ABSTRACT
Increasing evidence suggests that plasmacytoma variant translocation 1 (PVT1) plays a vital role in the development of multiple tumors including lung adenocarcinoma (LUAD). Eukaryotic initiation factor 4A-3 (EIF4A3) is considered a key factor in human cancers. However, the role and potential mechanism of PVT1 combined with EIF4A3 in LUAD remain unclear. This study investigated the effects and regulatory mechanisms of PVT1, EIF4A3, and circLMNB2 on the growth, migration, invasion, and epithelial-mesenchymal transition (EMT) of LUAD cells (H1299 and HCC827 cells) The expression level, diagnostic value and prognostic significance of PVT1, EIF4A3, and circLMNB2 were assessed, and enrichment analysis was performed using R package. Rescue experiments and a xenograft model were used to validate the PVT1/EIF4A3/circLMNB2 axis in LUAD. PVT1 and EIF4A3 were upregulated and indicated poor prognosis in LUAD. Knockdown of PVT1 and EIF4A3 suppressed LUAD cell proliferation, migration, invasion, and EMT. Mechanistically, PVT1 was stabilized by EIF4A3. PVT1 could recruit EIF4A3 to promote circLMNB2 expression. Rescue experiments indicated that circLMNB2 overexpression could reverse the reduced behavior caused by PVT1 or EIF4A3 knockdown. Enrichment analysis showed that PVT1/EIF4A3/circLMNB2 may regulate LUAD development by participating in ribosome biogenesis and spliceosome formation. Our findings demonstrate that PVT1/EIF4A3/circLMNB2 enhances the malignant behaviors of LUAD cells, providing a novel perspective for the clinical treatment of LUAD.
Graphical abstract
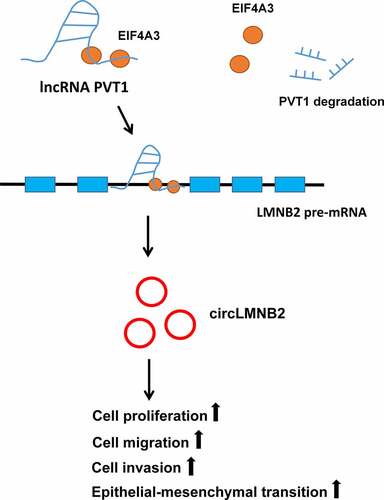
Highlights
PVT1 and EIF4A3 were upregulated and had prognostic significance in LUAD.
PVT1 and EIF4A3 knockdown suppressed LUAD cell proliferation, migration, invasion, and EMT.
PVT1 could be stabilized by EIF4A3.
PVT1 could recruit EIF4A3 to promote circLMNB2 expression in LUAD.
CircLMNB2 overexpression could reverse the reduced behavior by PVT1 or EIF4A3 knockdown.
Introduction
Lung cancer is one of the most common malignant tumors worldwide, with lung adenocarcinoma (LUAD) accounting for the most dominant subtype [Citation1,Citation2]. Although advancements in gene detection and targeted therapies have improved the quality of life of patients with cancer and 5-year overall survival rate, the treatment and prognosis of lung cancer remain unsatisfactory [Citation3–5]. Thus, it is necessary to investigate more effective biomarkers and underlying mechanisms to improve the diagnostic accuracy, clinical efficiency and prognosis of LUAD.
Long noncoding RNAs (lncRNAs) play key roles in various tumors [Citation6–9]. For instance, dysregulation of plasmacytoma variant translocation 1 (PVT1) is associated with multiple malignant cancers [Citation10]. In gastric cancer, PVT1 mediates angiogenesis by activating the signal transducer and activator of transcription 3/ vascular endothelial growth factor A axis [Citation11]. PVT1 promotes colorectal cancer progression by enhancing myc expression [Citation12]. Recent studies have shown that PVT1 plays an oncogenic role in lung cancer [Citation13–15], suggesting that PVT1 is vital in lung cancer development. Thus, conducting an in-depth study on the mechanisms of PVT1 to promote its clinical application in the diagnosis and prognosis of lung cancer is worthwhile.
RNA-binding proteins (RBPs) are a special set of proteins which can modify RNA processing, influence RNA localization and transport, or regulate RNA stability [Citation16,Citation17]. Eukaryotic translation initiation factor 4A3 (EIF4A3) is a putative RNA helicase and belongs to the DEAD box protein family. It is reported that EIF4A3 plays crucial roles in nonsense-mediated decay, RNA splicing, and RNA trafficking [Citation18,Citation19]. A study showed that EIF4A3 could regulate the TFEB-mediated transcriptional response via GSK3B to control autophagy [Citation20]. One research revealed that lncRNA CASC11 could promote HCC progression through EIF4A3-mediated E2F1 activation [Citation21]. Another report showed that EIF4A3 could facilitate glioblastoma tumorigenesis by stabilizing LINC00680 and TTN-AS1 [Citation22]. This study predicted that EIF4A3 is the most likely RBPs for PVT1 according to the UALCAN and ENCORI databases. Nevertheless, the regulatory mechanisms between EIF4A3 and PVT1 on the malignant biological behaviors of LUAD have not been elaborated.
Circular RNAs (circRNAs) are characterized by their covalent closed-loop structure and absence of 5’ end caps and 3’ poly (A) tail [Citation23–25]. Considering that their stability is not affected by RNA exonucleases, currently, their role in tumorigenesis has attracted increasing attention [Citation26]. For instance, the circRNA WHSC1 was reported to promote endometrial cancer development by regulating the miR-646/NPM1 pathway [Citation27]. Circular RNA hsa_circ_001783 was identified by sponging miR-200c-3p expression to regulate breast cancer progression [Citation28]. Circ-BNIP3 can aggravate hypoxia-induced injury in H9c2 cells by targeting miR-27a-3p/BNIP3 [Citation29]. Hsa_circ_0048409 (termed circLMNB2 in the remainder of this article) was a newly identified circRNA in our study. However, its role in LUAD has not yet been reported. In addition, it has been reported that circRNAs are generated by a non-canonical alternative splicing, and RBPs are important for the generation of circRNAs [Citation30]. In this study, whether EIF4A3 was involved in the biogenesis of circRNAs in LUAD remains to be explored.
This study aimed to investigate the regulatory mechanisms of PVT1 and EIF4A3 in LUAD cells. This study found that PVT1 and EIF4A3 were upregulated in LUAD and had diagnostic and prognostic values for LUAD. PVT1 could be stabilized by EIF4A3. PVT1 and EIF4A3 could promote the malignant behavior of LUAD in vitro and in vivo. Moreover, PVT1 could recruit EIF4A3 to regulate circLMNB2 biogenesis. Our findings reveal a novel regulatory mechanism of PVT1/EIF4A3/circLMNB2 in LUAD and provide a novel perspective on the potential use of PVT1 in LUAD clinical treatment.
Materials and methods
Data analysis
Expression profiles of EIF4A3, PVT1, and circLMNB2 in The Cancer Genome Atlas (TCGA, https://genome-cancer.ucsc.edu/)-Lung Adenocarcinoma (LUAD) dataset were analyzed using R software (version 3.6.3) ggplot2 package (version 3.3.3). The diagnostic values of PVT1, EIF4A3, and circLMNB2 in LUAD were analyzed using the pROC package (1.17.0.1). The overall survival (OS) of PVT1 in LUAD was analyzed using the Kaplan-Meier plotter (https://kmplot.com/analysis/index.php?p=service&cancer=lung) [Citation31]. The OS of EIF4A3 and circLMNB2 was analyzed using the survminer package (version 0.4.9). The correlation between EIF4A3 and PVT1 was analyzed using ggplot2 (version 3.3.3). The immunohistochemical images of EIF4A3 in LUAD tumor and normal tissues were obtained from the Human Protein Atlas (https://www.proteinatlas.org/). A Venn diagram was constructed using ggplot2 (version 3.3.3). The RNA-binding proteins (RBPs) of circLMNB2 were predicted by ENCORI (https://starbase.sysu.edu.cn/index.php) [Citation32] and the Cancer-Specific CircRNA Database (CSCD, http://gb.whu.edu.cn/CSCD/).
Clinical samples
Thirty-two paired LUAD tumor samples were obtained from the First Affiliated Hospital of Fujian Medical University from January 2019 to December 2020. Written informed consent was obtained from all patients involved. This study was approved by the First Affiliated Hospital of Fujian Medical University (MRCTA, ECFAH of FMU [2020]322). Patients’ clinical features are shown in .
Table 1. Clinical features of patients with lung adenocarcinoma (n = 32)
Cell culture
Human non-small cell lung cancer (NSCLC) cell lines H1299 and HCC827 were purchased from Procell Life Science&Technology Co.,Ltd. (Wuhan, China) and cultured in Roswell Park Memorial Institute-1640 (RPMI-1640) medium (Gibco, Grand Island, NY, USA) supplemented with 10% fetal bovine serum (FBS, Gibco) and 1% penicillin-streptomycin (Invitrogen, Carlsbad, CA, USA). Cells were maintained at 37°C with 5% CO2.
Cell transfection
H1299 and HCC827 cells (5 × 105 cells/well) were seeded in 6-well plates. EIF4A3/PVT1/circLMNB2 short hairpin RNAs (shRNAs) and scramble shRNA (2 μg/well) were transfected into H1299 or HCC827 cells using Lipofectamine® 2000 (Invitrogen, Carlsbad, CA, USA). Cells were harvested 48 h post-transfection for subsequent experiments. Stable cells expressing sh-PVT1, sh-EIF4A3, or their corresponding controls were selected by adding puromycin (500 μg/ml). The sequences were listed in Supplementary Table 1.
RNA extraction and reverse transcription-quantitative polymerase chain reaction
RNA was isolated from tissues or cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.). The extracted RNA (approximately 200 ng) was reverse-transcribed into cDNA using the EasyScript® First-Strand cDNA Synthesis SuperMix kit (Transgen Biotechnology Co., Ltd, Beijing, China) according to the manufacturer’s protocols. Quantitative polymerase chain reaction (qPCR) analysis was performed using the TransStart® Green qPCR SuperMix (Transgen, Beijing). The thermocycling conditions were as follows. 94°C for 5 min, followed by 40 cycles at 94°C for 10s, 58°C for 40s, and 72°C for 10s. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as an internal control. Relative gene expression was calculated using the 2−ΔΔCq method [Citation33]. The sequences of the PCR primers are listed in Supplementary Table 1.
Nuclear–cytoplasmic fractionation
The nucleus and cytoplasm of RNAs from H1299 or HCC827 cells were analyzed using the Nuclear/Cytosol Fractionation Kit (BioVision. lnc.) according to the manufacturer’s instructions [Citation34].
Actinomycin D treatment
For new RNA synthesis inhibition, H1299 or HCC827 cells were treated with 100 ng/ml actinomycin D (Cell Signaling Technology, Beverly, MA, USA) for 2, 4, and 6 h.
RNA pull-down
In brief, PVT1 transcripts were transcribed using T7 RNA polymerase (Ambion Life), then using the RNeasy Plus Mini Kit (QIAGEN) and treated with DNase I (QIAGEN) [Citation35]. Biotin-labeled wild-type PVT1 and antisense RNA were transcribed in vitro using T7 RNA polymerase (Ambion Life) and Biotin RNA Labeling Mix (Ambion Life) with RNase-free DNase I (QIAGEN) treatment. Subsequently, RNAs were purified using the RNeasy Plus Mini Kit (QIAGEN). H1299 or HCC827 cell lysates were incubated with biotinylated RNAs and streptavidin agarose beads (Invitrogen Life Technologies) at room temperature. After incubation for 2 h, the eluted proteins were detected by western blotting.
RNA immunoprecipitation
RNA immunoprecipitation (RIP) was performed using the Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, MA, USA) [Citation36]. The anti-EIF4A3 antibody (1:20, ab180573, Abcam, Cambridge, UK) was used for RIP. Cell lysates were incubated with EIF4A3 or IgG antibodies with A/G magnetic beads (Thermo Fisher Scientific) for 4 h at 4°C. qPCR analysis was performed to measure the enrichment of PVT1 and circLMNB2 in H1299 or HCC827 cells in different transfection groups.
Western blotting
Proteins were extracted from H1299 and HCC827 cells using RIPA lysis buffer, and the extracted proteins were quantified using the BCA protein assay kit (Beyotime). Proteins (40 μg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene difluoride (PVDF, EMD Millipore) membranes. The membranes were blocked with 5% nonfat milk at 4°C for 1 h. Subsequently, the membranes were incubated with primary antibodies against EIF4A3 (ab180573, 1:1000), vimentin (ab92547, 1:1000), N-cadherin (ab76011, 1:5000), E-cadherin (ab40772, 1:10000), and GAPDH (ab8245, 1:2000) at 4°C overnight. The next day, the membranes were incubated with horseradish peroxidase-conjugated secondary antibodies (ab6789, 1:2000; ab6721, 1:2000) at room temperature for 1 h. The blots were detected using an enhanced chemiluminescence kit (Pierce, Rockford, IL, USA).
Nascent RNA capture assay
Nascent RNAs were isolated using the Click-iT Nascent RNA Capture Kit (Invitrogen, Cat # C10365) according to the manufacturer’s protocol [Citation37]. Briefly, H1299 or HCC827 cells were incubated with 0.2 mM 5-ethynyl uridine (EU), subsequently, the total RNAs were extracted. The EU-labeled RNA was used for biotinylation in a Click-iT reaction buffer. The biotinylated RNA was collected using streptavidin magnetic beads.
Cell counting Kit-8
Cell viability was assessed using Cell Counting Kit-8 (CCK-8, Beyotime, Shanghai, China). H1299 or HCC827 cells (density, 6 × 103 cells/well) were seeded in 96-well plates. After transfection for 24, 48 or 72 h, cells were incubated with 10 μl CCK-8 solution at 37°C for 2 h. Absorbance was measured at 450 nm using a microplate reader (Bio-Rad Laboratories, Inc.).
Transwell assays
For cell migration analysis, cells were seeded at a density of 5 × 104 cells/well in the upper chamber with serum-free medium. Medium containing 10% FBS was added to the lower chamber. After incubation for 24 h, the cells on the bottom membrane were fixed with 4% paraformaldehyde at room temperature for 10 min and stained with 0.4% crystal violet solution. Cells were imaged using a microscopy (Nikon). The number of migratory tumor cells was counted in five randomly selected fields. For cell invasion analysis, the upper chamber was coated with Matrigel matrix (BD Biosciences) [Citation38]. Cells were seeded at a density of 5 × 104 cells/well in the upper chamber with serum-free medium. A medium containing 15% FBS was added to the lower chamber. After incubation for 24 h, the cells on the bottom surface were fixed with 4% paraformaldehyde at room temperature for 10 min and stained with 0.4% crystal violet solution. Cells were imaged using a microscopy (Nikon). Invasive tumor cells were counted in five randomly selected fields.
Xenograft tumor model
Animal experiments were conducted at Beijing Viewsolid Biotech Co., Ltd. Four-week-old female BALB/c nude mice were purchased from Sibf Biotechnology Co., Ltd (Beijing). The mice were maintained under the specific pathogen-free conditions. Stable PVT1 or EIF4A3 knockdown H1299 cells (2.5 × 106) were subcutaneously injected into the mice. Tumor size was measured every 7 days. The mice were sacrificed after 28 days. Tumor volume was calculated using the following formula (length × width2/2) [Citation39].
Statistical analyses
Statistical analyses were performed using GraphPad Prism V8 software. Data are presented as the mean ± standard deviation (SD). The Wilcoxon signed rank test or Student’s paired t-test was used for the analysis of clinical samples. Unpaired Student’s t-test was used for comparisons between cell groups. The correlation between EIF4A3 and PVT1 was analyzed using Pearson’s correlation coefficient. One-way analysis of variance was used for multiple comparisons. Statistical significance was set at P < 0.05.
Results
The aim of this study is to investigate the role and underlying mechanism of lncRNA PVT1 and EIF4A3 in regulating the tumor behavior of LUAD cells. We first analyzed the expression of lncRNA PVT1 between LUAD tumor tissues and their adjacent normal tissues, and identified the most possible interacting RBP EIF4A3. LncRNA PVT1 and EIF4A3 were both significantly upregulated in LUAD tumor tissues, which were associated with poorer overall survival of LUAD patients. Loss-of-function experiments showed the tumor-promoting role of lncRNA PVT1 and EIF4A3 in LUAD cells including cell proliferation, cell migration, cell invasion and EMT. Moreover, PVT1 could be stabilized by EIF4A3. CircLMNB2 was identified to be the target of PVT1. PVT1 could recruit EIF4A3 to promote circLMNB2 expression in LUAD cells. In conclusion, these results demonstrate that lncRNA PVT1/EIF4A3/ circLMNB2 axis plays an oncogenic role in regulating the malignant behavior of LUAD cells.
Plasmacytoma variant translocation 1 (PVT1) and eukaryotic initiation factor 4A-3 (EIF4A3) were upregulated and had a prognostic value in lung adenocarcinoma (LUAD)
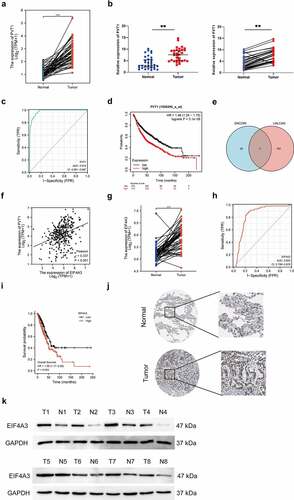
PVT1 has been reported to act as an ‘oncogene’ in LUAD. This study first analyzed the expression pattern of PVT1 in 57 paired LUAD tumor samples using the TCGA-LUAD dataset and observed that PVT1 was upregulated in LUAD tumor tissues compared to that in normal tissues (). Next, this study validated that PVT1 was highly expressed in LUAD tumor tissues (n = 32) using qRT-PCR (). Diagnostic value prediction results showed that PVT1 could predict lung cancer with high accuracy (, area under the curve [AUC] = 0.974, confidence interval [CI] = 0.961–0.987). Survival analysis showed that high PVT1 expression was associated with a poor prognosis in patients with NSCLC (). We predicted PVT1 RNA-binding and associated proteins using ENCORI (https://starbase.sysu.edu.cn/index.php) and UALCAN (http://ualcan.path.uab.edu/). The intersections were DGCR8, DHX9 and EIF4A3 (). A positive correlation was confirmed between PVT1 and EIF4A3 expressions in LUAD (, r=0.337, P < 0.001). Consistently, EIF4A3 expression was also higher in LUAD tumor tissues than in normal tissues (n = 57) (). EIF4A3 was highly expressed in random tumor tissues (). Moreover, EIF4A3 had a certain accuracy in the diagnosis of LUAD and a prognostic value for LUAD (). Taken together, our data revealed that high PVT1 and EIF4A3 expressions were associated with poor prognosis in patients with LUAD and might play a key role in LUAD tumorigenesis.
Figure 1. PVT1 and EIF4A3 were upregulated in LUAD. (a) The relative expression of PVT1 was analyzed in paired LUAD tumor samples (n = 57). *** p < 0.001. (b) The relative expression of PVT1 was analyzed in collected LUAD tumor samples (n = 32). ** p < 0.01. (c) The diagnostic value of PVT1 for LUAD was shown by ROC curve (AUC = 0.974, CI = 0.961–0.987). (d) The overall survival of lung cancer patients with PVT1 expression was analyzed by Kaplan-Meier plotter. (e) The intersection of PVT1 RNA-binding proteins predicted by ENCORI and associated proteins by UALCAN. (f) The correlation between PVT1 and EIF4A3 was analyzed by Pearson analysis. r = 0.337, P < 0.001. (g) The relative expression of EIF4A3 was analyzed in paired LUAD tumor samples (n = 57). *** p < 0.001. (h) The representative images of EIF4A3 expressing in lung cancer was obtained from human protein atlas. (i) The protein level of EIF4A3 was detected randomly in collected samples by western blot. (j) The diagnostic value of EIF4A3 for LUAD was analyzed using ROC curve (AUC = 0.832, CI = 0.788–0.876). (k) The overall survival analysis of lung cancer patients with EIF4A3 expression.
PVT1 and EIF4A3 served as oncogenes in LUAD
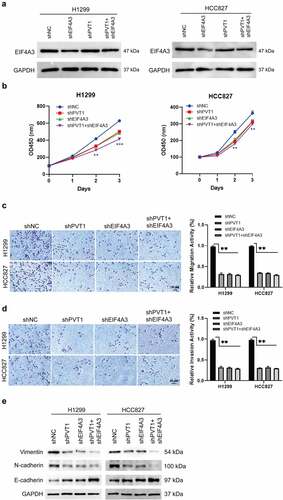
To investigate the biological role of PVT1 and EIF4A3 in LUAD, we performed loss-of-function assay in H1299 and HCC827 LUAD cells by knocking down PVT1 and EIF4A3. qPCR results showed that PVT1 shRNA #2 and EIF4A3 shRNA #1 were the most efficient shRNAs (Supplementary Figure 1A and 1B). EIF4A3 was significantly downregulated in sh-EIF4A3 cells but not in sh-PVT1 cells (). We constructed stable cell lines sh-PVT1, sh-EIF4A3, sh-PVT1 and sh-EIF4A3. Compared to sh-NC cells, knockdown of PVT1 or EIF4A3 could repress cell migration, invasion, proliferation, and epithelial-mesenchymal transition (EMT) (). Co-knockdown of PVT1 and EIF4A3 could reinforce the inhibitory effects by PVT1 knockdown or EIF4A3 knockdown alone. These data suggest that PVT1 and EIF4A3 can accelerate LUAD cell growth, migration, invasion and EMT in vitro.
Figure 2. PVT1 and EIF4A3 promoted lung adenocarcinoma cells proliferation, migration, invasion and EMT. H1299 and HCC827 cells were transfected with sh-EIF4A3, sh-PVT1, sh-EIF4A3 and sh-PVT1, or sh-NC. (a) EIF4A3 protein level was detected in the cells groups above mentioned by western blot. (b) Cell proliferation was analyzed by CCK-8 assay. (c and d) Cell migration and invasion abilities were analyzed by Transwell and Transwell-matrigel assays. (e) The EMT-related proteins were examined by western blot. *p < 0.05, **p < 0.01, ***p < 0.001. Scale bar,50 μM.
PVT1 was stabilized by EIF4A3 in LUAD
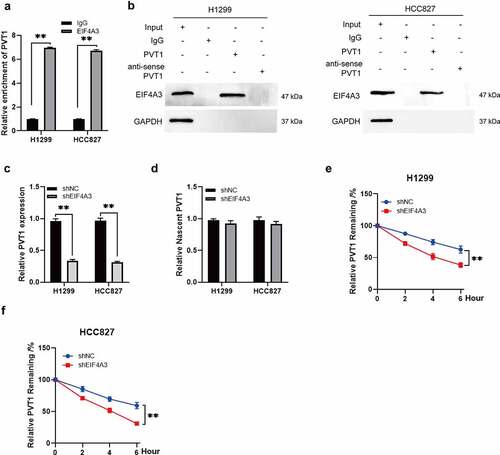
RIP assays were performed to verify the regulatory patterns of PVT1 and EIF4A3 in patients with LUAD. The results showed that PVT1 was more enriched in the anti-EIF4A3 complex compared to that in the anti-IgG complex (). The RNA pull-down assay showed that EIF4A3 could bind to PVT1 but not to mutant PVT1 (). PVT1 was downregulated in sh-EIF4A3 cells (), whereas, no significant difference was observed at nascent RNA of PVT1 between sh-EIF4A3 and sh-NC cells (). Furthermore, the half-life of PVT1 was reduced when EIF4A3 was knocked down (). These data suggest that PVT1 is stabilized by EIF4A3 in LUAD.
Figure 3. PVT1 was stabilized by EIF4A3. (a) The interaction between PVT1 and EIF4A3 was confirmed by RIP assay in H1299 and HCC827 cells. The relative enrichment of PVT1 was analyzed by qPCR. ** p < 0.01. (b) The interaction between PVT1 and EIF4A3 was performed by RNA-pull down. (c) The relative expression of PVT1 was analyzed in H1299 and HCC827 cells transfected with shEIF4A3 or shNC. **p < 0.01. (d) The relative nascent RNA level of PVT1 was detected in H1299 and HCC827 cells transfected with shEIF4A3 or shNC. (e and f) The relative expression of PVT1 was examined in H1299 and HCC827 cells with actinomycin D (ActD) treatment at different time. ** p < 0.01.
PVT1 recruited EIF4A3 to enhance circLMNB2 expression
LncRNAs can be considered a scaffold for the interaction between RBPs and target mRNAs. PVT1 was mainly localized in the nucleus (). PVT1 could bind to LMNB2 using ENCORI (https://starbase.sysu.edu.cn/index.php). Moreover, circLMNB2 was predicted to combine with EIF4A3 using ENCORI and CSCD (). qPCR analysis showed that circLMNB2 was downregulated when PVT1 was knocked down (). The binding motifs between EIF4A3 and LMNB2 are shown in . These data suggest that PVT1 might recruit EIF4A3 to regulate circLMNB2 expression. The RIP assay was performed to verify that EIF4A3 could interact with circLMNB2 as well as PVT1 (). EIF4A3 knockdown could reduce circLMNB2 expression (). However, circLMNB2 knockdown had no effect on the RNA and protein levels of EIF4A3 (). In addition, the interaction between EIF4A3 and circLMNB2 was obviously decreased when PVT1 was knocked down (). These results validate that PVT1 recruits EIF4A3 to promote circLMNB2 expression.
Figure 4. PVT1 recruited EIF4A3 to enhance circLMNB2 expression. (a) The nucleus and cytoplasm distribution of PVT1 was detected in H1299 and HCC827 cells. (b) The intersection of circRNAs regulated by EIF4A3 predicted by ENCORI and CSCD. (c) The relative expression of circLMNB2 was measured in H1299 and HCC827 cells transfected with shPVT1 or shNC. ** p < 0.01. (d) The graphic image showing the binding sites between EIF4A3 and LMNB2 mRNA. (e) The relative abundance of circLMNB2 with EIF4A3 was measured using RIP assay. *** p < 0.001. (f) The relative expression of circLMNB2 was detected in cells transfected with shEIF4A3 or shNC. ** p < 0.01. (g and h) EIF4A3 expression at mRNA and protein levels was detected by qPCR and western blot in cells transfected with shcircLMNB2 or shNC. (i) The interaction between EIF4A3 and circLMNB2 was detected in PVT1-knockdown H1299 and HCC827 cells by RIP. ***p < 0.001.
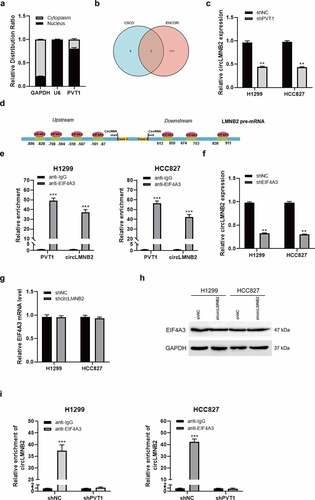
circLMNB2 promoted cell proliferation, migration, and invasion in LUAD cells
CircLMNB2 expression was upregulated in paired LUAD tumor samples (n = 57, ). The receiver operating characteristic (ROC) curve revealed that circLMNB2 had a certain accuracy for LUAD diagnosis (). OS analysis showed that high circLMNB2 expression was associated with a shorter survival time in patients with LUAD (). These analyses indicate that circLMNB2 may play an oncogenic role in LUAD. To verify the role of circLMNB2 in LUAD, we performed a loss-of-function assay using shRNAs. circLMNB2 shRNA #1 had the highest efficiency but it had no effect on LMNB2 expression and was used for subsequent research (Supplementary Figure 1C-1D). Moreover, circLMNB2 downregulation could suppress cell viability, migration and invasion (). Taken together, these data suggest that circLMNB2 may play an oncogenic role in LUAD.
Figure 5. circLMNB2 knockdown inhibited cell proliferation, migration and invasion in LUAD cells. (a) The relative expression of circLMNB2 was analyzed in paired LUAD tumor samples (n = 57). (b) The diagnostic value of circLMNB2 for LUAD was analyzed using ROC curve (AUC = 0.816, CI = 0.779–0.852). (c) The prognostic value of circLMNB2 for LUAD was analyzed. (d) The relative expression of LMNB2 and circLMNB2 was measured in H1299 and HCC827 cells transfected with sh-circLMNB2. (e) Cell viability was analyzed using CCK-8 assay. (f and g) Cell migration and invasion abilities were analyzed using Transwell and Transwell-matrigel assays. * p < 0.05, ** p < 0.01, *** p < 0.001. Scale bar,50 μM.
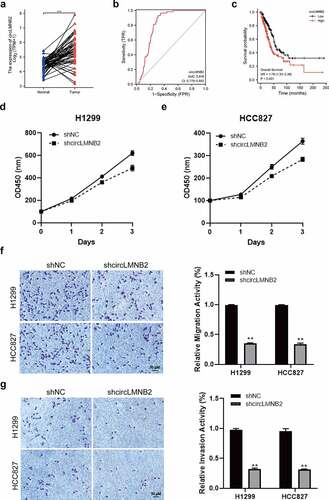
CircLMNB2 overexpression could block the reduced LUAD cells behavior induced by EIF4A3 or PVT1 knockdown
To further validate the functional role of PVT1-EIF4A3-circLMNB2 in LUAD, we performed rescue experiments. CircLMNB2 overexpression could reverse the reduced cell proliferation, migration, invasion and EMT resulting from PVT1 or EIF4A3 knockdown (). These results suggest that circLMNB2 is involved in PVT1/EIF4A3 regulated malignant behavior of LUAD.
Figure 6. CircLMNB2 overexpression could block the reduced LUAD cells behavior induced by EIF4A3 or PVT1 knockdown. H1299 and HCC827 cells were transfected with shPVT1, shEIF4A3, shPVT1 + circLMNB2, shEIF4A3 + circMNB2 or sh-NC + circNC. (a) Cell viability was analyzed by CCK-8. (b and c) Cell migration and cell invasion were analyzed using Transwell and Transwell-matrigel assays. (d) The EMT-related proteins were detected by western blot. Scale bar,50 μM.
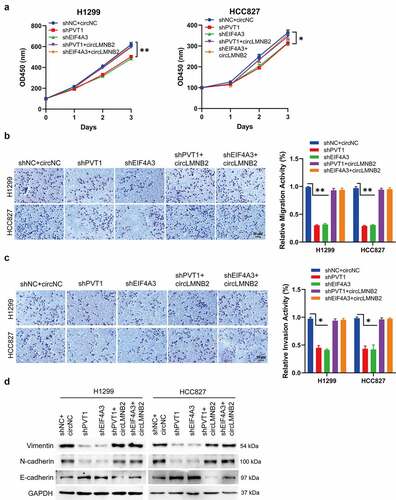
PVT1 and EIF4A3 promoted LUAD tumor growth in vivo
To further validate the effects of PVT1 and EIF4A3 on LUAD, we performed an in vivo assay. As shown in , PVT1 or EIF4A3 knockdown slowed tumor growth compared to the control. The co-silencing of PVT1 and EIF4A3 exacerbated these inhibitory effects. Consistently, the expression of circLMNB2 was downregulated upon PVT1 or EIF4A3 knockdown (). These results demonstrate that PVT1 and EIF4A3 play oncogenic roles in LUAD.
Figure 7. PVT1 and EIF4A3 knockdown suppressed LUAD tumor growth. (a) The representative pictures of xenograft tumor per group were displayed (n = 5). (b) The tumor volume was calculated every 7 days. (c) The relative expression of PVT1, EIF4A3, LMNB2 and circLMNB2 was measured by qPCR. * p < 0.05, ** p < 0.01, *** p < 0.001.
Pathways regulated by PVT1/EIF4A3/LMNB2 in LUAD
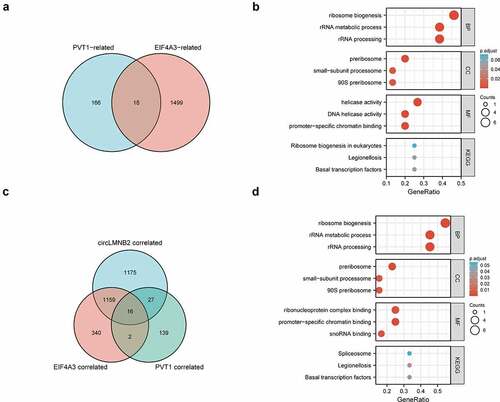
To further explore the potential oncogenic roles of PVT1/EIF4A3/circLMNB2 in LUAD, we analyzed the positively related genes with PVT1, EIF4A3 and/or circLMNB2 by Gene Ontology (GO)-Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis. The results showed that positively related genes were mainly involved in the ribosome biogenesis. KEGG pathway analysis revealed that the correlated genes were involved in ribosome biogenesis in eukaryotes and spliceosome formation (). These results demonstrate that the PVT1/EIF4A3/circLMNB2 axis might play an oncogenic role in LUAD by participating in ribosome biogenesis and spliceosome formation.
Figure 8. Pathways regulated by PVT1/EIF4A3/LMNB2 in LUAD. (a) The intersection between PVT1-positive-related genes and EIF4A3-positive-related genes in LUAD. (b) The bubble charts showed the enrichment results of the intersection genes of (A) in LUAD including biological processes (BP), cellular components (CC), molecular functions (MF) and KEGG pathway analysis. (c) The intersection among PVT1-positive-related genes, EIF4A3-positive-related genes and LMNB2-positive-related genes in LUAD. (d) The bubble charts showed the enrichment results of the intersection genes of (C) in LUAD including BP, CC, MF and KEGG pathway analysis.
Discussion
Lung cancer is one of the most common cancers worldwide. Despite recent advancements in diagnosis, classification and treatment, the OS rate of lung cancer remains low. Therefore, it is necessary to improve our understanding of the molecular pathology of lung cancer to better address these problems and develop personalized target therapies. This study uncovers an innovative mechanism of PVT1-mediated LUAD progression. The highlights of this study are as follows. First, PVT1 and EIF4A3 were upregulated and had diagnostic and prognostic significance in LUAD. Second, PVT1 stabilized by EIF4A3 could mediate LUAD tumor behaviors including cell proliferation, migration, invasion and EMT. Third, EIF4A3 recruited by PVT1 could mediate circLMNB2 generation. Fourth, circLMNB2 was a novel circRNA verified in our study, exerting an oncogenic effect on LUAD.
LncRNAs participate in various biological processes through chromatin, transcription or post-transcriptional regulation. LncRNA PVT1 has been involved in almost every type of cancer via multiple regulatory mechanisms. The most common way is to sponge miRNAs to regulate their target genes. For example, PVT1 acts as a miR-361-3p sponge to promote NSCLC [Citation40]. PVT1 competitively binds to miRNA-424-5p to regulate CARM1 in the radiosensitivity of NSCLC cells [Citation41]. PVT1 can promote cell migration in gastric cancer by functioning as a ceRNA of miR-30a [Citation42]. Some studies have revealed that PVT1 participates in cancer progression through genes or proteins. PVT1 interacts with MYC to synergistically promote tumorigenesis [Citation43]. PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2 [Citation44]. In this study, EIF4A3 was found to be a novel factor with PVT1 in LUAD. PVT1 and EIF4A3 were upregulated in LUAD and a positive association was confirmed using Pearson analysis. RIP assay and RNA pull-down experiments were performed to confirm the interaction between PVT1 and EIF4A3. Moreover, the RNA stability of PVT1 decreased when EIF4A3 was knocked down. These results suggest that PVT1 interacts with EIF4A3 and is stabilized by EIF4A3, both of which synergistically exert an oncogenic effect on LUAD.
Previous studies have shown that some RBPs participate in the biogenesis of circRNAs [Citation45–47]. For instance, EIF4A3 was reported to combine with MMP9 mRNA to promote circMMP9 circularization and expression in glioblastoma multiforme [Citation48]. Quaking plays a key role in circRNA biogenesis during human EMT [Citation49]. Consistent with previous findings, this study showed that EIF4A3 could bind to LMNB2 pre-mRNA to promote circLMNB2 generation, which was recruited by PVT1. In addition, bioinformatics analysis also showed that PVT1 could bind to the promoter region of LMNB2. Knockdown of PVT1 reduced circLMNB2 expression, suggesting that PVT1 regulates LMNB2 transcription. However, further detailed work is required to validate that EIF4A3 recruited by PVT1 regulates circLMNB2 biogenesis. Taken together, our results confirm our hypothesis that PVT1 recruits EIF4A3 to promote circLMNB2 biogenesis by binding to LMNB2 pre-mRNA. PVT1 regulates LMNB2 expression at the transcriptional level. Our study enriches the PVT1-involved LUAD regulatory network.
Increasing studies have shown that the role of circRNAs in cancers has been a focus of studies [Citation50,Citation51]. CircRNAs are involved in many biological processes associated with multiple diseases and cancers [Citation52–54]. Nevertheless, the exact functions and regulatory mechanisms of most circRNAs in lung cancer remain largely unknown. In this study, we identified a new circRNA, circLMNB2 in LUAD. CircLMNB2 was highly expressed in LUAD tissues compared to normal tissues, and was associated with poor prognosis and higher diagnostic efficiency of LUAD. Functionally, circLMNB2 knockdown reduced cell proliferation, migration and invasion. Overexpression of circLMNB2 rescued the blocking effects of PVT1 or EIF4A3 knockdown. These results demonstrate that circLMNB2 plays a critical role in promoting the oncogenesis and progression of LUAD.
These findings revealed new PVT1 functions in LUAD and extended the regulatory network of lncRNAs/RBPs/circRNA in cancer growth and development, thereby contributing to disease understanding and etiology, which is beneficial to the individualized treatment of cancer. On the view of potential diagnostic values, if a patient with pulmonary nodules is found to be with high expression of these molecular markers such as PVT1 and EIF4A3, he may be susceptible to lung cancer and could be better to do regular checkup. On the view of potential prognostic values, if PVT1, EIF4A3 or circLMNB2 were detected highly expressed in one LUAD patient, there is a very high probability that this patient has a poor prognosis. It might make a better therapeutic effect to use combined targeted drugs such as some agents targeting PVT1 or circLMNB2. Nowadays, treatments for lung cancer are available including thoracic surgery, radical radiotherapy, radiofrequency/microwave ablation, chemotherapy, targeted therapy, and immunologic therapy [Citation55,Citation56], the latter of which are emerging technologies. The advantage is that the targeted therapy is individually tailored to each patient but inevitably leading to drug resistance. It might be a good way to combine different treatments to minimize this limitation.
There are some limitations in our study. First, PVT1 and EIF4A3 could regulate circLMNB2 biogenesis, however, the detailed regulatory mechanism needs to be further investigated. Second, the downstream signaling pathways of PVT1/EIF4A3/circLMNB2-mediated LUAD tumorigenesis were unclear. Third, rescue experiments were validated using an in vivo study. The mechanism of circLMNB2 regulated LUAD development was not investigated and thus will be investigated in further studies.
Conclusions
In summary, our study demonstrates that PVT1 and EIF4A3 can facilitate LUAD progression in vitro and in vivo. PVT1 and EIF4A3 have diagnostic and prognostic values in LUAD. Mechanistically, PVT1 and EIF4A3 can regulate circLMNB2 biogenesis. PVT1 may be more stable by binding to EIF4A3. EIF4A3 can be recruited by PVT1 to mediate circLMNB2 generation. Our data provide powerful evidence for PVT1 as a clinical target in lung cancer.
Abbreviation
PVT1 Plasmacytoma variant translocation
EIF4A3 Eukaryotic Initiation factor 4A-3
CircLMNB2 Circular RNA LMNB2
LUAD Lung adenocarcinoma
EMT Epithelial-mesenchymal transition
ROC Receiver Operating Characteristic
CCK-8 Cell Counting Kit-8
Author contributions
MLQ designed the experiments. MLQ, MZC and ZPL performed the experiments, analyzed the data and wrote the manuscript. BL, JBX and XL collected the tumor tissues, performed the bioinformatics analysis. All authors read and approved the final manuscript.
Supplemental Material
Download Zip (634 KB)Acknowledgements
This study was funded by grants from Natural Science Foundation of Fujian Province (No. 2021J01717), Fujian Province Finance Project (No. BPB-2020QML) and Young and Middle-aged Key Personnel Training Project of Fujian Provincial Health Commission (No. 2020GGA047).
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424.
- Pascoe HM, Knipe HC, Pascoe D, et al. The many faces of lung adenocarcinoma: a pictorial essay. J Med Imaging Radiat Oncol. 2018;62(5):654–661.
- Chen Z, Teng X, Zhang J, et al. Molecular features of lung adenocarcinoma in young patients. BMC Cancer. 2019;19(1):777.
- Zhang C, Zhang G, Sun N, et al. Comprehensive molecular analyses of a TNF family-based signature with regard to prognosis, immune features, and biomarkers for immunotherapy in lung adenocarcinoma. EBioMedicine. 2020;59:102959.
- Moreira AL, Eng J. Personalized therapy for lung cancer. Chest. 2014;146(6):1649–1657.
- Boon RA, Jaé N, Holdt L, et al. Long noncoding RNAs: from clinical genetics to therapeutic targets. J Am Coll Cardiol. 2016;67(10):1214–1226.
- Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407.
- Rinn JL, Chang HY. Long noncoding RNAs: molecular modalities to organismal functions. Annu Rev Biochem. 2020;89(1):283–308.
- Choudhari R, Sedano MJ, Harrison AL, et al. Long noncoding RNAs in cancer: from discovery to therapeutic targets. Adv Clin Chem. 2020;95:105–147.
- Onagoruwa OT, Pal G, Ochu C, et al. Oncogenic Role of PVT1 and therapeutic implications. Front Oncol. 2020;10:17.
- Zhao J, Du P, Cui P, et al. LncRNA PVT1 promotes angiogenesis via activating the STAT3/VEGFA axis in gastric cancer. Oncogene. 2018;37(30):4094–4109.
- Shigeyasu K, Toden S, Ozawa T, et al. The PVT1 lncRNA is a novel epigenetic enhancer of MYC, and a promising risk-stratification biomarker in colorectal cancer. Mol Cancer. 2020;19(1):155.
- Pan Y, Liu L, Cheng Y, et al. Amplified LncRNA PVT1 promotes lung cancer proliferation and metastasis by facilitating VEGFC expression. Biochem Cell Biol. 2020;98(6):676–682.
- Xi Y, Shen W, Jin C, et al. PVT1 promotes the proliferation and migration of non-small cell lung cancer via regulating miR-148/RAB34 signal axis. Onco Targets Ther. 2020;13:1819–1832.
- Wu J, Ma C, Tang X, et al. The regulation and interaction of PVT1 and miR181a-5p contributes to the repression of SP1 expression by the combination of XJD decoction and cisplatin in human lung cancer cells. Biomed Pharmacother. 2020;121:109632.
- Gerstberger S, Hafner M, Ascano M, et al. Evolutionary conservation and expression of human RNA-binding proteins and their role in human genetic disease. Adv Exp Med Biol. 2014;825:1–55.
- Gerstberger S, Hafner M, Tuschl T. A census of human RNA-binding proteins. Nat Rev Genet. 2014;15(12):829–845.
- Chan CC, Dostie J, Diem MD, et al. eIF4A3 is a novel component of the exon junction complex. RNA. 2004;10(2):200–209.
- Lu WT, Wilczynska A, Smith E, et al. The diverse roles of the eIF4A family: you are the company you keep. Biochem Soc Trans. 2014;42(1):166–172.
- Sakellariou D, Tiberti M, Kleiber TH, et al. eIF4A3 regulates the TFEB-mediated transcriptional response via GSK3B to control autophagy. Cell Death Differ. 2021;28(12):3344–3356.
- Song H, Liu Y, Li X, et al. Long noncoding RNA CASC11 promotes hepatocarcinogenesis and HCC progression through EIF4A3-mediated E2F1 activation. Clin Transl Med. 2020;10(7):e220.
- Tang W, Wang D, Shao L, et al. LINC00680 and TTN-AS1 stabilized by EIF4A3 promoted malignant biological behaviors of glioblastoma cells. Mol Ther Nucleic Acids. 2020;19:905–921.
- Kristensen LS, Andersen MS, Stagsted L, et al. The biogenesis, biology and characterization of circular RNAs. Nat Rev Genet. 2019;20(11):675–691.
- Yang Q, Li F, He AT, et al. Circular RNAs: expression, localization, and therapeutic potentials. Mol Ther. 2021;29(5):1683–1702.
- Lei M, Zheng G, Ning Q, et al. Translation and functional roles of circular RNAs in human cancer. Mol Cancer. 2020;19(1):30.
- Xiao MS, Ai Y, Wilusz JE. Biogenesis and functions of circular RNAs come into focus. Trends Cell Biol. 2020;30(3):226–240.
- Liu Y, Chen S, Zong ZH, et al. CircRNA WHSC1 targets the miR-646/NPM1 pathway to promote the development of endometrial cancer. J Cell Mol Med. 2020;24(12):6898–6907.
- Liu Z, Zhou Y, Liang G, et al. Circular RNA hsa_circ_001783 regulates breast cancer progression via sponging miR-200c-3p. Cell Death Dis. 2019;10(2):55.
- Li Y, Ren S, Xia J, et al. EIF4A3-Induced circ-BNIP3 aggravated hypoxia-induced injury of H9c2 cells by targeting miR-27a-3p/BNIP3. Mol Ther Nucleic Acids. 2020;19:533–545.
- Mumtaz PT, Taban Q, Dar MA, et al. Deep insights in circular RNAs: from biogenesis to therapeutics. Biol Proced Online. 2020;22(1):10.
- Lánczky A, Győrffy B. Web-Based survival analysis tool tailored for medical research (KMplot): development and implementation. J Med Internet Res. 2021;23(7):e27633.
- Li JH, Liu S, Zhou H, et al. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res. 2014;42(D1):D92–97.
- Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25(4):402–408.
- Seo M, Lee SO, Kim JH, et al. MAP4-regulated dynein-dependent trafficking of BTN3A1 controls the TBK1-IRF3 signaling axis. Proc Natl Acad Sci U S A. 2016;113(50):14390–14395.
- Frara N, Abdelmagid SM, Sondag GR, et al. Transgenic expression of osteoactivin/gpnmb enhances bone formation in vivo and osteoprogenitor differentiation ex Vivo. J Cell Physiol. 2016;231(1):72–83.
- Zhao X, Jie X, Gao YK, et al. Long non-coding RNA CACNA1G-AS1 promotes proliferation and invasion and inhibits apoptosis by regulating expression of miR-205 in human keloid fibroblasts. Biosci Rep. 2020;40(6):BSR20192839.
- Chutake YK, Costello WN, Lam C, et al. Altered nucleosome positioning at the transcription start site and deficient transcriptional initiation in Friedreich ataxia. J Biol Chem. 2014;289(22):15194–15202.
- Yao S, Hu M, Hao T, et al. MiRNA-891a-5p mediates HIV-1 Tat and KSHV Orf-K1 synergistic induction of angiogenesis by activating NF-κB signaling. Nucleic Acids Res. 2015;43(19):9362–9378.
- Uno M, Saitoh Y, Mochida K, et al. NF-κB inducing kinase, a central signaling component of the non-canonical pathway of NF-κB, contributes to ovarian cancer progression. PLoS One. 2014;9(2):e88347.
- Qi G, Li L. Long non-coding RNA PVT1 contributes to cell growth and metastasis in non-small-cell lung cancer by regulating miR-361-3p/SOX9 axis and activating Wnt/β-catenin signaling pathway. Biomed Pharmacother. 2020;126:110100.
- Wang D, Hu Y. Long non-coding RNA PVT1 competitively binds MicroRNA-424-5p to Regulate CARM1 in radiosensitivity of non-small-cell lung cancer. Mol Ther Nucleic Acids. 2019;16:130–140.
- Wang L, Xiao B, Yu T, et al. lncRNA PVT1 promotes the migration of gastric cancer by functioning as ceRNA of miR-30a and regulating Snail. J Cell Physiol. 2021;236(1):536–548.
- Jin K, Wang S, Zhang Y, et al. Long non-coding RNA PVT1 interacts with MYC and its downstream molecules to synergistically promote tumorigenesis. Cell Mol Life Sci. 2019;76(21):4275–4289.
- Guo J, Hao C, Wang C, et al. Long noncoding RNA PVT1 modulates hepatocellular carcinoma cell proliferation and apoptosis by recruiting EZH2. Cancer Cell Int. 2018;18(1):98.
- Bach DH, Lee SK, Sood AK. Circular RNAs in cancer. Mol Ther Nucleic Acids. 2019;16:118–129.
- Conn SJ, Pillman KA, Toubia J, et al. The RNA binding protein quaking regulates formation of circRNAs. Cell. 2015;160(6):1125–1134.
- Ivanov A, Memczak S, Wyler E, et al. Analysis of intron sequences reveals hallmarks of circular RNA biogenesis in animals. Cell Rep. 2015;10(2):170–177.
- Wang R, Zhang S, Chen X, et al. EIF4A3-induced circular RNA MMP9 (circMMP9) acts as a sponge of miR-124 and promotes glioblastoma multiforme cell tumorigenesis. Mol Cancer. 2018;17(1):166.
- Chen X, Liu Y, Xu C, et al. QKI is a critical pre-mRNA alternative splicing regulator of cardiac myofibrillogenesis and contractile function. Nat Commun. 2021;12(1):89.
- Chen Y, Yang F, Fang E, et al. Circular RNA circAGO2 drives cancer progression through facilitating HuR-repressed functions of AGO2-miRNA complexes. Cell Death Differ. 2019;26(7):1346–1364.
- Zhong Y, Du Y, Yang X, et al. Circular RNAs function as ceRNAs to regulate and control human cancer progression. Mol Cancer. 2018;17(1):79.
- Ng WL, Mohd MTB, Shukla K. Functional role of circular RNAs in cancer development and progression. RNA Biol. 2018;15(8):995–1005.
- Wang C, Tan S, Li J, et al. CircRNAs in lung cancer - Biogenesis, function and clinical implication. Cancer Lett. 2020;492:106–115.
- Huang G, Li S, Yang N, et al. Recent progress in circular RNAs in human cancers. Cancer Lett. 2017;404:8–18.
- Jones GS, Baldwin DR. Recent advances in the management of lung cancer. Clin Med (Lond). 2018;18(Suppl 2):s41–s46.
- Bade BC, Dela Cruz CS. Lung Cancer 2020: epidemiology, etiology, and prevention. Clin Chest Med. 2020;41(1):1–24.
