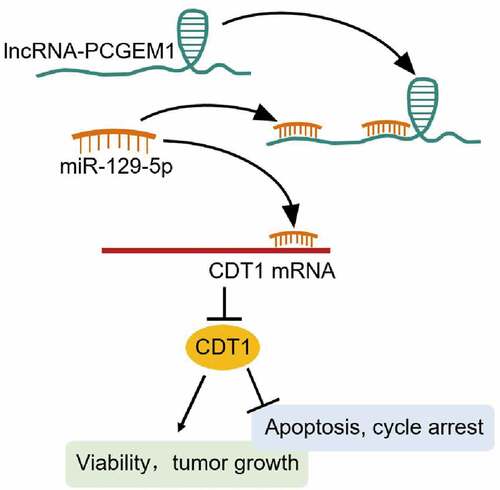
ABSTRACT
PCGEM1 facilitates prostate cancer (PCa) progression. This study aimed to elucidate the mechanism of action of PCGEM1 in PCa. The expression of PCGEM1, microRNA miR-129-5p, chromatin licensing, and DNA replication factor 1 (CDT1) was detected by quantitative reverse transcription-PCR (qRT-PCR). A series of function experiments including cell counting kit-8 (CCK-8), caspase-3 activity, and cell cycle assays were performed to evaluate the influence of PCGEM1, miR-129-5p, and CDT1 on the biological processes of PCa cells. CyclinD1, cyclin dependent kinase 4 (CDK4), Bax, and Bcl-2 protein levels were measured by western blotting. Subcellular isolation revealed the distribution of PCa cells. The connections between PCGEM1, miR-129-5p, and CDT1 were evaluated by luciferase, RIP assay, and Pearson correlation analysis. Both PCGEM1 and CDT1 were upregulated in PCa, while miR-129-5p was downregulated and negatively correlated with PCGEM1 and CDT1. Downregulation of PCGEM1 or CDT1 inhibited the viability, promoted apoptosis and cycle arrest of PCa cells in vitro, and controlled tumor growth in vivo. PCGEM1 plays a crucial role in the progression of PCa by sponging miR-129-5p as a ceRNA of CDT1. PCGEM1 is a CDT1-dependent PCa promoter site that absorbs miR-129-5p.
GRAPHICAL ABSTRACT
Introduction
There were more than 1.2 million new cases of prostate cancer (PCa) in 2018, resulting in approximately 300,000 deaths, which accounted for 3.8% of all cancer-related deaths in men [Citation1,Citation2]. The diversity of PCa monitoring methods and the asymptomatic nature of the early stages of cancer make early detection extremely difficult [Citation3]. Furthermore, alterations in genetic composition have been shown to significantly increase the risk of family PCa worldwide [Citation4]. Therefore, there is an urgent need to identify PCa risk genes and develop effective screening and treatment methods.
Long non-coding RNAs (lncRNAs), transcripts larger than 200 bp, are a class of RNAs that do not translate into proteins [Citation5]. Accumulating evidence suggests that lncRNAs participate in the regulation of gene expression in the cytoplasm and nucleus through a variety of pathways. In addition, differential expression of lncRNA has been found to play a critical role in tumors, including PCa [Citation6]. PCGEM1 is highly prostate-specific and attractive biomarker [Citation7]. Several studies have shown that PCGEM1 is upregulated in PCa tissues, indicating malignant progression of tumors [Citation8,Citation9]. Nevertheless, the mechanism of PCGEM1 in PCa remains unknown.
MicroRNAs participate in a variety of physiological processes that influence cancer occurrence through complex cross-regulation between lncRNAs and mRNAs [Citation10]. As a key inhibitor involved in the occurrence of various cancers, miR-129-5p is expressed at low levels in the lung [Citation11], colon [Citation12], and PCa [Citation13]. Previous studies have shown that miR-129-5p serves as a downstream target of PCGEM1, regulating the occurrence and development of gastric cancer [Citation14] and endometrial cancer [Citation15]. However, the potential role of PCGEM1/miR-129-5p in PCa progression has not yet been studied.
Chromatin licensing and DNA replication factor 1 (CDT1), located at the centromere in an Ndc80 complex-dependent manner, has an independent mitotic role in addition to its prototypic function in the origin licensing of DNA replication [Citation16]. Overexpression of CDT1 was revealed to be a predictor of low survival in hepatocellular carcinoma patients [Citation17] and a prognostic marker for breast cancer patients [Citation18]. Importantly, abnormal expression of CDT1 was reported to predict the initiation risk of DNA replication in PCa [Citation19]. However, the function and mechanism of CDT1 in PCa have not yet been explored.
Herein, the specific relationship between PCGEM1/miR-129-5p/CDT1 in PCa was verified, and its effect on cancer cell biological function was discussed. We hypothesized that PCGEM1 is upregulated in PCa, promoting the malignant behavior of PCa by sponging miR-129-5p and releasing CDT1. The aim of this study was to develop an effective PCa screening and treatment strategy.
Methods
Tissue collection
All studies involving patient samples were approved by the ethics committee of the Third Hospital of Wuhan (approval number: 武三医伦 KY2022-012). PCa tissue and adjacent normal tissue from 26 patients with radical prostatectomy excised PCa were immediately frozen at −80°C. All patients voluntarily participated in the study and provided written informed consent. None of the patients received neoadjuvant chemotherapy or endocrine therapy before surgery. The clinical characteristics of the patients with PCa are listed in Supplementary Table 1.
Cell Culture
The human prostate cell line (LNCAP, 22RV1, and MDA-PCA-2B) and human prostate epithelial cell line RWPE1 were obtained from ATCC (USA). RWPE-1 was stored in K-SFM medium (Gibco, USA). 22RV1 and LNCaP cells were stored in DMEM, and MDA-PCA-2B cells were cultured in F-12 K medium. All cell lines were supplemented with 1% P/S (Sangon, China) and 10% fetal bovine serum (FBS, Gibco) and placed in an incubator at 37°C and 5% CO2.
Nuclear cytoplasmic fractionation
The position of PCGEM1 in the cells was mapped using nuclear cytoplasmic separation. NE PER Nuclear and Cytoplasmic Extraction Reagents (Thermo Fisher Scientific) were used to isolate the nucleus and cytoplasm of LNCaP and 22RV1 cells. Nuclear control transcript (Uracil6, U6), cytoplasmic control transcript (glyceraldehyde-3-phosphate dehydrogenase, GAPDH), and PCGEM1 levels in the nucleus and cytoplasm were determined by qRT-PCR [Citation20].
Cell transfection
GenePharma (China) provided siRNA for PCGEM1 or CDT1 (si-PCGEM1 or si-CDT1) and its negative control siRNA(si-NC). MiR-129-5p inhibitor and inhibitor NC were collected from Switchgear Genomics (USA). LNCaP and 22RV1 cells were seeded into 6-well plates (5 × 104 cells/mL) and incubated overnight. Lipofectamine2000 (Thermo Fisher Scientific, USA) was used to transfect 50 nM siRNA or 100 nM inhibitor into cells, which were collected after 48 h transfection at 37°C. The transfection efficiency was then determined.
Quantitative reverse transcription-PCR (qRT-PCR) assay
RNA was extracted from whole-cell lysates using the Eastep Super RNA Extract Reagent Kit (Promega, USA). The first-strand cDNA was inverted using a cDNA synthesis kit (Invitrogen). SYBR qPCR Master Mix (Vazyme, USA) was used for PCR in the Prism 7000 Real-Time PCR system, with GAPDH serving as the internal standard. In addition, the NucleoSpin® miRNA kit (Macherey Nagel, France) was used to extract miRNAs. Small nuclear U6B (RNU6B) RNA was used as a housekeeping gene. cDNA synthesis was performed using an miRScript II RT kit. miRNAs were quantified using the Miscript SYBR Green PCR Kit. mRNA and miRNA expression was measured by 2−ΔΔCt [Citation21]. The primer sequences are listed in .
Table 1. The sequences of the primers in this study
Cell counting kit-8 (CCK-8) assay
A CCK-8 kit (Roche, Basel, Switzerland) was used to measure cell viability. Briefly, 22RV1 and LNCaP cells were seeded into 96-well plates (3 × 103 cells per well). After transfection for 24, 48, 72, and 96 h, 10 μL CCK-8 solution was added to each well at 37°C for 2 h. Absorbance was measured at 450 nm using an FLx800 fluorescence microplate reader (BioTek, USA) [Citation22].
Caspase-3 activity assay
The apoptosis of LNCaP and 22RV1 cells was detected using a caspase-3 activity assay kit (Beyotime, China). LNCaP and 22RV1 cells (1 × 105) were lysed in cell lysis buffer for 15 min at 4°C, and protein concentration was determined using Pierce bicinchoninic acid (BCA) (Thermo Scientific). After incubating the reaction mixture system composed of cell lysate (10 μL), detection buffer solution (80 μL), and Ac-DEVD-pNA (10 μL, 2 mM) in 96-well plates at 37°C for 2 h, the optical density was measured using a microplate reader at 405 nm [Citation23].
Cell cycle assay
After transfection for 72 h, LNCaP and 22RV1 cells (1 × 106 cells) were collected, centrifuged, washed twice with phosphate-buffered saline (PBS), and precipitated. The precipitates were fixed in 70% ethanol at 4°C for 4 h. Next, the cells were stained with 500 µL of staining solution (staining buffer 465 µL, 50× RNase A 10 µL, 20× propidium iodide 25 µL) at 37°C for 1 h according to the manufacturer’s instructions. Finally, the cell cycle distribution was determined using flow cytometry (FACS CantoTM II Flow Cytometer, BD Biosciences, USA) [Citation24].
Tumor growth assay
The Shanghai Experimental Animal Center (China) provided BALB/c nude mice (aged 5 weeks, two mice). LNCaP cells (5 × 106/mL) treated with PCGEM1 shRNA or sh-NC (GenePharma, China) were subcutaneously injected into the right side of the posterior flank of mice, and the mice were divided into sh-lnc (n = 5) and sh-NC groups (n = 5). Tumor size was measured with calipers every week. After feeding for 5 weeks, nude mice were euthanized with excessive CO2, and tumors were removed using a scalpel, photographed, and weighed. The animal procedures were approved by our animal committee [Citation25].
Luciferase assay
The 3’-untranslated region (UTR) products of PCGEM1 and CDT1 containing miR-129-5p target sequences were obtained by PCR and delivered into pmirGLO (Promega) to construct wild-type PCGEM1 and CDT1 reporter vectors (PCGEM1-WT and CDT1-WT). Similarly, PCGEM1 (PCGEM1-MUT) and CDT1 mutant vectors (MUT1, MUT2, and Co-MUT) were generated by site-directed mutagenesis. LNCaP and 22RV1 cells cultured for 24 h were co-transfected with wild-type or mutant vectors and miR-129-5p mimic for 48 h, using Lipofectamine 2000 (Invitrogen). Luciferase activity was analyzed using a dual-luciferase reporter assay system (Promega) [Citation26].
RNA immunoprecipitation (RIP) assay
The BersinBioTM RIP kit (Ca# Bes5101, BersinBio, China) was used for RIP, following the manufacturer’s instructions. Briefly, LNCaP and 22RV1 cells (1 × 107 cells) were lysed using RNA lysis buffer for 20 min on ice. Cell lysates were incubated with DNase for 10 min at 37°C and then centrifuged at 16,100 g for 10 min. Then, samples were conjugated with magnetic beads with 5 μg Ago2 or negative control IgG antibody and incubated at 4°C for 4 h. Then, 40 μL protein A-Sepharose was added to each sample, and the mixture was incubated at 4°C for 4 h. After washing with PBS, the precipitated RNA was resuspended and analyzed by qRT-PCR [Citation27].
Western blot analysis
LNCaP and 22RV1 cells were treated with RIPA buffer (Beyotime) to obtain proteins, which were quantified using BCA (Pierce, USA). A 10% sulfate-polyacrylamide gel electrophoresis was utilized to separate the equivalent amount of protein, which was then delivered to a polyvinylidene fluoride membrane. The membrane was sealed with PBST containing 5% skim milk, and then coexisted with CDT1 (ab202067; Abcam, UK), Bax (ab3191; Abcam), Bcl-2 (ab218123; Abcam), CyclinD1 (ab134175; Abcam), CDK4 (ab68266; Abcam), or GAPDH antibody (ab181602; Abcam) at 4°C overnight. The membrane was incubated with the secondary antibody (ab205718; Abcam) for 1 h at 37°C. Protein bands were observed using enhanced chemiluminescence (ECL) substrates (Pierce, USA) [Citation28].
Statistical analysis
Data are expressed as the mean ± standard deviation and were analyzed using SPSS Statistics 21 software (SPSS, USA). Statistical significance was set at P < 0.05. Statistical significance was analyzed using one-way analysis of variance (ANOVA) or Student’s t-test.
Results
Herein, we illustrated the regulatory network of PCGEM1/miR-129-5p/CDT1 in PCa and hypothesized that PCGEM1promoted malignant behavior of PCa cells by sponging miR-129-5p and releasing CDT1. Clinical specimen analysis showed that both PCGEM1 and CDT1 were upregulated, while miR-129-5p was downregulated in PCa. In addition, loss-of-function analysis revealed that PCGEM1 or CDT1 knockdown inhibited viability and tumor growth and promoted apoptosis and cycle arrest in PCa cells. PCGEM1 is a CDT1-dependent PCa promoter site that absorbs miR-129-5p.
PCGEM1/miR-129-5p/CDT1 axis was confirmed in PCa
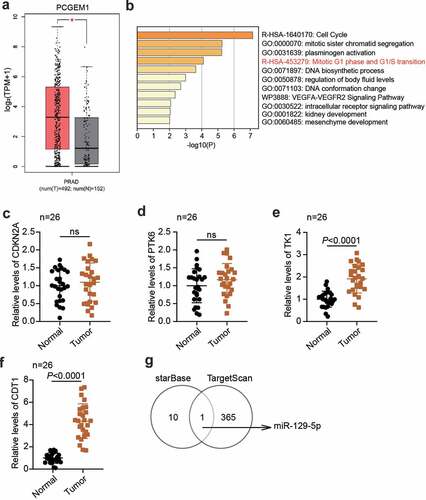
According to the data from GEPIA, PCGEM1 was upregulated in PCa (). GSE38241 was the mRNA expression profile including PCa samples and non-cancer samples, which was utilized to screen 63 upregulated genes with adj.P < 0.01 and logFC>2. Then, 63 genes were uploaded to Metascape for biological analysis, showing that four genes (CDKN2A, PTK6, TK1, and CDT1) were related to the mitotic G1 phase and G1/S transition (). Next, qRT-PCR was used to detect differences between these four genes in PCa tissues and normal tissues, and it was found that CDT1 was the most upregulated mRNA in cancer tissues (). Therefore, CDT1was selected as an interesting mRNA. To identify key miRNAs, starBase was performed to identify the downstream miRNAs of PCGEM1, and TargetScan was used to predict the upstream miRNAs of CDT1. Finally, miR-129-5p was confirmed to be able to bind to PCGEM1 and CDT1 ().
Figure 1. The key regulators were confirmed in PCa. (a) the PCGEM1 expression in PCa was confirmed by GEPIA database. (b) the key biological process involving the key genes in GSE38241 was enriched by metascape. (c-f) analysis of CDKN2A (c), PTK6 (d), TK1 (e) and CDT1 (f)level in PCa as well as normal samples based on qRT-PCR assay. (n = 26). (g) miR-129-5p was the only miRNA binding to PCGEM1 and CDT1 based on the prediction of starBase and TargetScan.
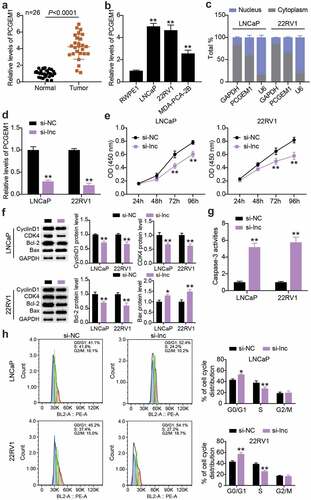
PCGEM1 was up-regulated in PCa and affected the malignant phenotypes of cancer cells
We determined PCGEM1 expression in PCa and normal tissues. The results showed that PCGEM1 levels in tumor tissues were approximately 4-fold higher than those in normal tissues (). PCGEM1 expression was detected in the three PCa cells. As illustrated in (), the expression of PCGEM1 in LNCaP, 22RV1, and MDA-PCA-2B cells was 5, 4.6, and 2.5 times higher than that in RWPE1 cells, respectively. The nucleoplasmic regionalization of PCGEM1 was analyzed using the subcellular classification of LNCaP and 22RV1 cells. We found that PCGEM1 expression was higher in the cytoplasm than in the nucleus, indicating that PCGEM1 is mainly localized in the cytoplasm and may play a role through transcription (). Subsequently, this study clarified PCGEM1ʹs role in the function of PCa cells. First, 22RV1 and LNCaP cells were treated with siRNA to downregulate PCGEM1 expression, which showed that the PCGEM1 level in the si-lnc group was 20% of that in the si-NC group (). Second, in in vitro experiments, CCK-8 analysis revealed that inhibition of PCGEM1 reduced cell viability by approximately 25% (). In addition, western blotting revealed down-regulation of CyclinD1, CDK4, and Bcl-2 protein levels and upregulation of Bax protein levels in the si-lnc group compared to the si-NC group (). Caspase-3 activity analysis revealed that the apoptosis rates of LNCaP and 22RV1 cells in the si-lnc group were increased by about 5 and 5.8 times, respectively, compared with those in the si-NC group (). In addition, PCGEM1 silencing enhanced the proportion of G0/G1 phase cells and decreased the proportion of S phase cells ().
Figure 2. PCGEM1 was up-regulated in PCa and affected the malignant phenotypes of cancer cells. (a) analysis of PCGEM1 expression in PCa as well as normal samples based on qRT-PCR. (b) PCGEM1 level in prostate cell lines was uncovered using qRT-PCR. **P < 0.001 vs. RWPE1. (c) Subcellular fractionation for PCGEM1 in 22RV1 and LNCaP cells. (d) knockdown efficiency of PCGEM1 was evaluated utilizing qRT-PCR assay. (e) viability of LNCaP and 22RV1 cells treated with si-lnc was uncovered utilizing CCK-8. (f) bax, Bcl-2, CyclinD1 and CDK4 protein levels were evaluated using western blotting assay in LNCaP and 22RV1 cells treated by si-lnc. (g) caspase-3 activity of LNCaP and 22RV1 cells delivered si-lnc were assessed using the caspase-3 activity assay. (h) Cell cycle of LNCaP as well as 22RV1 delivered si-lnc was assessed using flow cytometry assay. *P < 0.05; **P < 0.001 vs. si-NC.
Interference with PCGEM1 inhibited the growth of PCa cells in vivo
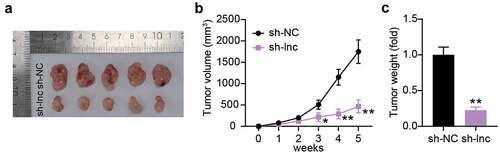
To clarify the impact of PCGEM1 on PCa cell growth in vivo, we injected LNCaP cells with PCGEM1 knockdown into nude mice. In contrast to the sh-NC group, the tumor morphology and volume of nude mice in the low PCGEM1 expression group were smaller (). The tumor was weighed, showing that PCGEM1 silencing attenuated the tumor weight. These results suggest that the knockdown of PCGEM1 may inhibit PCa cell growth in vivo ().
Figure 3. Interference with PCGEM1 inhibited the growth of PCa cells in vivo. (a) images of tumor in si-lnc group. (b) the difference of the growth curve of tumor volume in si-lnc groups were portrayed. (c) the difference of tumor weight in si-lnc groups were portrayed. *P < 0.05; **P < 0.001 vs. sh-NC.
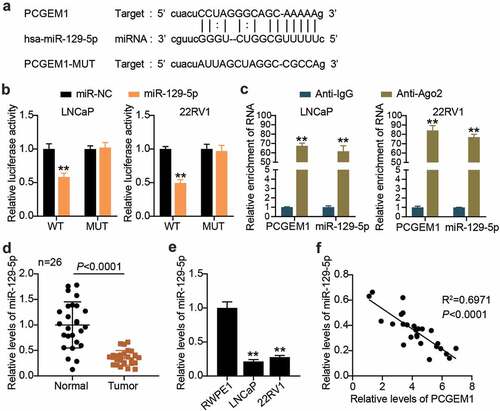
PCGEM1 sponged miR-129-5p
Using bioinformatics analysis, we found that there were binding sites between the pCGEM1 and miR-129-5p sequences (). Next, the binding relationship between pCGEM1 and miR-129-5p was further explored. Luciferase assay results showed that the miR-129-5p ectopic expression only reduced PCGEM1-WT luciferase activity, whereas the luciferase activity of PCGEM1-MUT did not respond to the overexpression of miR-129-5p (). In addition, it was revealed that miR-129-5p and PCGEM1 were enriched in compounds precipitated by anti-Ago2 antibodies in the RIP experiment (). The interaction between PCGEM1 and miR-129-5p has been demonstrated. By measuring miR-129-5p levels in clinical tissues, it was determined that miR-129-5p levels were lower in cancer tissues than in paracancerous specimens (). Similarly, we also found that miR-129-5p expression was significantly lower in both types of cancer cells compared with that in RWPE1 cells (). Pearson correlation analysis also confirmed a negative correlation between PCGEM1 and miR-129-5p in cancer (). In conclusion, the PCGEM1 sponge absorbed miR-129-5p.
Figure 4. PCGEM1 sponged miR-129-5p. (a) the wild-type and mutant binding site of PCGEM1 for miR-129-5p were constructed. (b) interaction of PCGEM1 and miR-129-5p was ascertained through luciferase assay. **P < 0.001 vs. miR-NC. (c) Relationship of PCGEM1 and miR-129-5p was validated using RIP assays. **P < 0.001 vs. Anti-IgG. (d) Analysis of miR-129-5p level in PCa as well as normal samples based on qRT-PCR. (e) miR-129-5p expression in prostate cell lines was uncovered utilizing qRT-PCR. **P < 0.001 vs. RWPE1. (f) Pearson was utilized for exploring the connection of PCGEM1 with miR-129-5p.
PCGEM1/miR-129-5p accelerated the development of PCa
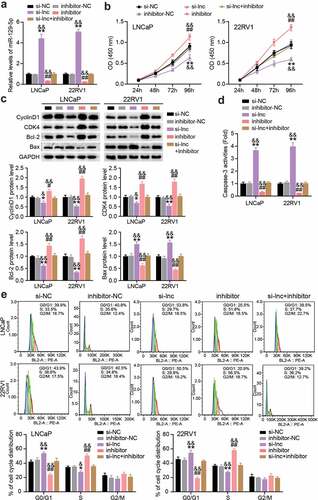
In a follow-up study, we performed a rescue trial to verify the role of PCGEM1/miR-129-5p in PCa. First, we reduced the expression of PCGEM1 and miR-129-5p in LNCaP and 22RV1 cells, indicating that miR-129-5p increased after PCGEM1 knockdown, but decreased when miR-129-5p was knocked down (). In functional analysis, as shown in (), CCK-8 showed that cell viability was enhanced by miR-129-5p interference, and miR-129-5p interference counteracted PCGEM1 silencing on cell viability inhibition. In addition, western blotting analysis showed that the protein levels of CyclinD1, CDK4, and Bcl-2 were upregulated and the protein level of Bax was downregulated after miR-129-5p interference, resulting in a reversal of protein levels after PCGEM1 silencing (). Furthermore, miR-129-5p inhibitor inhibited the caspase-3 activity of cancer cells and reversed the enhanced apoptosis effect of si-lnc (). Moreover, flow cytometry revealed that the PCGEM1 knockdown effect on G0/G1 phase cell promotion was caused by the low expression of miR-129-5p (). In other words, we conclude that PCGEM1 modulates miR-129-5p to achieve its functional effects on PCa cells.
Figure 5. PCGEM1/miR-129-5p accelerated the development of PCa. (a) The miR-129-5p level was uncovered utilizing qRT-PCR assay in LNCaP and 22RV1 delivered si-lnc or inhibitor. (b) The viability of LNCaP as well as 22RV1 treated by si-lnc or miR-129-5p inhibitor was uncovered utilizing the CCK-8 assay. (c) Bax, Bcl-2, CyclinD1 and CDK4 protein levels were evaluated using western blotting assay in LNCaP and 22RV1 cells treated by si-lnc or miR-129-5p inhibitor. (d) The caspase-3 activity of LNCaP and 22RV1 cells treated with si-lnc or miR-129-5p inhibitor was assessed using the caspase-3 activity assay. (e) Cell cycle of LNCaP as well as 22RV1 delivered by si-lnc as well as miR-129-5p inhibitor were uncovered utilizing flow cytometry assay. *P < 0.05, **P < 0.001 vs. si-NC; #P < 0.05, ##P < 0.001 vs. inhibitor-NC; &P < 0.05, &&P < 0.001 vs. si-lnc+inhibitor.
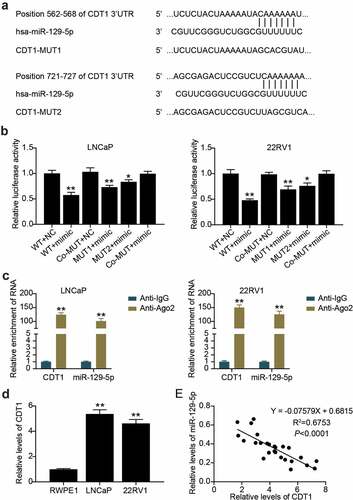
CDT1 was a downstream target of miR-129-5p
Targetscan predicted that there were two binding sites between CDT1 and miR-129-5p (). The luciferase assay showed that compared with the WT + NC group, the luciferase activity of cancer cells in WT + mimic, MUT1 + mimic, and MUT2 + mimic groups decreased by 50%, 30% and 20%, respectively (). In addition, the RIP assay revealed that miR-129-5p and CDT1 were enriched in Anti-Ago2 antibody-precipitated compounds compared with anti-IgG (). We further found that CDT1 in LNCaP and 22RV1 cells was 5 and 4.5 times higher than that in RWPE1 cells, respectively (). Additionally, miR-129-5p levels were inversely correlated with CDT1 levels (). miR-129-5p targets CDT1.
Figure 6. CDT1 was a downstream target of miR-129-5p. (a) The wild-type and mutant binding site of CDT1 for miR-129-5p were constructed. (b)The interaction between CDT1 and miR-129-5p was confirmed by luciferase reporter assay. *P < 0.05; **P < 0.001 vs. WT-NC. (c) Relationship of CDT1 and miR-129-5p was validated using RIP assays. **P < 0.001 vs. Anti-IgG. (d) The CDT1 expression in prostate cell lines was measured using qRT-PCR. **P < 0.001 vs. RWPE1. (e) Pearson was utilized for exploring the connection of CDT1 with miR-129-5p.
miR-129-5p restrained PCa cell survival and facilitated apoptosis by regulating CDT1
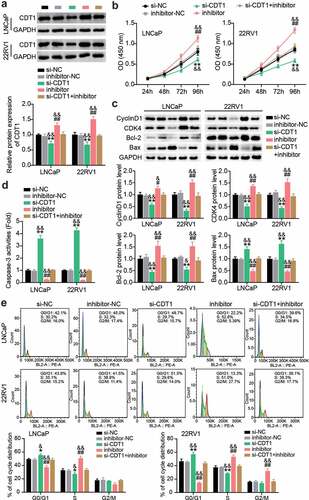
siRNAs targeting CDT1 and miR-129-5p inhibitor were transfected into LNCaP and 22RV1 cells. Western blotting showed that miR-129-5p inhibitor treatment upregulated CDT1 protein levels, while si-CDT1 treatment downregulated CDT1 protein levels (). The CCK-8 assay revealed that the viability of LNCaP and 22RV1 cells was significantly inhibited after CDT1 knockdown, and the pro-cell viability effect of miR-129-5p inhibitor was largely mitigated (). In addition, western blotting showed that CDT1 knock-down inhibited CyclinD1, CDK4, and Bcl-2 protein levels; promoted Bax; and reversed the regulatory effect of miR-129-5p inhibitor (). Caspase-3 activity analysis showed that the level of apoptosis was increased after si-CDT1 transfection, and the inhibition of apoptosis by miR-129-5p inhibitor was eliminated (). In addition, after downregulation of CDT1, the proportion of G0/G1 cells increased, and the proportion of S decreased, while knockdown of miR-129-5p reversed this effect ().
Figure 7. miR-129-5p restrained PCa cells survival whereas facilitated apoptosis by regulating CDT1. (a) The CDT1 protein expression was evaluated using western blotting assay in LNCaP and 22RV1 cells treated by si-CDT1. (b) The viability of LNCaP and 22RV1 cells treated by si-CDT1 was assessed using the CCK-8 assay. (c) Bax, Bcl-2, CyclinD1 and CDK4 protein levels were evaluated using western blotting assay in LNCaP and 22RV1 cells treated by si-CDT1 inhibitor. (d) The caspase-3 activity of LNCaP and 22RV1 delivered si-CDT1 was uncovered utilizing the caspase-3 activity assay. (e) Cell cycle of LNCaP as well as 22RV1 delivered si-CDT1 was assessed using flow cytometry assay. *P < 0.05, **P < 0.001 vs. si-NC; #P < 0.05, ##P < 0.001 vs. inhibitor-NC; &P < 0.05, &&P < 0.001 vs. si-CDT1+ inhibitor.
Discussion
lncRNAs play a crucial role in multiple processes that modulate gene expression, and their transcription can lead to gene silencing or gene activation [Citation29,Citation30]. It has been revealed that lncRNAs participate in the biological processes of many diseases, including cancer, regulating cell proliferation, apoptosis, differentiation, and metastasis [Citation31,Citation32]. For example, elevated levels of lncRNA ROR in PCa tissue are associated with late tumor staging, lymph node or distant metastasis, and modulation of the AKT pathway to promote malignant proliferation of PCa cells [Citation33]. Overexpression of lncRNA PVT1 in PCa tissues and cells is associated with low overall and disease-free survival and tumor staging [Citation34]. Down-regulated lncRNA CRNDE inhibits the proliferation and migration of PCa cells and induces apoptosis [Citation28].
Although many lncRNAs have been discovered, their exact functions and potential mechanisms in cancer remain to be further investigated. Here, we investigated the role of PCGEM1 in PCa. PCGEM1 was found to be upregulated in PCa, which was similar to previous studies [Citation35]. In addition, in previous studies, the silencing of PCGME1 by small interfering RNA significantly restrained PCa cell proliferation and growth, and induced early apoptosis [Citation36,Citation37]. Based on these results, we further found that knockdown of PCGME1 restrained the viability of PCa cells and resulted in cell cycle arrest. In addition, subcellular localization experiments showed that PCGME1 plays a role in the cytoplasm of PCa cells.
IncRNAs can exert miRNA sponge effects through the ceRNA mechanism [Citation38]. In this study, we found that miR-129-5p was negatively regulated by PCGEM1 and negatively correlated with PCGEM1 in PCa. Using a bioinformatics website and luciferase assay, PCGEM1 was revealed as a sponge of miR-129-5p. In addition, RNA immunoprecipitation with Ago2 revealed that miR-129-5p and PCGEM1 were highly enriched in PCa cells. This result is consistent with the findings of Zhang et al. [Citation14] and Li et al. [Citation15] that PCGEM1 targets and negatively regulates miR-129-5p. Additionally, we further revealed that miR-129-5p was under-expressed in PCa, and miR-129-5p knockdown promoted cancer cell viability and inhibited apoptosis and cycle arrest. This is consistent with previous studies that have reported miR-129-5p as an anticancer agent in PCa [Citation13].
DNA replication licensing factor CDT1, a basic protein that replicates the corresponding origin licensing in G1, is degraded in the S phase and reaccumulates in the G2 phase [Citation39]. The accumulation of CDT1 regulates the DNA replication phenotype, thus affecting tumor cell replication and apoptosis [Citation40]. Accumulated literature suggests that CDT1 may be a new marker for cancer diagnosis and prognosis [Citation41]. CDT1 was upregulated in both hepatocellular carcinoma [Citation17] and breast cancer [Citation18]. However, in a study of lung cancer, CDT1 expression was shown to be suppressed in cancer tissues and cells [Citation42]. It was speculated that the differential expression of CDT1 may be related to the tissue type. In this study, we uncovered for the first time that CDT1 was expressed more in PCa, and its knockdown restrained LNCaP and 22RV1 cell viability. This conclusion was similar to that found by Takeo et al. [Citation19] in LNCaP cells with increased proliferation and increased CDT1 expression. In addition, this study found that silencing CDT1 promoted apoptosis and cell cycle arrest in PCa cells, which had similar properties to the initiation of CDT1 replication in G1 phase cells in yeast strains [Citation43]. Furthermore, we found that CDT1 was the downstream target gene of miR-129-5p, and CDT1 knockdown eliminated the effect of interfering miR-129-5p on PCa cells.
Although solid studies have been conducted on the role of PCGEM1/miR-129-5p/CDT1 in PCa, there are still limitations to our study. On the one hand, we did not study the effect of PCGEM1 on tumor metastasis in vivo. In addition, the correlation between PCGEM1 and survival prognosis and cancer metastasis in PCa patients requires further study.
Conclusion
Our study showed that PCGEM1 promoted the progression of PCa by acting on miR-129-5p and modulating CDT1 through sponges. It is suggested that PCGEM1/miR-129-5p/CDT1 could be used as a biomarker for PCa screening and as a therapeutic target.
Ethics approval
Ethics Committee of The third hospital of Wuhan (Wuhan, China) approved the present study. Clinical analysis complied with the ethical standards of the Declaration of Helsinki.
Consent to participate
Written informed consent acquired from all patients.
Consent for publication
Participants offered consent for publication.
Availability of data and material
The datasets used and/or analyzed in this study are available from the corresponding author on reasonable request.
Code availability
Not available.
Authors’ contributions
QF and FFW conducted the experiments and analyzed data. JY and WS offered the experimental consideration. ZH and LX collected data. HC and XW and WZ did the analysis and interpretation of data. The manuscript was read and approved through all authors.
Supplemental Material
Download Zip (24.9 MB)Acknowledgements
None.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here.
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Pernar CH, Ebot EM, Wilson KM, et al. The epidemiology of prostate cancer. Cold Spring Harb Perspect Med. 2018;8(12):a030361.
- Rawla P. Epidemiology of prostate Cancer. World J Oncol. 2019;10(2):63–89.
- Eeles R, Goh C, Castro E, et al. The genetic epidemiology of prostate cancer and its clinical implications. Nat Rev Urol. 2014;11(1):18–31.
- Ali T, Grote P. Beyond the RNA-dependent function of LncRNA genes. eLife. 2020;9. DOI:10.7554/eLife.60583
- Hua JT, Chen S, He HH. Landscape of noncoding RNA in prostate Cancer. Trends Genet. 2019;35(11):840–851.
- Walsh AL, Tuzova AV, Bolton EM, et al. Long noncoding RNAs and prostate carcinogenesis: the missing ‘linc’? Trends Mol Med. 2014;20(8):428–436.
- Ho TT, Huang J, Zhou N, et al. Regulation of PCGEM1 by p54/nrb in prostate cancer. Sci Rep. 2016;6(1):34529.
- Fu X, Ravindranath L, Tran N, et al. Regulation of apoptosis by a prostate-specific and prostate cancer-associated noncoding gene, PCGEM1. DNA Cell Biol. 2006;25(3):135–141.
- Wang JY, Yang Y, Ma Y, et al. Potential regulatory role of lncRNA-miRNA-mRNA axis in osteosarcoma. Biomed Pharmacothe. 2020;121:109627.
- Xu C, Du Z, Ren S, et al. MiR-129-5p sensitization of lung cancer cells to etoposide-induced apoptosis by reducing YWHAB. J Cancer. 2020;11(4):858–866.
- Wu Q, Meng WY, Jie Y, et al. LncRNA MALAT1 induces colon cancer development by regulating miR-129-5p/HMGB1 axis. J Cell Physiol. 2018;233(9):6750–6757.
- Gao G, Xiu D, Yang B, et al. miR-129-5p inhibits prostate cancer proliferation via targeting ETV1. Onco Targets Ther. 2019;12:3531–3544.
- Zhang T, Piao HY, Guo S, et al. LncRNA PCGEM1 enhances metastasis and gastric cancer invasion through targeting of miR-129-5p to regulate P4HA2 expression. Exp Mol Pathol. 2020;116:104487.
- Li Q, Shen F, Zhao L. The relationship between lncRNA PCGEM1 and STAT3 during the occurrence and development of endometrial carcinoma. Biomed Pharmacothe. 2018;107:918–928.
- Agarwal S, Smith KP, Zhou Y, et al. Cdt1 stabilizes kinetochore-microtubule attachments via an aurora B kinase-dependent mechanism. J Cell Biol. 2018;217(10):3446–3463.
- Karavias D, Maroulis I, Papadaki H, et al. Overexpression of CDT1 Is a predictor of poor survival in patients with hepatocellular carcinoma. J Gastrointest Surg. 2016;20(3):568–579.
- Mahadevappa R, Neves H, Yuen SM, et al. The prognostic significance of cdc6 and cdt1 in breast cancer. Sci Rep. 2017;7(1):985.
- Kosaka T, Yasumizu Y, Miyazaki Y, et al. Potent increased risk of the initiation of DNA replication in human prostate cancer with the use of 5α-reductase inhibitors. Am J Clin Exp Urol. 2014;2(2):136–144.
- Guan H, Zhu T, Wu S, et al. Long noncoding RNA LINC00673-v4 promotes aggressiveness of lung adenocarcinoma via activating WNT/β-catenin signaling. Proc Natl Acad Sci U S A. 2019;116(28):14019–14028.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta C(T)) Method. Methods (San Diego. Calif). 2001;25(4):402–408.
- Cheng Q, Zhang M, Zhang M, et al. Long non-coding RNA LOC285194 regulates vascular smooth muscle cell apoptosis in atherosclerosis. Bioengineered. 2020;11(1):53–60.
- Ming-Yan Y, Jing Z, Shu-Qin G, et al. Liraglutide inhibits the apoptosis of human nucleus pulposus cells induced by high glucose through PI3K/Akt/caspase-3 signaling pathway. Biosci Rep. 2019;39(8). DOI:10.1042/BSR20190109
- Liang Z, Xu J, Ma Z, et al. MiR-187 suppresses non-small-cell lung cancer cell proliferation by targeting FGF9. Bioengineered. 2020;11(1):70–80.
- Ding M, Jiang CY, Zhang Y, et al. SIRT7 depletion inhibits cell proliferation and androgen-induced autophagy by suppressing the AR signaling in prostate cancer. J Exp Clin Cancer Res. 2020;39(1):28.
- Han B, Ge Y, Cui J, et al. Down-regulation of lncRNA DNAJC3-AS1 inhibits colon cancer via regulating miR-214-3p/LIVIN axis. Bioengineered. 2020;11(1):524–535.
- Jiang Y, Liu H, Yu H, et al. Circular RNA calm4 regulates hypoxia-induced pulmonary arterial smooth muscle cells pyroptosis via the Circ-Calm4/miR-124-3p/PDCD6 axis. Arterioscler Thromb Vasc Biol. 2021;41(5):1675–1693.
- Liu K, Zhao D, Wang D. LINC00528 regulates myocardial infarction by targeting the miR-143-3p/COX-2 axis. Bioengineered. 2020;11(1):11–18.
- Jathar S, Kumar V, Srivastava J, et al. Technological developments in lncRNA Biology. Adv Exp Med Biol. 2017;1008:283–323.
- Xu J, Bai J, Zhang X, et al. A comprehensive overview of lncRNA annotation resources. Brief Bioinform. 2017;18(2):236–249.
- Peng WX, Koirala P, Mo YY. LncRNA-mediated regulation of cell signaling in cancer. Oncogene. 2017;36(41):5661–5667.
- Liu S, Li J, Kang L, et al. Degradation of long non-coding RNA-CIR decelerates proliferation, invasion and migration, but promotes apoptosis of osteosarcoma cells. Cancer Cell Int. 2019;19(1):349.
- Zhai XQ, Meng FM, Hu SF, et al. Mechanism of LncRNA ROR promoting prostate cancer by regulating Akt. Eur Rev Med Pharmacol Sci. 2019;23(5):1969–1977.
- Yang J, Li C, Mudd A, et al. LncRNA PVT1 predicts prognosis and regulates tumor growth in prostate cancer. Biosci Biotechnol Biochem. 2017;81(12):2301–2306.
- Prensner JR, Sahu A, Iyer MK, et al. The IncRNAs PCGEM1 and PRNCR1 are not implicated in castration resistant prostate cancer. Oncotarget. 2014;5(6):1434–1438.
- Zhang S, Li Z, Zhang L, et al. MEF2‑activated long non‑coding RNA PCGEM1 promotes cell proliferation in hormone‑refractory prostate cancer through downregulation of miR‑148a. Mol Med Rep. 2018;18(1):202–208.
- He JH, Zhang JZ, Han ZP, et al. Reciprocal regulation of PCGEM1 and miR-145 promote proliferation of LNCaP prostate cancer cells. J Exp Clin Cancer Res. 2014;33(1):72.
- Luo Y, Li H, Huang H, et al. Integrated analysis of ceRNA network in hepatocellular carcinoma using bioinformatics analysis. Medicine (Baltimore). 2021;100(22):e26194.
- Varma D, Chandrasekaran S, Sundin LJ, et al. Recruitment of the human Cdt1 replication licensing protein by the loop domain of hec1 is required for stable kinetochore-microtubule attachment. Nat Cell Biol. 2012;14(6):593–603.
- Milhollen MA, Narayanan U, Soucy TA, et al. Inhibition of NEDD8-activating enzyme induces rereplication and apoptosis in human tumor cells consistent with deregulating CDT1 turnover. Cancer Res. 2011;71(8):3042–3051.
- Petropoulou C, Kotantaki P, Karamitros D, et al. Cdt1 and geminin in cancer: markers or triggers of malignant transformation? Front Biosci. 2008;13(13):4485–4494.
- Cao Y, Li P, Wang H, et al. SIRT3 promotion reduces resistance to cisplatin in lung cancer by modulating the FOXO3/CDT1 axis. Cancer Med. 2021;10(4):1394–1404.
- Zhai Y, Cheng E, Wu H, et al. Open-ringed structure of the Cdt1-Mcm2-7 complex as a precursor of the MCM double hexamer. Nat Struct Mol Biol. 2017;24(3):300–308.
