species has extensively been used to engineer lipid accumulation to enhance the biosynthesis of biodiesel and other lipid-derived products [Citation163,Citation164]. The subcellular engineering of lipase-dependent pathways in Y. lipolytica resulted from 14-folds increased lipid titer [Citation165]. Yook et al. [Citation166] developed an efficient xylose-utilizing Y. lipolytica strain through CRISPR–Cas9-mediated rational and evolutionary engineering. This isomerase-based xylose xylose-utilizing strain, YSXID, produced 12.01 g/L lipids with a maximum yield of 0.16 g/g, the highest ever reported, from lignocellulosic hydrolysates. In the oleaginous yeast domain, Rhodosporidium toruloides can also utilize carbon sources (apart from sugars) available in lignocellulosic hydrolysates, such as organic acids lignin-derived phenolic compounds [Citation167,Citation168]. Some engineering has been implemented on this species to improve lipid conversion and productivity. Diaz et al. [Citation169] implemented combining evolutionary and metabolic engineering to R. toruloides and reported high lipid yield (0.179 g/g) with non-detoxified lignocellulosic hydrolysates. Increased lipid production was also observed in R. Toruloides strains by overexpressing gene encodes Δ9 fatty acid desaturase (Δ9FAD), which synthesize palmitoleic and oleic acids [Citation170]. Recently, Chopra et al. [Citation171] reported increased lipid production via applying diverse cultivation practices to R. toruloides strains.
In addition, the robustness of yeast strains against toxic fermentation inhibitors (i.e., lignin-derived phenolics, organic acids, and sugar degradation products) present in lignocellulosic hydrolysates is crucial for the sustainability of biofuels. The robustness of model yeast, S. cerevisiae, has been extensively improved through several metabolic and evolutionary engineering approaches [Citation172]. The tolerance of other ethanol-produced yeasts such as S. passalidarum and S. stipites to fermentation inhibitors has not been as extensively studied as that of S. cerevisiae. Although their robustness relies heavily on the type and level of inhibitors present in the hydrolysates [Citation173,Citation174], these species display substantial tolerance. The robustness of Y. lipolytica to lignocellulosic fermentation inhibitors has also been improved through metabolic engineering approaches [Citation175,Citation176]. The overpressing of the native genes XR, XDH, and XK also improved tolerance against formic acid, furfural, and coniferyl aldehyde [Citation177]. The engineering efforts were performed to improve the overall biofuel production capacity of diverse yeast strains ().
Table 4. Summary of metabolic/genetic engineering approaches to advancing biofuel production from lignocellulosic hydrolyzates by yeasts strain
4.4. Fungi
Unsaturated fatty acids are desirable starting material for production of biodiesel. Ascomycota and Mucoromycota are major phyla comprising of oleaginous filamentous fungi capable of production of single-cell oils approximately 15–36% cell dry weight. Single-cell oils rich in monounsaturated fatty acids and saturated fatty acids can be utilized for production of third generation biodiesel. So, lots of work has been done to optimize the culture conditions for maximum production of single-cell oils from Mortierella, Mucor, and Aspergillus Cunninghamella [Citation178]. Several metabolic engineering strategies for increased accumulation of lipid from oleaginous mucors have been adopted by several workers. 340 mg/l steariodonic acid was achieved by overexpression of fad3 gene (coding for fatty acid desaturase 3) in Mucor circinelloides [Citation179] Ashbya gossypii mainly used for industrial production of riboflavin, is being engineered for increased production of lipid droplets also. Lipid production can be increased by providing lipid precursor and blocking the other competing pathways of lipid accumulation. A knock out in AgPOX1 gene resulted in blocking of β-oxidation pathway and increase the yield (approximately 70% of its total cell dry weight) of total lipids, additionally the culture medium was supplied with 1% glucose and 2% oleic acid to prevent the above pathway completely [Citation180]. In the direction to increase the lipid accumulation some lignocellulosic hydrolysates, were also tested with 11 oleaginous fungal strains mainly including Mortierella, Aspergillus, and Cunninghamella. In the form of lignocellulosic hydrolysate substrate, mild sulphuric acid treated wheat straw provided sufficient xylose, on which Mortierella isabelline can grow and accumulate high amount of lipid (39.4%) [Citation181]. The fungus mainly accumulates long-chain fatty acids, while the medium chain fatty acids are desirable starting material for biofuel production. To overcome this problem one of the fungi Mucor circinelloides has been genetically modified. The β- oxidation pathway was modified by integrating heterologous acyl-ACP thioesterase into Fatty acid synthetase complex with a sequential knockout of acyl-CoA thioesterase and/or acyl-Co-A oxidase genes. A total increase in accumulation of medium chain fatty acids was reported around 47.45% in compare to wild strain, which can accumulate only 2.25% [Citation182,Citation183].
Nanostructures are very much important for immobilization of various fungal strain in packed bed reactors, to improve the yield of hydrolyzing enzymes. More than 90% yield of biodiesel was reported in Candida rugosa, Rhizomucor miehei, etc., through the immobilization of these strains in nanostructures [Citation184]. Besides Yeast, being a good eukaryotic system for metabolic engineering, other fungi such as Trichoderma spp. and Aspergillus spp. are main source of cellulase enzyme production at industrial scale. T. reesei has higher cellulase content but due to lack of β-glucosidase production co-culture with A. phoenicis gave a 2.5-fold increase β-glucosidase production [Citation185]. In a different investigation, repertoire of proteins in the secretome of a catabolite repressor-deficient strain of Penicillium funiculosum, PfMig1 88, was successfully evaluated to enhance the saccharification of sugarcane bagasse [Citation186].
5. Recent advances in microbial cell surface engineering for biofuel process
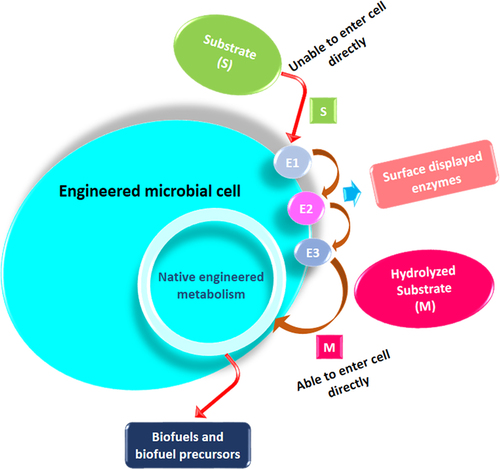
Microbial cell surface engineering is an innovative technique to endow novel functions on host cells through displaying functional proteins or enzymes on the cell surface [Citation187]. This approach has diverse strategies that involve communicating hydrolytic enzymes on the surface of microbial strains to degrade the LCS. Metabolically convert the degraded sugars directly into biofuels and biofuel precursors, thus elevating the status of microorganisms from immobilization stuff to a novel whole-cell biocatalyst (). Yeast, mainly S. cerevisiae, is the major mainstay of this approach. Its surface display was first explored to develop ‘arming yeast’ that can be a good platform for self-immobilized biocatalysts [Citation188]. Soon after, similar display systems were also employed to transform other yeast species, such as P. pastoris and Y. Lipolytica [Citation189,Citation190]. In the last couple of years, various studies demonstrated the significance of microbial cell surface display employed to design and optimize biofuel production from the LCS [Citation191–193]. It has been shown that the use of yeast surface display to the lignocellulolytic enzymes, such as endoglucanase, cellobiohydrolase, β-glucosidase, etc., exhibited better hydrolytic activities than of their free enzyme counterpart [Citation194,Citation195]. Substantial development has also been reported in the degree of synergy if these surface-displayed enzymes are used in combination for substrate hydrolysis [Citation196].
Interestingly, certain microbial clostridia can produce cellulosome, a complex of scaffolding proteins that requires multiple enzymes for efficient hydrolysis. Goyal et al. [Citation197] developed a functional display of trifunctional minicellulosomes on the S. cerevisiae. They obtained 3-fold higher ethanol production than a similar yeast consortium secreting only the three cellulases on phosphoric acid swollen cellulose. Similarly, enhancement of hydrolytic activity with improved ethanol titer has been reported while a bi-functional minicellulosome was accomplished on S. Cerevisiae [Citation198]. A cellulosome with a multiple-component assembly system was also developed through disulfide bonds. This cellulosome complex enabled yeast to ferment cellulose into ethanol proficiently [Citation192] directly. Recently, Dong et al. [Citation199] engineered P. pastoris to display minicellulosomes. The incubation of the protein-displaying yeast with three recombinant cellulases, including endoglucanase, an exoglucanase, and a β-glucosidase fused with a carbohydrate-binding module led to a cellulosome displaying P. pastoris able to direct conversion of carboxymethyl cellulose to bioethanol, observing an impressive ethanol titer of 5.1 g/L. The surface display engineering approaches employed on various microbial consortiums have been attempted to hydrolyze other lignocellulosic components, including xylan and lignin [Citation200–202]. summarizes the applications of microbial cell surface display during biofuel production.
Table 5. Applications of microbial cell surface display during biofuel production
6. Recently enabled technological advancement for biofuel processes
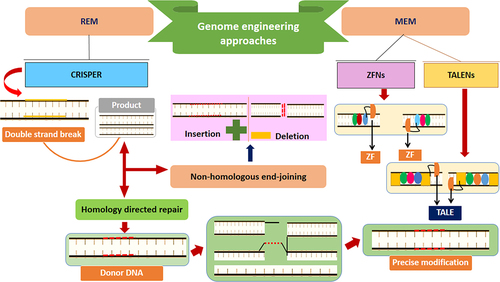
Quantification and regulation of the metabolic flux and metabolic pathway are crucial for optimizing the microbial biofuel production processes [Citation203]. Biosensors, genetically encoded compounds that convert input signals to a measurable output (i.e., gene expression, fluorescence, etc.), have been included recently among persuasive tools in the metabolic engineering field [Citation204,Citation205]. They have more comprehensive applications in biofuel production processes, from increasing substrate utilization and precursor accessibility to optimizing the product titers and activating pathways to in vivo monitoring of the target compounds [Citation206]. Several studies regarding metabolic engineering exploited biosensors to optimize the biofuel products, such as 2-ketoisovalerate [Citation207], isoprene [Citation208], fatty acids [Citation209], butanol and alkenes [Citation210], and fatty alcohol [Citation211]. More advanced gains in optimizing these processes can be achieved through efficient genetic engineering/editing tools, including CRISPR, to improve cell metabolism [Citation212]. There are two key genome editing tools/methods: modified endonuclease-mediated (MEM) engineering and RNA-guided endonuclease-mediate (REM) engineering (). Researchers in this field have experienced a dramatic revolution through such methods, and numerous challenges in the metabolic issues of biofuel production have been positively revolutionized during the last few years [Citation25,Citation213,Citation214]. Recent efforts have also been made to express new predictable and controllable multiplex genes and optimize such tools with high-throughput editing and efficiency of the genes [Citation215,Citation216].
Figure 5. Overview of genome engineering strategies for microbial biofuel production, MEM = modified en-donuclease-mediated engineering, REM = RNA-guided endonuclease-mediate, TALENs = transcription activator-like effector nucleases, ZFNs = zinc-finger nucleases, ZF = zinc-finger, TALE = transcription activator-like effector.
7. Recent patents on metabolic engineering of microorganism for biomass conversion to biofuels
Protection of intellectual property (IP) related to the strategies and methods to improve biofuel production by modifying microorganisms is under intense development. Several recombinant Z. mobilis or their allied species express xylanases that enable pentose sugars’ catabolism with improved ethanol production [Citation217,Citation218] and synthesize butanol and/or isomers of butanol [Citation29] have been patented. Recombinant yeast that can ferment pentose sugars [Citation219] restrained xylitol [Citation220] and that synthesize ethanol and fatty alcohols [Citation221,Citation222] have also been patented. The University of California, United States, recently filed a patent describing biofuel production by recombinant microorganisms [Citation223]. This patent describes metabolically modified microorganisms helpful in producing biofuels, more specifically 1-butanol, 1-propanol, 2-methyl-1-butanol, 3-methyl-1-butanol, and 2-phenylethanol from a suitable substrate. More recently, the trustees of three universities of the US (Princeton University, Massachusetts Institute of Technology, Whitehead Institute for Biomedical Research), and the Kyoto University of Japan filed a patent application [Citation224] that describes recombinant yeast that overproduces the heavy alcohol, mainly branched-chain alcohols. This yeast strain contains at least one deletion, disruption, or mutation from the individual GLN, VPS, GNP, AVT, GCN, or YDR391C gene families and combinations thereof. Some of the recent patents on the modification of microorganisms for advancing biofuel production (). In addition, Algenol Biofuels and Joule Unlimited, US biofuels companies, have made significant contributions in photoautotrophic ethanol production using Synechococcus 7942 and Synechocystis 6803 systems. Algenol has filled various patent starting from 1998, publication number US6306639B1, followed by US8163516B2 (filed 9 February 2009), which was in itself first patent showing increased ethanol yields via overexpression of an alcohol dehydrogenase [Citation225] Further, in recent years filed a patent dealing with Cynobacteria (US20140113342A1) for production of 1, 2-propanediol with yields of ~0.011 g L-1[Citation226]. The Joule Unlimited patent space target wider metabolite profile than Algenol Biofuels, comprising of supplier photosynthetic framework for production of various industrially important metabolites. In their one of the patents using JCC1581_B isolate (patent application US20120164705A1, (filed 13 November 2011), yield of 5.62 g/l after 13.7 days was achieved, which was the higher ever reported yield for the Synechococcus 7002 framework [Citation227].
Table 6. Recent patents on modification of microorganisms for advancing biofuel production
8. Safety and hazard concerns
Recombinant microorganisms have opened new alternatives for cleaner and more efficient production of various metabolites including biofuels. Still, high cost, low efficiency of raw material conversion and unintentionally release of genetically modified microorganisms in the environment have limited the use of bioprocess using living cells and cell-free metabolic engineering becoming popular to harnesses the metabolic activities of cell lysates in vitro [Citation228]. Similarly, compound toxicity in bacteria becoming a major concern due to accumulation of toxic compounds during expression of heterologous biosynthetic pathway. The production of desired compound can be limited, on the other hand, the toxic intermediates can be useful as important antimicrobial compounds [Citation229]. The genetically modified microorganisms can have effect on indigenous community of natural microorganisms by creating more vigorous strains, overpowering the existing strains by recombination and potent gene exchange, loss of genetic pool, narrowing genetic diversity, permanent loss of primary metabolic pathway that can lead to loss of accumulation/production of important secondary metabolites [Citation230]. Though the gene exchange by natural recombination process is occurring at its own pace, still the unknown consequences are expected by foreign gene expression and results in altered metabolic pathway, growth rate, and response to external environmental factors [Citation231,Citation232].
Environmental risk assessment of various genetically modified organisms (GMO) has been done time to time [Citation233–235] but still there is a lack of specific rules and regulatory bodies regarding production of value added by products through genetically modified microorganisms (GMMO). There is a need to emphasize and differentiate the specific assessment criteria for both GMMOs and GMOs. The environmental risk assessment area is vast and open to explore the overall performance of GMMOs.
9. Conclusions and future prospects
Through the application of metabolic and bioprocess engineering approaches, numerous challenges associated with the production of lignocellulosic biofuels have been successfully revolutionized during the past few years. In brief, improvements in the biosynthesis of polysaccharides, lignin, and lipids in the lignocellulosic biomass have led to limited success, and further progress is required for sustainable biofuels. Metabolic pathways of microbial strain from diverse domains, such as yeasts, bacteria, and cyanobacteria have been successfully engineered to produce bioethanol from lignocellulose with a high yield near its maximum theoretical yield (0.51 g/g glucose). Yeasts, mainly S. cerevisiae, are already being engineered and commercialized to ferment simple sugars to isobutanol and other alcohols. Some non-conventiona,l such as S. passalidarum, P. stipites, etc., and a few oleaginous yeasts, i.e., Y. lipolytica, R. toruloides are being engineered to produce both bioethanol and biodiesel. Yeast strains engineered to surface display hydrolytic enzymes appear to be much more promising and have a broader prospect for commercial-scale bioethanol production directly from LCB in upcoming years. The bacterium Z. mobilis, E. coli, and other thermophiles are being engineered to ferment the xylose and could be used in commercial processes for biofuels production from lignocellulosic hydrolysates. Cyanobacteria and other photosynthetic bacteria also seem to generate commercializable biofuels in the future potentially. Although metabolic engineering is a well-established strategy for transforming microorganisms into efficient cell factories, there is a high possibility that novel robust industrial phenotypes could emerge from the coupling of more database mining, omics technologies, and advanced genome engineering strategies. Futuristic, metabolic engineering of microorganisms to discover new enzymes catalyzing unknown reactions, engineered enzymes with enhanced performances, and new synthetic pathways designed for higher metabolic flux towards target biofuels and other value-added products may enable the production of more sustainable biofuels.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Additional information
Funding
References
- Kazemi Shariat Panahi H, Dehhaghi M, Kinder JE, et al. A review on green liquid fuels for the transportation sector: a prospect of microbial solutions to climate change. Biofuel Res J. 2019;6(3):995–1024.
- Joshi A, Kanthaliya B, Arora J. Current scenario of potential renewable energy sources for sustainable development in India. J Plant Sci Res. 2019;35(2):205–214.
- Javed SA, Zhu B, Liu S. Forecast of biofuel production and consumption in top CO2 emitting countries using a novel grey model. J Clean Prod. 2019;276:123997.
- Santika WG, Anisuzzaman M, Bahri PA, et al. From goals to joules: a quantitative approach of interlinkages between energy and the sustainable development goals. Energy Res Soc Sci. 2019;50:201–214.
- Arora NK, Mishra I. COP26: more challenges than achievements. Environ Sustain. 2021;8(1):1–4.
- Pryshliak N, Tokarchuk D. Socio-economic and environmental benefits of biofuel production development from agricultural waste in Ukraine. Enviro Socio-Econ Stud. 2020;8(1):18–27.
- Subramaniam Y, Masron TA, Azman NHN. Biofuels, environmental sustainability, and food security: a review of 51 countries. Energy Res Soc Sci. 2020;68:101549.
- Isikgor FH, Becer CR. Lignocellulosic biomass: a sustainable platform for the production of bio-based chemicals and polymers. Polym Chem. 2015;6:4497–4559.
- Joshi A, Kanthaliya B, Arora J. Halophytes of Thar Desert: potential source of nutrition and feedstuff. Int J Bioassays. 2018;8:5674–5683.
- Sharma V, Joshi A, Ramawat KG, et al. Bioethanol production from halophytes of thardesert: a “green gold. In: Basu SK, Zandi P, Chalaras SK, editors. Environment at crossroads: Challenges, dynamics and Solutions. Guilan Prov., Iran: Haghshenass Publishing; 2017. p. 219–235.
- Vu HP, Nguyen LN, Vu MT, et al. A comprehensive review on the framework to valorise lignocellulosic biomass as biorefinery feedstocks. Sci Total Environ. 2020;743:140630.
- Joshi A, Kanthaliya B, Arora J. Halophytes: The nonconventional crops as source of biofuel production. In: Grigore MN, editor. Handbook of halophytes: From molecules to ecosystems towards biosaline agriculture. Cham: Springer; 2020. p. 1–28.
- Oliveira DM, Mota TR, Salatta FV, et al. Designing xylan for improved sustainable biofuel production. Plant Biotechnol J. 2019;17(12):2225. DOI:10.1111/pbi.13150.
- Martarello DC, Almeida AM, Sinzker RC, et al. The known unknowns in lignin biosynthesis and its engineering to improve lignocellulosic saccharification efficiency. Biomass Conv Bioref. 2021. DOI:10.1007/s13399-021-01291-6.
- Balan V. Current challenges in commercially producing biofuels from lignocellulosic biomass. Int Sch Res Notices, 2014 2014 32 ; Article ID 463074
- Joshi A, Kanthaliya B, Meena S, et al. Process consolidation approaches for cellulosic ethanol production. In: Ray RC, Eds. Sustainable biofuels. United Kingdom: Academic Press, Elsevier BV; 2021. p. 43–72.
- Dionisi D, Anderson JA, Aulenta F, et al. The potential of microbial processes for lignocellulosic biomass conversion to ethanol: a review. J Chem Technol Biotechnol. 2015;90(3):366–383. DOI:10.1002/jctb.4544.
- Tsegaye B, Balomajumder C, Roy P. Microbial delignification and hydrolysis of lignocellulosic biomass to enhance biofuel production: an overview and future prospect. Bull Natl Res Cent. 2019;43(1):1–16.
- Lu H, Yadav V, Zhong M, et al. Bioengineered microbial platforms for biomass-derived biofuel production–A review. Chemosphere. 2022;288:132528.
- Zhu P, Abdelaziz OY, Hulteberg CP, et al. New synthetic approaches to biofuels from lignocellulosic biomass. Curr Opin Green Sustain Chem. 2020;21:16–21.
- Brandt BA, García-Aparicio MD, Görgens JF, et al. Rational engineering of saccharomyces cerevisiae towards improved tolerance to multiple inhibitors in lignocellulose fermentations. Biotechnol Biofuels. 2021;14(1):1–18. DOI:10.1186/s13068-021-02021-w.
- Arun J, Gopinath KP, SundarRajan P, et al. Upgradation of nostoc punctriforme under subcritical conditions into liquid hydrocarbons (bio-oil) via hydro-deoxygenation: optimization and engine tests. J Environ Chem Eng. 2021;9(4):105230. DOI:10.1016/j.jece.2021.105230.
- Liang F, Englund E, Lindberg P, et al. Engineered cyanobacteria with enhanced growth show increased ethanol production and higher biofuel to biomass ratio. Metab Eng. 2018;46:51–59.
- Cunha JT, Gomes DG, Romaní A, et al. Cell surface engineering of Saccharomyces cerevisiae for simultaneous valorization of corn cob and cheese whey via ethanol production. Energy Convers Manag. 2021;243:114359.
- Cai P, Gao J, Zhou Y. CRISPR-mediated genome editing in non-conventional yeasts for biotechnological applications. Microb Cell Fact. 2019;18(1):1–12.
- Liu X, Xie H, Roussou S, et al. Current advances in engineering cyanobacteria and their applications for photosynthetic butanol production. Curr Opin Biotechnol. 2022;2022(73):143–150.
- Lubieniechi S, Peranantham T, Levin BD. Recent patents on genetic modification of plants and microbes for biomass conversion to biofuels. Rec Patent DNA Gene Seq. 2013;7(1):25–35.
- Infante EG, Secchi AR, Leite LF, et al. Scientific articles and patent applications on biodiesel production from lignocellulosic biomass. J Environ Protect. 2021;12(6):371–390.
- Zhang M, Chou YC, Franden MA, et al. Engineered Zymomonas for the production of 2, 3-butanediol. United States. 2021. https://patents.google.com/patent/US20190153483A1. 25 10 2021
- Salehi Jouzani G, Taherzadeh MJ. Advances in consolidated bioprocessing systems for bioethanol and butanol production from biomass: a comprehensive review. Biofuel Res J. 2015;2:152–195.
- Karimi K, Taherzadeh MJ. A critical review of analytical methods in pretreatment of lignocelluloses: composition, imaging, and crystallinity. Bioresour Technol. 2016;200:1008–1018.
- Bhatia SK, Jagtap SS, Bedekar AA, et al. Recent developments in pretreatment technologies on lignocellulosic biomass: effect of key parameters, technological improvements, and challenges. Bioresour Technol. 2020 300 ;122724.
- Brandon AG, Scheller HV. Engineering of bioenergy crops: dominant genetic approaches to improve polysaccharide properties and composition in biomass. Front Plant Sci. 2020;11:282.
- Jung SJ, Kim SH, Chung IM. Comparison of lignin, cellulose, and hemicellulose contents for biofuels utilization among 4 types of lignocellulosic crops. Biomass Bioen. 2015;83:322–327.
- Lao J, Oikawa A, Bromley JR, et al. The plant glycosyltransferase clone collection for functional genomics. The Plant J. 2014;79(3):517–529.
- Hu H, Zhang R, Feng S, et al. Three AtCesA6-like members enhance biomass production by distinctively promoting cell growth in Arabidopsis. Plant Biotechnol J. 2018;16(5):976–988.
- Stein O, Granot D. An overview of sucrose synthases in plants. Front Plant Sci. 2019;10:95.
- Yang Y, Yoo CG, Guo HB, et al. Overexpression of a domain of unknown function 266-containing protein results in high cellulose content, reduced recalcitrance, and enhanced plant growth in the bioenergy crop populus. Biotechnol Biofuels. 2017;1:1–3.
- Li M, Wang S, Liu Y, et al. Overexpression of PsnSuSy1, 2 genes enhances secondary cell wall thickening, vegetative growth, and mechanical strength in transgenic tobacco. Plant Mol Bio. 2019;100(3):215–230.
- Vega‐Sánchez ME, Loqué D, Lao J, et al. Engineering temporal accumulation of a low recalcitrance polysaccharide leads to increased C6 sugar content in plant cell walls. Plant Biotechnol J. 2015;13(7):903–914. DOI:10.1111/pbi.12326.
- Aznar A, Chalvin C, Shih PM, et al. Gene stacking of multiple traits for high yield of fermentable sugars in plant biomass. Biotechnol Biofuels. 2018;11(1):1–4.
- Gao Y, Lipton AS, Wittmer Y, et al. A grass-specific cellulose–xylan interaction dominates in sorghum secondary cell walls. Nat Commun. 2020;11(1):1–10.
- Tsai CJ, Xu P, Xue LJ, et al. Compensatory guaiacyl lignin biosynthesis at the expense of syringyl lignin in 4CL1 -knockout poplar. Plant Physiol. 2020;183(1):123–136. DOI:10.1104/pp.19.01550.
- Schultz JA, Coleman HD. Pectin and xylan biosynthesis in poplar: Implications and opportunities for biofuels production. Front Plant Sci. 2021 12 ;1772.
- Li G, Jones KC, Eudes A, et al. Overexpression of a rice BAHD acyltransferase gene in switchgrass (Panicum virgatum L.) enhances saccharification. BMC Biotechnol. 2018;18(1):54. DOI:10.1186/s12896-018-0464-8.
- Tian Y, Lin CY, Park JH, et al. Overexpression of the rice BAHD acyltransferase AT10 increases xylan-bound p-coumarate and reduces lignin in sorghum bicolor. Biotechnol Biofuels. 2021;14(1):1–9. DOI:10.1186/s13068-021-02068-9.
- Umezawa T. Lignin modification in planta for valorization. Phytochem Rev. 2018;17(6):1305–1327.
- Ralph J, Lapierre C, Boerjan W. Lignin structure and its engineering. Curr Opin Biotechnol. 2019;56:240–249.
- Park JJ, Yoo CG, Flanagan A, et al. Defined tetra-allelic gene disruption of the 4-coumarate: coenzyme A ligase 1 (Pv4CL1) gene by CRISPR/Cas9 in switchgrass results in lignin reduction and improved sugar release. Biotechnol Biofuels. 2017;10(1):1–11. DOI:10.1186/s13068-017-0972-0.
- Wu S, Takata N, Sakamoto S, et al. Simultaneous manipulation of lignin structure and secondary cell wall formation in transgenic poplar. J Wood Sci. 2020;66:1–9.
- Qin S, Fan C, Li X, et al. LACCASE14 is required for the deposition of guaiacyl lignin and affects cell wall digestibility in poplar. Biotechnol Biofuel. 2020;13(1):1–14. DOI:10.1186/s13068-020-01843-4.
- Liu Y, Yan J, Wang K, et al. MiR396- GRF module associates with switchgrass biomass yield and feedstock quality. Plant Biotechnol J. 2021;19(8):1523–1536.
- Singh R, Arora A, Singh V. Biodiesel from oil produced in vegetative tissues of biomass-A review. Biores Technol. 2021;326:124772.
- Yuan L, Li R. Metabolic engineering a model oilseed camelina sativa for the sustainable production of high-value designed oils. Front Plant Sci. 2020;12:11.
- Correa SM, Alseekh S, Atehortúa L, et al. Model-assisted identification of metabolic engineering strategies for Jatropha curcas lipid pathways. Plant J. 2020;104(1):76–95. DOI:10.1111/tpj.14906.
- Vanhercke T, El Tahchy A, Liu Q, et al. Metabolic engineering of biomass for high energy density: oilseed‐like triacylglycerol yields from plant leaves. Plant Biotechnol J. 2014;12(2):231–239. DOI:10.1111/pbi.12131.
- Chapman KD, Dyer JM, Mullen RT. Commentary: why don’t plant leaves get fat? Plant Sci. 2013;207:128–134.
- Napier JA, Haslam RP, Beaudoin F, et al. Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr Opin Plant Biol. 2014;19:68–75.
- Gao Y, Sun Y, Gao H, et al. Ectopic overexpression of a type-II DGAT (CeDGAT2-2) derived from oil-rich tuber of cyperus esculentus enhances accumulation of oil and oleic acid in tobacco leaves. Biotechnol Biofuels. 2021;14(1):1–16. DOI:10.1186/s13068-020-01854-1.
- Kalinger RS, Williams D, Ahmadi Pirshahid A, et al. Production of C6–C14 Medium-chain fatty acids in seeds and leaves via overexpression of single hotdog-fold acyl-lipid thioesterases. Lipids. 2021;56(3):327–344. DOI:10.1002/lipd.12299.
- Yee S, Rolland V, Reynolds KB, et al. Sesamum indicum oleosin l improves oil packaging in nicotiana benthamiana leaves. Plant Direct. 2021;5(9):e343. DOI:10.1002/pld3.343.
- Parajuli S, Kannan B, Karan R, et al. Towards oilcane: Engineering hyperaccumulation of triacylglycerol into sugarcane stems. GCB Bioener. 2020;12(7):476–490. DOI:10.1111/gcbb.12684.
- Okada S, Taylor M, Zhou XR, et al. Producing cyclopropane fatty acid in plant leafy biomass via expression of bacterial and plant cyclopropane fatty acid synthases. Front Plant Sci. 2020;11:30.
- Vanhercke T, Belide S, Taylor MC, et al. Up‐regulation of lipid biosynthesis increases the oil content in leaves of sorghum bicolor. Plant Biotechnol J. 2019 17 1 ;220–232.
- Gao CY, Mao X, Shang HQ, et al. Enhanced Oil accumulation in tobacco (nicotiana tabacum l.) leaves by ectopic overexpression of vgdgat1a for renewable production of biofuels. Curr Sci. 2018;114(6):1234–1240. DOI:10.18520/cs/v114/i06/1234-1240.
- Ji XJ, Mao X, Hao QT, et al. Splice variants of the castor WRI1 gene upregulate fatty acid and oil biosynthesis when expressed in tobacco leaves. Int J Molecul Sci. 2018;19(1):146. DOI:10.3390/ijms19010146.
- Vanhercke T, Divi UK, El Tahchy A, et al. Step changes in leaf oil accumulation via iterative metabolic engineering. Metabol Eng. 2017;39:237–246.
- An D, Kim H, Ju S, et al. Expression of Camelina WRINKLED1 isoforms rescue the seed phenotype of the Arabidopsis wri1 mutant and increase the triacylglycerol content in tobacco leaves. Front Plant Sci. 2017;8:34.
- Zale J, Jung JH, Kim JY, et al. Metabolic engineering of sugarcane to accumulate energy‐dense triacylglycerols in vegetative biomass. Plant Biotechnol J. 2016;14(2):661–669. DOI:10.1111/pbi.12411.
- Song JM, Zhang Y, Zhou ZW, et al. Oil plant genomes: current state of the science. J Exp Bot. 2021. DOI:10.1093/jxb/erab472.
- Ojha R, Kaur S, Sinha K, et al. Characterization of oleosin genes from forage sorghum in Arabidopsis and yeast reveals their role in storage lipid stability. Planta. 2021;254(5):1–2. DOI:10.1007/s00425-021-03744-8.
- Chu KL, Koley S, Jenkins LM, et al. Metabolic flux analysis of the non-transitory starch tradeoff for lipid production in mature tobacco leaves. Metab Eng. 2021;69:231–248.
- Muthulakshmi C, Sivaranjani R, Selvi S. Modification of sesame (Sesamum indicum L.) for Triacylglycerol accumulation in plant biomass for biofuel applications. Biotechnol Rep. 2021;32:e00668.
- Viswanathan MB, Cheng MH, Clemente TE, et al. Economic perspective of ethanol and biodiesel coproduction from industrial hemp. J Cleaner Prod. 2021;299:126875.
- Liu H, Sun J, Chang JS, et al. Engineering microbes for direct fermentation of cellulose to bioethanol. Crit Rev Biotechnol. 2018;38(7):1089–1105. DOI:10.1080/07388551.2018.1452891.
- Cheon S, Kim HM, Gustavsson M, et al. Recent trends in metabolic engineering of microorganisms for the production of advanced biofuels. Curr Opin Chem Biol. 2016;35:10–21.
- Bilal M, Iqbal HM, Hu H, et al. Metabolic engineering and enzyme-mediated processing: a biotechnological venture towards biofuel production–a review. Renewable Sustainable Energy Rev. 2018;82:436–447.
- Macedo N, Brigha CJ. From beverages to biofuels: the journeys of ethanol-producing microorganisms. Int J Biotechnol Wellness Ind. 2014;3(3):79–87.
- Choi YJ, Lee J, Jang YS, et al. Metabolic engineering of microorganisms for the production of higher alcohols. MBio. 2014;5(5):e01524–14. DOI:10.1128/mBio.01524-14.
- Kumar R, Kumar P. Future microbial applications for bioenergy production: a perspective. Front Microbiol. 2017;8:450.
- Majidian P, Tabatabaei M, Zeinolabedini M, et al. Metabolic engineering of microorganisms for biofuel production. Renewable Sustainable Energy Rev. 2018;82:3863–3885.
- Jiang Y, Dong W, Xin F, et al. Designing synthetic microbial consortia for biofuel production. Trends Biotechnol. 2020;38(8):828–831. DOI:10.1016/j.tibtech.2020.02.002.
- Cai W, Zhang W. Engineering modular polyketide synthases for production of biofuels and industrial chemicals. Curr Opin Biotechnol. 2018;50:32–38.
- Kazemi Shariat Panahi H, Dehhaghi M, Dehhaghi S, et al. Engineered bacteria for valorizing lignocellulosic biomass into bioethanol. Biores Technol. 2022;344:126212.
- Zhao N, Bai Y, Liu CG, et al. Flocculating Zymomonas mobilis is a promising host to be engineered for fuel ethanol production from lignocellulosic biomass. Biotechnol J. 2014;9(3):362–371. DOI:10.1002/biot.201300367.
- Chou YC, Linger J, Yang S, et al. Genetic engineering and improvement of a Zymomonas mobilis for arabinose utilization and its performance on pretreated corn stover hydrolyzate. J Biotechnol Biomat. 2015;28:5(NREL/JA-5100-64639.
- Lou J, Wang J, Yang Y, et al. Development and characterization of efficient xylose utilization strains of Zymomonas mobilis. Biotechnol Biofuels. 2021;14(1):1–8. DOI:10.1186/s13068-021-02082-x.
- Sarkar P, Mukherjee M, Goswami G, et al. Adaptive laboratory evolution induced novel mutations in Zymomonas mobilis ATCC ZW658: a potential platform for co-utilization of glucose and xylose. J Indust Microbiol Biotechnol. 2020;47(3):329–341.
- Ren C, Chen T, Zhang J, et al. An evolved xylose transporter from zymomonas mobilis enhances sugar transport in Escherichia coli. Microbial Cell Factor. 2009;8(1):1–9.
- Dunn KL, Rao CV. Expression of a xylose-specific transporter improves ethanol production by metabolically engineered zymomonas mobilis. Appl Microbiol Biotechnol. 2014;98(15):6897–6905.
- Sarkar P, Goswami G, Mukherjee M, et al. Heterologous expression of xylose specific transporter improves xylose utilization by recombinant Zymomonas mobilis strain in presence of glucose. Process Biochem. 2021;102:190–198.
- Shui ZX, Qin H, Wu B, et al. Adaptive laboratory evolution of ethanologenic zymomonas mobilis strain tolerant to furfural and acetic acid inhibitors. Appl Microbiol Biotechnol. 2015;99(13):5739–5748.
- Yang Y, Hu M, Tang Y, et al. Progress and perspective on lignocellulosic hydrolysate inhibitor tolerance improvement in Zymomonas mobilis. Bioresources Bioprocess. 2018;5(1):1–2.
- Nouri H, Moghimi H, Marashi SA, et al. Impact of hfq and sigE on the tolerance of Zymomonas mobilis ZM4 to furfural and acetic acid stresses. PloS one. 2020;15(10):e0240330.
- Yi X, Mei J, Lin L, et al. Overexpression of dioxygenase encoding gene accelerates the phenolic aldehyde conversion and ethanol fermentability of zymomonas mobilis. Appl Biochem Biotechnol. 2021;193(9):3017–3027.
- Yang S, Mohagheghi A, Franden MA, et al. Metabolic engineering of zymomonas mobilis for 2,3-butanediol production from lignocellulosic biomass sugars. Biotechnol Biofuels. 2016;9(1):1–5.
- Subramanian V, Lunin VV, Farmer SJ, et al. Phylogenetics-based identification and characterization of a superior 2,3-butanediol dehydrogenase for Zymomonas mobilis expression. Biotechnol Biofuels. 2020;13(1):1–20.
- He MX, Wu B, Qin H, et al. Zymomonas mobilis: a novel platform for future biorefineries. Biotechnol Biofuels. 2014;7(1):1–5.
- Qiu M, Shen W, Yan X, et al. Metabolic engineering of Zymomonas mobilis for anaerobic isobutanol production. Biotechnol Biofuels. 2020;13(1):1–4. DOI:10.1186/s13068-020-1654-x.
- Zhou S, Iverson AG, Grayburn WS. Engineering a native homoethanol pathway in Escherichia coli B for ethanol production. Biotechnol Lett. 2008;30(2):335–342.
- Xu P, Koffas MA. Metabolic engineering of Escherichia coli for biofuel production. Biofuels. 2010;1(3):493–504.
- Cotta MA. Ethanol production from lignocellulosic biomass by recombinant Escherichia coli strain FBR5. Bioengineered. 2012;3(4):197–202.
- Huerta‐Beristain G, Cabrera‐Ruiz R, Hernandez‐Chavez G, et al. Metabolic engineering and adaptive evolution of Escherichia coli KO11 for ethanol production through the Entner-doudoroff and the pentose phosphate pathways. J Chemical Technol Biotechnol. 2017;92(5):990–996.
- Kashiwagi FM, Ojima Y, Taya M. Metabolic Engineering of Escherichia coli KO11 with the NADH regeneration system for enhancing ethanol production. J Chem Eng Jpn. 2018;51(3):264–268.
- Vinuselvi P, Lee SK. Engineered Escherichia coli capable of co-utilization of cellobiose and xylose. Enzyme Microbial Technol. 2012;50(1):1–4.
- Kim SM, Choi BY, Ryu YS, et al. Simultaneous utilization of glucose and xylose via novel mechanisms in engineered Escherichia coli. Metabol Eng. 2015;30:141–148.
- Sun J, Tian K, Wang J, et al. Improved ethanol productivity from lignocellulosic hydrolysates by Escherichia coli with regulated glucose utilization. Microbial Cell Factor. 2018;17(1):1–8.
- Cao H, Wei D, Yang Y, et al. Systems-level understanding of ethanol-induced stresses and adaptation in E. coli. Sci Rep. 2017;7(1):1–5.
- Tan Z, Khakbaz P, Chen Y, et al. Engineering Escherichia coli membrane phospholipid head distribution improves tolerance and production of biorenewables. Metabol Eng. 2017;44:1–2.
- Lupino KM, Romano KA, Simons MJ, et al. A recurrent silent mutation implicates fecA in ethanol tolerance by Escherichia coli. BMC Microbiol. 2018;18(1):1–7.
- Kurgan G, Panyon LA, Rodriguez-Sanchez Y, et al. Bioprospecting of native efflux pumps to enhance furfural tolerance in ethanologenic Escherichia coli. Appl Environ Microbiol. 2019;85(6):e02985–18.
- Zheng Y, Kong S, Luo S, et al. Improving furfural tolerance of Escherichia coli by integrating adaptive laboratory evolution with crispr-enabled trackable genome engineering (CREATE). ACS Sust Chem Eng. 2022;10(7):2318–2330.
- Liang L, Liu R, Freed EF, et al. Synthetic biology and metabolic engineering employing Escherichia coli for c2–c6 bioalcohol production. Front Bioeng Biotechnol. 2020;8:710.
- Gu P, Liu L, Ma Q, et al. Metabolic engineering of Escherichia coli for the production of isobutanol: a review. World J Microbiol Biotechnol. 2021;37(10):1–9. DOI:10.1007/s11274-021-03140-0.
- Soma Y, Takahashi M, Fujiwara Y, et al. Design of synthetic quorum sensing achieving induction timing-independent signal stabilization for dynamic metabolic engineering of E. coli. ACS Synth Biol. 2021;10(6):1384–1393. DOI:10.1021/acssynbio.1c00008.
- Zhiqiang WE, Xiaoman SU, Qingzhuo WA, et al. Recent advances in metabolic engineering of clostridia for n-butanol production. Synthetic Biol J. 2021 2 2 :194.
- Du G, Che J, Wu Y, et al. Disruption of hydrogenase gene for enhancing butanol selectivity and production in Clostridium acetobutylicum. Biochem Eng J. 2021;171:108014.
- Bao T, Hou W, Wu X, et al. Engineering Clostridium cellulovorans for highly selective n -butanol production from cellulose in consolidated bioprocessing. Biotechnol Bioeng. 2021;118(7):2703–2718.
- Wong YM, Wu TY, Ling TC, et al. Evaluating new bio-hydrogen producers: Clostridium perfringens strain JJC, clostridium bifermentans strain WYM and clostridium sp. strain ade. TY J Biosci Bioengin. 2018;25:590–598.
- Son YS, Jeon JM, Kim DH, et al. Improved bio-hydrogen production by overexpression of glucose-6-phosphate dehydrogenase and FeFe hydrogenase in Clostridium acetobutylicum. Int J Hydrogen Energy. 2021;46(74):36687–36695.
- Zhang Y, Li Z, Liu Y, et al. Systems metabolic engineering of Vibrio natriegens for the production of 1, 3-propanediol. Metab Eng. 2021;65:52–65.
- Panich J, Fong B, Singer SW. Metabolic Engineering of Cupriavidus necator H16 for sustainable biofuels from CO2. Trends Biotechnol. 2021;39(4):412–424.
- Jia D, He M, Tian Y, et al. Metabolic engineering of gas-fermenting clostridium ljungdahlii for efficient co-production of isopropanol, 3-hydroxybutyrate, and ethanol. ACS Synth Biol. 2021;10(10):2628–2638.
- Nguyen AD, Lee EY. Engineered methanotrophy: a sustainable solution for methane-based industrial biomanufacturing. Trends Biotechnol. 2021;39(4):381–396.
- Gęsicka A, Oleskowicz-Popiel P, Łężyk M. Recent trends in methane to bioproduct conversion by methanotrophs. Biotechnol Adv. 2021;53:107861.
- Pathania R, Srivastava A, Srivastava S, et al. Metabolic systems biology and multi-omics of cyanobacteria: perspectives and future directions. Biores Technol. 2022;343:126007.
- Deng MD, Coleman JR. Ethanol synthesis by genetic engineering in cyanobacteria. Appl Environ Microbiol. 1999;65(2):523–528.
- Dexter J, Fu P. Metabolic engineering of cyanobacteria for ethanol production. Energy Environ Sci. 2009;2(8):857–864.
- Chow TJ, Su HY, Tsai TY, et al. Using recombinant cyanobacterium (Synechococcus elongatus) with increased carbohydrate productivity as feedstock for bioethanol production via separate hydrolysis and fermentation process. Biores Technol. 2015;184:33–41.
- Namakoshi K, Nakajima T, Yoshikawa K, et al. Combinatorial deletions of glgC and phaCE enhance ethanol production in Synechocystis sp. PCC 6803. J Biotechnol. 2016;239:13–19.
- Yoshikawa K, Toya Y, Shimizu H. Metabolic engineering of Synechocystis sp. PCC 6803 for enhanced ethanol production based on flux balance analysis. Bioprocess Biosyst Eng. 2017;40(5):791–796.
- Wang M, Luan G, Lu X. Engineering ethanol production in a marine cyanobacterium Synechococcus sp. PCC7002 through simultaneously removing glycogen synthesis genes and introducing ethanolgenic cassettes. J Biotechnol. 2020;317:1–4.
- Roussou S, Albergati A, Liang F, et al. Engineered cyanobacteria with additional overexpression of selected Calvin-Benson-Bassham enzymes show further increased ethanol production. Metabolic Eng Commun. 2021;12:e00161.
- Chou HH, Su HY, Chow TJ, et al. Engineering cyanobacteria with enhanced growth in simulated flue gases for high-yield bioethanol production. Biochem Eng J. 2021;165:107823.
- Miao R, Liu X, Englund E, et al. Isobutanol production in Synechocystis PCC 6803 using heterologous and endogenous alcohol dehydrogenases. Metabolic Engineer Commun. 2017;5:45–53.
- Liu X, Miao R, Lindberg P. Modular engineering for efficient photosynthetic biosynthesis of 1-butanol from CO 2 in cyanobacteria. Energy & Environmental Sci. 2019;12(9):2765–2777.
- Kobayashi S, Atsumi S, Ikebukuro K, et al. Light-induced production of isobutanol and 3-methyl-1-butanol by metabolically engineered cyanobacteria. Microb Cell Fact. 2022;21(1):1.
- Purdy HM, Pfleger BF, Reed JL. Introduction of NADH-dependent nitrate assimilation in Synechococcus sp. PCC 7002 improves photosynthetic production of 2-methyl-1-butanol and isobutanol. Metab Eng. 2022;69:87–97.
- Wu L XX, JW XSF, Chen HT, et al. Establishment of a resource recycling strategy by optimizing isobutanol production in engineered cyanobacteria using high salinity stress. Biotechnol Biofuels. 2021;14(1):1–5.
- Srivastava V, Amanna R, Rowden SJ, et al. Adaptive laboratory evolution of the fast-growing cyanobacterium Synechococcus elongatus PCC 11801 for improved solvent tolerance. J Biosci Bioeng. 2021;131(5):491–500.
- Yunus IS, Wang Z, Sattayawat P, et al. Improved bioproduction of 1-octanol using engineered synechocystis sp. PCC 6803. PCC 6803. ACS Synthetic Biol. 2021;10(6):1417–1428. DOI:10.1021/acssynbio.1c00029.
- Mustila H, Kugler A, Stensjö K. Isobutene production in Synechocystis sp. PCC 6803 by introducing α-ketoisocaproate dioxygenase from Rattus norvegicus. 12:Metabol Engineer Commun 2021 . e00163.
- Rodrigues JS, Lindberg P. Metabolic engineering of Synechocystis sp. PCC 6803 for improved bisabolene production. Metab Eng Commun. 2021;12:e00159.
- Masukawa H, Kitashima M, Inoue K, et al. Genetic engineering of cyanobacteria to enhance biohydrogen production from sunlight and water. Ambio. 2012;41(S2):169–173.
- Khetkorn W, Khanna N, Incharoensakdi A, et al. Metabolic and genetic engineering of cyanobacteria for enhanced hydrogen production. Biofuels. 2013;4(5):535–561.
- Zhang D, Dechatiwongse P, Del Rio-chanona EA, et al. Modelling of light and temperature influences on cyanobacterial growth and biohydrogen production. Algal Res. 2015;9:263–274.
- Malek Shahkouhi A, Motamedian E, Virolle M-J. Reconstruction of a regulated two-cell metabolic model to study biohydrogen production in a diazotrophic cyanobacterium anabaena variabilis ATCC 29413. PloS one. 2020;15(1):e0227977.
- Rather MA, Srivastav AK. A study on biohydrogen production based on biophotolysis from cyanobacteria. Ann Roman Soc Cell Biol. 2021;25:12500–12509.
- Lian J, Mishra S, Zhao H. Recent advances in metabolic engineering of saccharomyces cerevisiae: new tools and their applications. Metab Eng. 2018;50:85–108.
- Ko JK, Lee JH, Jung JH, et al. Recent advances and future directions in plant and yeast engineering to improve lignocellulosic biofuel production. Renewable Sustainable Energy Rev. 2020;134:110390.
- Lee SM, Jellison T, Alper HS. Systematic and evolutionary engineering of a xylose isomerase-based pathway in Saccharomyces cerevisiae for efficient conversion yields. Biotechnol Biofuels. 2014;7(1):1–8.
- Hoang PTN, Ko JK, Gong G, et al. Genomic and phenotypic characterization of a refactored xylose-utilizing Saccharomyces cerevisiae strain for lignocellulosic biofuel production. Biotechnol Biofuels. 2018;11:1–13.
- Jagtap RS, Mahajan DM, Mistry SR, et al. Improving ethanol yields in sugarcane molasses fermentation by engineering the high osmolarity glycerol pathway while maintaining osmotolerance in Saccharomyces cerevisiae. Appl Microbiol Biotechnol. 2019;103(2):1031–1042. DOI:10.1007/s00253-018-9532-1.
- Zhu L, Li P, Sun T, et al. Overexpression of SFA1 in engineered Saccharomyces cerevisiae to increase xylose utilization and ethanol production from different lignocellulose hydrolysates. Bioresour Technol. 2020;313:123724.
- Sloothaak J, Tamayo-Ramos JA, Odoni DI, et al. Identification and functional characterization of novel xylose transporters from the cell factories aspergillus niger and trichoderma reesei. Biotechnol Biofuels. 2016;9(1):1–15. DOI:10.1186/s13068-016-0564-4.
- Wang M, Yu C, Zhao H. Directed evolution of xylose specific transporters to facilitate glucose‐xylose co‐utilization. Biotechnol Bioeng. 2016;113(3):484–491.
- Nijland JG, Shin HY, Boender LG, et al. Improved xylose metabolism by a cyc8 mutant of saccharomyces cerevisiae. Appl Environ Microbiol. 2017;83(11):e00095–17. DOI:10.1128/AEM.00095-17.
- Zheng L, Wei S, Wu M, et al. Improving xylose fermentation in saccharomyces cerevisiae by expressing nuclear-localized hexokinase 2. Microorganisms. 2020;8(6):856. DOI:10.3390/microorganisms8060856.
- Dzanaeva L, Kruk B, Ruchala J, et al. The impact of transcription factors Znf1, Sip4, Adr1, Tup1, and Hap4 on xylose alcoholic fermentation in the engineered yeast Saccharomyces cerevisiae. Antonie van Leeuwenhoek. 2021;114(9):1373–1385. DOI:10.1007/s10482-021-01607-6.
- Rodrussamee N, Sattayawat P, Yamada M. Highly efficient conversion of xylose to ethanol without glucose repression by newly isolated thermotolerant Spathaspora passalidarum CMUWF1–2. BMC Microbial. 2018;18:1–11.
- Selim KA, Easa SM, El-Diwany AI. The xylose metabolizing yeast Spathaspora passalidarum is a promising genetic treasure for improving bioethanol production. Fermentation. 2020;6(1):33.
- Ryu S, Trinh CT, Elliot MA. Understanding functional roles of native pentose-specific transporters for activating dormant pentose metabolism in Yarrowia lipolytica. Appl Environ Microbial. 2018;84(3):e02146–17.
- Qiao K, Wasylenko TM, Zhou K, et al. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35(2):173–177.
- Liu H, Marsafari M, Wang F, et al. Engineering acetyl-CoA metabolic shortcut for eco-friendly production of polyketides triacetic acid lactone in Yarrowia lipolytica. Metab Eng. 2019;56:60–68.
- Yang K, Qiao Y, Li F, et al. Subcellular engineering of lipase dependent pathways directed towards lipid related organelles for highly effectively compartmentalized biosynthesis of triacylglycerol derived products in Yarrowia lipolytica. Metab Eng. 2019;55:231–238.
- Yook SD, Kim J, Gong G, et al. High-yield lipid production from lignocellulosic biomass using engineered xylose-utilizing Yarrowia lipolytica. GCB Bioenergy. 2020;12(9):670–679. DOI:10.1111/gcbb.12699.
- Zhang S, Skerker JM, Rutter CD, et al. Engineering Rhodosporidium toruloides for increased lipid production. Biotechnol Bioeng. 2016;113(5):1056–1066.
- Wang C, Yoon SH, Shah AA, et al. Farnesol production from Escherichia coli by harnessing the exogenous mevalonate pathway. Biotechnol Bioeng. 2010;107(3):421–429. DOI:10.1002/bit.22831.
- Díaz T, Fillet S, Campoy S, et al. Combining evolutionary and metabolic engineering in Rhodosporidium toruloides for lipid production with non-detoxified wheat straw hydrolysates. Appl Microbiol Biotechnol. 2018;102(7):287–3300. DOI:10.1007/s00253-018-8810-2.
- Tsai YY, Ohashi T, Wu CC, et al. Delta-9 fatty acid desaturase overexpression enhanced lipid production and oleic acid content in Rhodosporidium toruloides for preferable yeast lipid production. J Biosci Bioeng. 2019;127(4):430–440. DOI:10.1016/j.jbiosc.2018.09.005.
- Chopra J, Tiwari BR, Dubey BK, et al. Environmental impact analysis of oleaginous yeast based biodiesel and bio-crude production by life cycle assessment. J Clean Prod. 2020;271:122349.
- Qiu Z, Jiang R. Improving Saccharomyces cerevisiae ethanol production and tolerance via RNA polymerase II subunit Rpb7. Biotechnol Biofuels. 2017;10(1):1–13.
- Soares LB, Bonan CIDG, Biazi LE, et al. Investigation of hemicellulosic hydrolysate inhibitor resistance and fermentation strategies to overcome inhibition in non-saccharomyces species. Biomass Bioen. 2020;137:105549.
- Pacheco TF, Machado BR, de Moraisjúnior WG, et al. Enhanced tolerance of spathaspora passalidarum to sugarcane bagasse hydrolysate for ethanol production from xylose. Appl Biochem Biotechnol. 2021;193(7):2182–2197. DOI:10.1007/s12010-021-03544-6.
- Sitepu IR, Garay LA, Sestric R, et al. Oleaginous yeasts for biodiesel: current and future trends in biology and production. Biotechnol Adv. 2014;32(7):1336–1360. DOI:10.1016/j.biotechadv.2014.08.003.
- Wang Z, Zhou L, Lu M, et al. Adaptive laboratory evolution of Yarrowia lipolytica improves ferulic acid tolerance. Appl Microbiol Biotechnol. 2021;105(4):1745–1758. DOI:10.1007/s00253-021-11130-3.
- Konzock O, Zaghen S, Norbeck J. Tolerance of Yarrowia lipolytica to inhibitors commonly found in lignocellulosichydrolysates. BMC Microbial. 2021;21:1–10.
- Mhlongo SI, Ezeokoli OT, Roopnarain A, et al. The potential of single-cell oils derived from filamentous fungi as alternative feedstock sources for biodiesel production. Front Microbiol. 2021;12:57.
- Yang J, Khan MAK, López-García S, et al. Improved SDA production in high lipid accumulating strain of mucor circinelloides WJ11 by genetic modification. Am J Biochem Biotechnol 2020 . 16:147.
- Ledesma-Amaro R, Lozano-Martínez P, Jiménez A, et al. Engineering ashbya gossypii for efficient biolipid production. Bioengineered. 2015;6(2):119–123.
- Zheng Y, Yu X, Zeng J, et al. Feasibility of filamentous fungi for biofuel production using hydrolysate from dilute sulfuric acid pretreatment of wheat straw. Biotechnol Biofuels. 2012;5(1):1.
- Hussain SA, Hameed A, Khan M, et al. Engineering of fatty acid synthases (FASs) to boost the production of medium-chain fatty acids (MCFAs) in mucor circinelloides. Int J Molecular Sci. 2019;20(3):786.
- Hussain SA, Garcia A, Khan M, et al. Increased accumulation of medium-chain fatty acids by dynamic degradation of long-chain fatty acids in Mucor circinelloides. Genes (Basel). 2020;11(8):890.
- Santos S, Puna J, Gomes J. A review on bio-based catalysts (immobilized enzymes) used for biodiesel production. Energies. 2020;13(11):3013.
- Liu Y, Tang Y, Gao H, et al. Challenges and future perspectives of promising biotechnologies for lignocellulosic biorefinery. Molecules. 2021;26(17):5411.
- Ogunyewo OA, Upadhyay P, Rajacharya GH, et al. Accessory enzymes of hypercellulolytic Penicillium funiculosum facilitate complete saccharification of sugarcane bagasse. Biotechnol Biofuels. 2021;14(1):1–7.
- Tanaka T, Kondo A. Cell surface engineering of industrial microorganisms for biorefining applications. Biotechnol Adv. 2015;33(7):1403–1411.
- Hasunuma T, Kondo A. Development of yeast cell factories for consolidated bioprocessing of lignocellulose to bioethanol through cell surface engineering. Biotechnol Adv. 2012;30(6):1207–1218.
- Kuroda K, Ueda M. Arming technology in yeast—novel strategy for whole-cell biocatalyst and protein engineering. Biomolecules. 2013;3(4):632–650.
- Yamada R, Kimoto Y, Ogino H. Combinatorial library strategy for strong overexpression of the lipase from Geobacillustherm ocatenulatus on the cell surface of yeast Pichia pastoris. Biochem Eng J. 2016;113:7–11.
- Chen X. Yeast cell surface display: an efficient strategy for improvement of bioethanol fermentation performance. Bioengineered. 2017;8(2):115–119.
- Tang H, Wang J, Wang S, et al. Efficient yeast surface-display of novel complex synthetic cellulosomes. Microb Cell Fact. 2018;17(1):1–13. DOI:10.1186/s12934-018-0971-2.
- Gutiérrez-García AK, Alvarez-Guzmán CL, De Leon-Rodriguez A. Autodisplay of alpha amylase from bacillus megaterium in E. coli for the bioconversion of starch into hydrogen, ethanol and succinic acid. Enzyme Microb Technol. 2020;134:109477.
- Liu Z, Inokuma K, Ho SH, et al. Combined cell-surface display-and secretion-based strategies for production of cellulosic ethanol with Saccharomyces cerevisiae. Biotechnol Biofuels. 2015;8(1):1–12. DOI:10.1186/s13068-015-0344-6.
- Claes A, Deparis Q, Foulquié-Moreno MR, et al. Simultaneous secretion of seven lignocellulolytic enzymes by an industrial second-generation yeast strain enables efficient ethanol production from multiple polymeric substrates. Metab Eng. 2020;59:131–141.
- Chin WC, Lin KH, Liu CC, et al. Improved n-butanol production via co-expression of membrane-targeted tilapia metallothionein and the clostridial metabolic pathway in Escherichia coli. BMC Biotechnol. 2017;17(1):1–14. DOI:10.1186/s12896-017-0356-3.
- Goyal G, Tsai SL, Madan B, et al. Simultaneous cell growth and ethanol production from cellulose by an engineered yeast consortium displaying a functional mini-cellulosome. Microb Cell Fact. 2011;10(1):1–8. DOI:10.1186/1475-2859-10-89.
- Fan LH, Zhang ZJ, Yu XY, et al. Self-surface assembly of cellulosomes with two miniscaffoldins on Saccharomyces cerevisiae for cellulosic ethanol production. Proc Natl Acad Sci. 2012;109(33):13260–13265. DOI:10.1073/pnas.1209856109.
- Dong C, Qiao J, Wang X, et al. Engineering Pichia pastoris with surface-display minicellulosomes for carboxymethyl cellulose hydrolysis and ethanol production. Biotechnol Biofuels. 2020;13:1–9.
- Guirimand G, Sasaki K, Inokuma K, et al. Cell surface engineering of Saccharomyces cerevisiae combined with membrane separation technology for xylitol production from rice straw hydrolysate. Appl Microbiol Biotechnol. 2016;100(8):3477–3487. DOI:10.1007/s00253-015-7179-8.
- Prasad RK, Chatterjee S, Mazumder PB, et al. Bioethanol production from waste lignocelluloses: a review on microbial degradation potential. Chemosphere. 2019;231:588–606.
- Celińska E, Nicaud JM, Białas W. Hydrolytic secretome engineering in Yarrowia lipolytica for consolidated bioprocessing on polysaccharide resources: review on starch, cellulose, xylan, and inulin. Appl Microbiol Biotechnol. 2021 105 ;975–989.
- Morgan SA, Nadler DC, Yokoo R, et al. Biofuel metabolic engineering with biosensors. Curr Opin Chem Biol. 2016;35:150–158.
- Liu Y, Liu Y, Wang M. Design, optimization and application of small molecule biosensor in metabolic engineering. Front Microbiol. 2012;2017:8.
- Ko YS, Kim JW, Lee JA, et al. Tools and strategies of systems metabolic engineering for the development of microbial cell factories for chemical production. Chem Soc Rev. 2020;49(14):4615–4636. DOI:10.1039/D0CS00155D.
- Lalwani MA, Zhao EM, Avalos JL. Current and future modalities of dynamic control in metabolic engineering. Curr Opin Biotechnol. 2018;52:56–65.
- Yu H, Wang N, Huo W, et al. Establishment of BmoR-based biosensor to screen isobutanol overproducer. Microb Cell Fact. 2019;18(1):1–11. DOI:10.1186/s12934-019-1084-2.
- Kim SK, Kim SH, Subhadra B, et al. A genetically encoded biosensor for monitoring isoprene production in engineered Escherichia coli. ACS Synth Biol. 2018;7(10):2379–2390. DOI:10.1021/acssynbio.8b00164.
- ShabbirHussain M, Wheeldon I, Blenner MA. A strong hybrid fatty acid inducible transcriptional sensor built from yarrowia lipolytica upstream activating and regulatory sequences. Biotechnol J. 2017;12(10):1700248.
- Shi S, Choi YW, Zhao H, et al. Discovery and engineering of a 1-butanol biosensor in Saccharomyces cerevisiae. Bioresour Technol. 2017;245:1343–1351.
- Dabirian Y, Gonçalves Teixeira P, Nielsen J, et al. FadR-based biosensor-assisted screening for genes enhancing fatty acyl-coa pools in saccharomyces cerevisiae. ACS Synth Biol. 2019;8(8):1788–1800. DOI:10.1021/acssynbio.9b00118.
- Shapiro RS, Chavez A, Collins JJ. CRISPR-based genomic tools for the manipulation of genetically intractable microorganisms. Nat Rev Microbiol. 2018;16(6):333–339.
- Cho S, Shin J, Cho BK. Applications of CRISPR/Cas system to bacterial metabolic engineering. Int J Mol Sci. 2018;19(4):1089.
- Shanmugam S, Ngo HH, Wu YR. Advanced CRISPR/Cas-based genome editing tools for microbial biofuels production: a review. Renew Energy. 2020;149:1107–1119.
- Yang LI, Xiaolin SHEN, Xinxiao SUN, et al. Advances of CRISPR gene editing in microbial synthetic biology. Synth Biol. 2021;2(1 106–125).
- Liu X, Xie H, Roussou S, et al. Current advances in engineering cyanobacteria and their applications for photosynthetic butanol production. Curr Opin Biotechnol. 2022;73:143–150.
- Viitanen PV, Mccutchen CM, Chou YC, et al. Zymomonas xylitol synthesis mutant that uses xylose for ethanol production. 2015. Spain. https://patents.google.com/patent/ES2554805T3/en. 11 11 2021
- Eliot AC, Tao L, Viitanen PV. Enhancing d-xylose and l-arabinose utilization in Zymomonas cells”. 2018. European Patent. https://patents.google.com/patent/EP3160987B1/en. 12 11 2021
- Xu H, Satory D, Jackson C. Xylose metabolizing yeast. 2021. United States. https://patents.google.com/patent/US20210017526A1/en. 12 11 2021
- Konishi H, Mutaguchi K, Fukuda A, et al. Yeast with inhibited accumulation of xylitol. 2020. Japan. https://patents.google.com/patent/JP2020115827A/en. 11 11 2021
- Konishi H, Mutaguchi K, Fukuda A, et al. Yeast producing ethanol from xylose. 2019. Japan. https://patents.google.com/patent/JP6616311B2/en. 11 11 2021
- D’espaux L, Keasling JD. Yeast host cells and methods for producing fatty alcohols. 2020. United States. https://patents.google.com/patent/US10557152B2/en. 12 11 2021
- Onishi T, Tada N. Recombinant yeast, and method for producing ethanol using same. Japan. https://patents.google.com/patent/JP2020025493A/en. 12 11 2021
- Liao JC, Atsumi S, Cann AF. Biofuel production by recombinant microorganisms”. United States; 2017. https://www.osti.gov/servlets/purl/1368211 12 11 2021
- Dehring U, Kramer D, Ziegler K. Selection of ADH in genetically modified cyanobacteria for the production of ethanol, official gazette of the United States patent and trademark office patents, 2012, Patent Publication Number: US8163516B2.
- Ziegler K, Weissert C, Duehring U, et al. Production of 1,2-propanediol in cyanobacteria, 2014, Patent Publication Number: US20140113342A1
- Reppas NB. Metabolic Switch, 2012, Patent Publication Number: US20120164705A1
- Lim HJ, Kim DM. Cell-free metabolic engineering: recent developments and future prospects. Meth Protocols. 2019;2(2):33.
- Planson AG, Carbonell P, Paillard E, et al. Compound toxicity screening and structure-activity relationship modeling in Escherichia coli. Biotechnol Bioeng. 2012;109(3):846–850.
- Lee H, Kim DU, Son J, et al. Environmental Risk assessment of living modified microorganisms (LMM) on the indigenous microbial community. Sustainability. 2020;12(14):5566.
- Phillips T. Genetically modified organisms (GMOs): transgenic crops and recombinant DNA technology. Nat Educ. 2008;1:213.
- Azizoglu U, Jouzani GS, Yilmaz N, et al. Genetically modified entomopathogenic bacteria, recent developments, benefits and impacts: a review. Sci Total Environ. 2020;734:139169.
- Bauer-Panskus A, Miyazaki J, Kawall K, et al. Risk assessment of genetically engineered plants that can persist and propagate in the environment. Environ Sci Eur. 2020;32(1):1–5.
- Kolseth AK, D’Hertefeldt T, Emmerich M, et al. Influence of genetically modified organisms on agro-ecosystem processes. Agric Ecosyst Environ. 2015;214:96–106.
- Snow AA, Andow DA, Gepts P, et al. Genetically engineered organisms and the environment: Current status and recommendations 1. Ecol Appl. 2005;15(2):377–404.
- Cha M, Chung D, Elkins JG, et al. Westpheling J. metabolic engineering of caldicellulosiruptor bescii yields increased hydrogen production from lignocellulosic biomass. Biotechnol Biofuels. 2013;6(1):85. DOI:10.1186/1754-6834-6-85.
- Das M, Patra P, Ghosh A. Metabolic engineering for enhancing microbial biosynthesis of advanced biofuels. Renewable Sustainable Energy Rev. 2020;119:109562.
- Liew F, Henstra AM, K&pke M, et al. Metabolic engineering of Clostridium autoethanogenum for selective alcohol production. Metab Eng. 2017;40:104–114.
- Gaida SM, Liedtke A, Jentges AHW, et al. Metabolic engineering of Clostridium cellulolyticum for the production of n-butanol from crystalline cellulose. Microb Cell Fact. 2016;15(1). DOI:10.1186/s12934-12015-10406-12932
- Yu L, Xu M, Tang IC, et al. Metabolic engineering of Clostridium tyrobutyricum for n-butanol production through co-utilization of glucose and xylose. Biotechnol Bioeng. 2015;112(10):2134–2141. DOI:10.1002/bit.25613.
- Argyros DA, Tripathi SA, Barrett TF, et al. High ethanol titers from cellulose by using metabolically engineered thermophilic, anaerobic microbes. Appl Environ Microbiol. 2011;77(23):8288–8294. DOI:10.1128/AEM.00646-11.
- Chen Z, Huang J, Wu Y, et al. Metabolic engineering of Corynebacterium glutamicum for the production of 3-hydroxypropionic acid from glucose and xylose. Metab Eng. 2017;39:151–158.
- Li L, Li K, Wang Y, et al. Metabolic engineering of Enterobacter cloacae for high-yield production of enantiopure (2R, 3R)-2, 3-butanediol from lignocellulose-derived sugars. Metab Eng. 2015;28:19–27.
- Wargacki AJ, Leonard E, Win MN, et al. An engineered microbial platform for direct biofuel production from brown macroalgae. Science. 2012;335(6066):308–313. DOI:10.1126/science.1214547.
- Steen EJ, Kang Y, Bokinsky G, et al. Microbial production of fatty-acid derived fuels and chemicals from plant biomass. Nature. 2010;463(7280):559–562. DOI:10.1038/nature08721.
- Chen Z, Wu Y, Huang J, et al. Metabolic engineering of Klebsiella pneumoniae for the de novo production of 2-butanol as a potential biofuel. Biores Technol. 2015;197:260–265.
- Gibbons J, Gu L, Zhu H, et al. Identification of two genes required for heptadecane production in a N2-fixing cyanobacterium Anabaena sp. strain PCC 7120. AMB Express. 2018;8(1):1–13. DOI:10.1186/s13568-018-0700-6.
- Hu B, Lidstrom ME. Metabolic engineering of Methylobacterium extorquens AM1 for 1-butanol production. Biotechnol Biofuels. 2014;7(1):156.
- Varman AM, Xiao Y, Pakrasi HB. Metabolic Engineering of synechocystis sp. Strain PCC 6803 for isobutanol production. Appl Environ Microbiol. 2013;79(3):908–914.
- Verhoeven MD, Lee M, Kamoen L, et al. Mutations in PMR1 stimulate xylose isomerase activity and anaerobic growth on xylose of engineered Saccharomyces cerevisiae by influencing manganese homeostasis. Sci Rep. 2017;7(1):1–11. DOI:10.1038/srep46155.
- Xu H, Kim S, Sorek H, et al. PHO13 deletion-induced transcriptional activation prevents sedoheptulose accumulation during xylose metabolism in engineered Saccharomyces cerevisiae. Metab Eng. 2016;34:88–96.
- Xie CY, Yang BX, Wu Y-J. Wu YJ, et al.Construction of industrial xylose-fermenting Saccharomyces cerevisiae strains through combined approaches. Process Biochem. 2020;96:80–89.
- Shin HY, Nijland JG, de Waal PP, et al. The amino‐terminal tail of Hxt11 confers membrane stability to the Hxt2 sugar transporter and improves xylose fermentation in the presence of acetic acid. Biotechnol Bioeng. 2017;114(9):1937–1945.
- Wu R, Chen D, Cao S, et al. Enhanced ethanol production from sugarcane molasses by industrially engineered Saccharomyces cerevisiae via replacement of the PHO4 gene. RSC Adv. 2020;10(4):2267–2276. DOI:10.1039/C9RA08673K.
- Papapetridis I, van Dijk M, Dobbe AP, et al. Improving ethanol yield in acetate-reducing Saccharomyces cerevisiae by cofactor engineering of 6-phosphogluconate dehydrogenase and deletion of ALD6. Microb Cell Fact. 2016;15(1):1–16. DOI:10.1186/s12934-016-0465-z.
- Henningsen BM, Hon S, Covalla SF, et al. Increasing anaerobic acetate consumption and ethanol yields in Saccharomyces cerevisiae with NADPH-specific alcohol dehydrogenase. Appl Environ Microbiol. 2015;81(23):8108–8117. DOI:10.1128/AEM.01689-15.
- Swinnen S, Henriques SF, Shrestha R, et al. Improvement of yeast tolerance to acetic acid through Haa1 transcription factor engineering: towards the underlying mechanisms. Microb Cell Fact. 2017;16(1):1–15. DOI:10.1186/s12934-016-0621-5.
- Niehus X, Crutz-Le Coq AM, Sandoval G, et al. Engineering Yarrowia lipolytica to enhance lipid production from lignocellulosic materials. Biotechnol Biofuels. 2018;11(1):1–10. DOI:10.1186/s13068-018-1010-6.
- Li H, Alper HS. Enabling xylose utilization in Yarrowia lipolytica for lipid production. Biotechnol J. 2016;11(9):1230–1240.
- Daskalaki A, Perdikouli N, Aggeli D, et al. Laboratory evolution strategies for improving lipid accumulation in Yarrowia lipolytica. Appl Microbiol Biotechnol. 2019;103(20):8585–8596.
- Qiao K, Wasylenko M, Zhou K, et al. Lipid production in Yarrowia lipolytica is maximized by engineering cytosolic redox metabolism. Nat Biotechnol. 2017;35(2):173–177. DOI:10.1038/nbt.3763.
- Shen H, Li Q, X Y, et al. Lipid production by rhodotorula glutinis in continuous cultivation with a gravity sedimentation system. Ind J Microbial. 2020;60(2):246–250. DOI:10.1007/s12088-019-00849-3.
- Guo M, Cheng S, Chen G, et al. Improvement of lipid production in oleaginous yeast Rhodosporidium toruloides by ultraviolet mutagenesis. Eng Life Sci. 2019;19(8):548–556.
- Li YJ, Lu YY, Zhang ZJ, et al. Co-fermentation of cellulose and sucrose/xylose by engineered yeasts for bioethanol production. Energy Fuels. 2017;31(4):4061–4067. DOI:10.1021/acs.energyfuels.7b00032.
- Chen X, Xiao Y, Shen W, et al. Display of phytase on the cell surface of Saccharomyces cerevisiae to degrade phytate phosphorus and improve bioethanol production. Appl Microbial Biotechnol. 2016;100(5):2449–2458. DOI:10.1007/s00253-015-7170-4.
- Inokuma K, Hasunuma T, Kondo A. Efficient yeast cell-surface display of exo-and endo-cellulase using the SED1 anchoring region and its original promoter. Biotechnol Biofuels. 2014;7(1):1–11.
- Nakatani Y, Yamada R, Ogino C, et al. Synergetic effect of yeast cell-surface expression of cellulase and expansin-like protein on direct ethanol production from cellulose. Microb Cell Fact. 2013;12(1):1–7.
- Xu Y, Li F, Yang K, et al. A facile and robust non-natural three enzyme biocatalytic cascade based on Escherichia coli surface assembly for fatty alcohol production. Energy Convers Manag. 2019;181:501–506.
- Muñoz-Gutiérrez I, Oropeza R, Gosset G, et al. Cell surface display of a β-glucosidase employing the type V secretion system on ethanologenic Escherichia coli for the fermentation of cellobiose to ethanol. J Ind Microbiol Biotechnol. 2012;39(8):1141–1152.
- Soma Y, Inokuma K, Tanaka T, et al. Direct isopropanol production from cellobiose by engineered Escherichia coli using a synthetic pathway and a cell surface display system. J Biosci Bioeng. 2012;114(1):80–85. DOI:10.1016/j.jbiosc.2012.02.019.
- Avalos JL, Hammer SK, Kuroda K, et al. System and method for increased alcohol tolerance and production in yeast. 2021 8 12 2021. https://patentscope.wipo.int/search/en/detail.jsf?docId=WO2021062082.
- Stephanopoulos G, Qiao K, Xu P. Strain and bioprocess engineering for high lipid production. 2017. United States. https://patents.google.com/patent/US20170183670A1/en. 8 12 2021
- Song Y, Zhang H. Method for raising Yarrowia lipolytica lipid content by molecular modification. 2016 8 12 2021. China. https://patents.google.com/patent/CN105886524A/en.
- Dühring ULF, Enke H, Ziegler K, et al. Metabolically enhanced cyanobacterial cell for the production of ethanol. 2015. United States. https://patents.google.com/patent/US91t27297B2/en. 10 12 2021