ABSTRACT
Myocardial infarction (MI) is the leading cause of sudden death. Long non-doing RNAs (lncRNAs) were demonstrated to play crucial roles in multiple diseases, including cancer and cardiovascular diseases. Nevertheless, the molecular mechanism of lncNRAs in MI is unclear. In this study, we integrated bioinformatics and molecular biological experiments to identify the novel lncRNA transcripts and elucidated its regulatory mechanism in MI. First, we identified 10 dysregualted lncRNAs and found that lncRNA Gm47283 was the top risk factor in MI. Bioinformatics analysis predicted that lncRNA Gm47283 exerted function via targeting miR-706 and Ptgs2. Ptgs2 was also the known regulator of ferroptosis. Inhibition or overexpression of lncRNA Gm47283 could regulate Ptgs2 expression and downstream ferroptosis activity. Overexpression of miR-706 could inhibit the expression of Ptgs2 and the activity of ferroptosis, thereby attenuated cellular injury. Mechanically, co-transfection experiments showed that overexpression of miR-706 could reverse the damage effect that was caused by lncRNA Gm47283 overexpression, via inhibiting Ptgs2 and ferroptosis. Additionally, inhibition of lncRNA Gm47283 by stem cell membrane coated siRNA could attenuate MI in vivo. Our study elucidated a novel mechanism containing lncRNA Gm47283/miR-706/Ptgs2/ferroptosis in MI, which provided a potential therapeutic for MI.
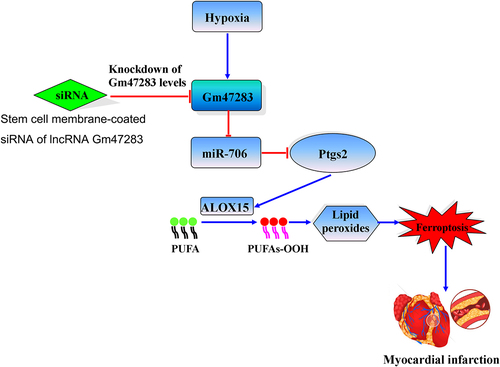
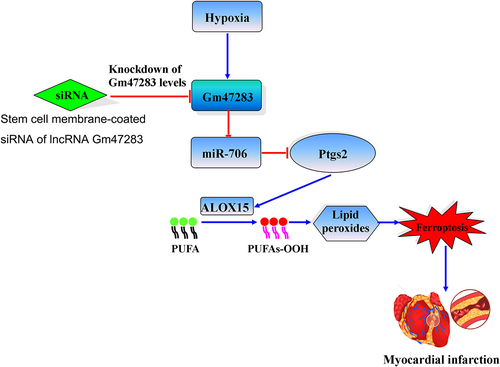
Graphical Abstract. Stem cell membrane coated siRNA of lncRNA Gm47283 inhibits cardiomyocyte ferroptosis in myocardial infarction rat. Stem cell membrane-coated siRNA of lncRNA Gm47283 increases miR-706, and then miR-706 suppresses the expression of Ptgs2 to reduce lipid peroxidation toxicity, and then inhibits cardiomyocyte ferroptosis. PUFA: polyunsaturated fatty acid.
Graphical Abstract
Highlights
It was confirmed that ferroptosis was related to myocardial infarction.
We identified lncRNA Gm47283 was the top risk factor in myocardial infarction.
We found that lncRNA Gm47283 exerted function via targeting miR-706 and Ptgs2 in myocardial infarction.
For the first time, we used stem cell membrane coated siRNA of lncRNA Gm47283 to treat myocardial infarction.
Introduction
Acute myocardial infarction (AMI) is mainly caused from coronary artery diseases, which is considered as the leading cause of morbidity and mortality worldwide [Citation1]. Recent studies have focused on identifying the early diagnosis markers and discovering novel clinical treatments for patients of AMI to improve the survival levels [Citation2]. However, the molecular mechanism of AMI is complex. Previous studies have found that some biological processes, such as apoptosis, fibrosis and inflammation are involved in the pathological processes of AMI [Citation3–6]. Therefore, it is urgent to find the novel regulator and its regulatory axes of AMI and provide more information for clinical treatment.
Non-coding RNAs (ncRNAs), especially micro RNAs (miRNAs) and long non-coding RNAs (lncRNAs) have been reported to play key roles in the physiological and pathological processes of MI, such as miR-21, miR-133, and lncRNA ZFAS1 [Citation7–9]. Mechanically, miRNAs were demonstrated to exert function via targeting the 3’ untranslated region (3ʹUTR) of protein coding genes and gene body of lncRNAs [Citation10]. Additionally, the newly proposed lncRNA is also demonstrated as the crucial regulators of complex pathological processes in cardiovascular diseases. Briefly, lncRNAs are defined as the transcript with more than 200 nucleotides and without non-protein coding ability, which involve in regulatory processes mainly via regulating gene transcription, translation, and alternative splicing in transcriptional and post-transcriptional levels [Citation11]. Importantly, lncRNAs, miRNAs, and downstream protein coding genes can compose the close regulatory axes, which are named as competitive endogenous RNAs (ceRNAs). In the model of ceRNAs, activation of lncRNAs can protect the degradation of downstream protein coding genes via sponging common miRNAs. Studies have found that lncRNAs regulate MI through ceRNA regulatory mechanisms. For instance, Bin et al. have demonstrated that lncRNA CRRL suppresses cardiomyocyte regeneration via functioning as the endogenous decoy of miR-199a-3p and thereby activate the expression of Hopx, which leads to increased cardiac injury [Citation12]. Wang et al. found that lncRNA APF can regulate miR-188-3p, and thus affects ATG7 expression, autophagic cell death, and MI [Citation13]. Hu et al. found that inhibition of MALAT1 can attenuate myocardial apoptosis through suppressing Pten expression by sponging miR-320 in AMI mouse [Citation14]. All these studies inspire us to investigate the mechanism of AMI through lncRNA mediated ceRNAs perspective.
The loss of cardiomyocytes is the typical phenomenon during AMI, which is validated from numerous necrocytosis processes, like apoptosis, pyroptosis, autophagy, and ferroptosis [Citation5,Citation15–17]. Ferroptosis is a novel form of regulated cell death identified as iron-dependence, lipid peroxidation collection, and reactive oxygen species (ROS) generation [Citation18–20]. It is remarkably distinct from other types of classical regulated cell death, such as apoptosis, necrosis, and necroptosis. Ferroptosis participated in various diseases, including cancer, Parkinson’s disease, stroke, and IRI [Citation21–24]. However, its effect in the progression of AMI remains unclear.
In this study, we firstly used a bioinformatics pipeline to identify risk lncRNAs in MI based on microarray data. Results showed that 10 lncRNAs were dysregulated in MI. In vivo and in vitro models validated that lncRNA Gm47283 was the high risk factor of MI. In addition, TNF and NF-κB signaling pathways were interpreted as activated regulatory processes in MI. And four genes were both enriched in the two pathways were annotated, including ferroptosis-related gene Ptgs2. LncRNA Gm47283 and Ptgs2 were also activated in MI models in vivo and in vitro. Bioinformatics analysis predicted that lncRNA Gm47283 could regulate Ptgs2 via endogenous sponging miR-706. Inhibition or overexpression of lncRNA Gm47283 could regulate Ptgs2 expression and ferroptosis activity. Mechanically, we found that overexpression of miR-706 could attenuate the Gm47283-induced damage effects, via regulating cardiomyocyte ferroptosis activity by targeting Ptgs2. Moreover, inhibition of lncRNA Gm47283 by nanometer materials coated siRNA could attenuate MI in vivo. Our study revealed a novel role for lncRNA Gm47283 in regulating MI via ferroptosis pathway and lncRNA Gm47283/miR-706/Ptgs2 regulatory axis could provide more information for therapeutic target design of MI.
Methods
Ethics statement
The procedures for the use of animals (c57bl/6 mouse; 20–25 g) in this work were in accordance with the regulations of the Ethic Committees of Xuzhou Cancer Hospital (ethics approval number: 2022–03-002-K01) and conformed to the National Research Council (NRC) Guide for the Care and Use of Laboratory Animals (2011, 8th edition).
Bioinformatics analysis
We downloaded the raw expression data matrix and processed the gene expression of MI from GEO database with the accession number of GSE153485. We only reserved the samples of four groups: Sham_6 h group, Sham_24 h group, MI_6 h group and MI_24 h group (20 samples). We merged the groups into two groups, Sham group and MI group. Limma method was used to identify differentially expressed lncRNAs and genes with the threshold of 1.5 fold change or P-value <0.05. Protein–protein interaction network was extracted from STRING database by mapping the up-regulated genes [Citation25]. Degree analysis of the network was performed via Cytoscape.
We used miRanda software to identify the downstream miRNAs of lncRNA Gm47283. Furthermore, we also used TargetScan software to identify the targeting relationship between miRNAs and downstream up-regulated genes [Citation26].
Mouse model of MI
Male mice were used in this research. The mice were housed in a colony room at a controlled temperature (22°C) and humidity, under a 12-h light/dark cycle, and with food and water freely available. All surgical procedures were carried out in accordance with the National Institutes of Health Guide for the Care and Use of Laboratory Animals. In brief, the mice were anesthetized by 3% pentobarbital sodium and then ligated the anterior descending branch of the left coronary artery (LAD) for 48 h to establish the in vivo MI models [Citation27]. The sham group mice were opened the chest bur not ligated with LAD. The mice were randomly divided into three groups as follows: (1) the sham group, which underwent sham operation and received vehicle (PBS, caudal vein injection); (2) the model group, which was subjected to LAD and received vehicle (PBS, caudal vein injection); and (3) the siRNA group, which were subjected to LAD and treated with siRNA of lncRNA Gm47283 (30 nM siRNA dose per mice every day for one week, caudal vein injection).
Measurement of infarct size
The heart was stained with TTC (triphenyltetrazolium chloride, Sigma-Aldrich, USA), and the size of the infarct was measured. After the remaining blood was washed away, the heart was cut into 2-mm thick sections below the ligature and stained with 1% TTC at 37°C for 15 min. Infarct area is stainless while the non-infarct area is stained red. The left ventricle was separated, and the ischemic area of the ventricle was dissected. The infarct area was measured by the weight ratio of infarct area and left ventricle.
HE and Masson staining
Frozen sections of heart tissue were performed according to Dang’s method [Citation28]. The mice hearts were rapidly harvested, washed, and stored in 4% paraformaldehyde for 48 h. Then the heart was cross-cut into 2 mm thick sections for hematoxylin and eosin (HE) staining and Masson staining via staining kit (HE: G1120, Masson: G1340, Solarbio, Beijing, China). Stained sections were photographed by the light microscope (Hitachi, Tokyo, Japan) at 100x magnification.
Cell culture and in vitro cell models
The HL-1 cell lines were cultured in 25 cm2 cell culture flask (Corning, NYC, USA) with Dulbecco’s Modified Eagle Medium (DMEM) (Invitrogen, Waltham, USA) containing 15% fetal bovine serum (Invitrogen, Waltham, USA) at 37°C, 5% CO2 environment. In vitro MI models were established by H2O2 stimulation or hypoxia for 48 h.
Vector construction and transfection
The full-length of lncRNA Gm47283 was synthesized by PCR and inserted into pcDNA3.1 vector. Besides, pcDNA3.1 empty vector was used as a negative control. HL-1 was washed with serum-free medium and then incubated in 5 ml serum-free medium for 4–6 h for transfection. lncRNA Gm47283 or control vectors (1 µg/mL) and x-treme GENE siRNA (Invitrogen, Carlsbad, USA) were separately mixed with 300 μl serum-free medium for 5 min. Then, two mixtures were combined and incubated at room temperature for 18 min. Finally, the plasmid mixture and x-treme GENE siRNA were added to the HL-1 and incubated at 37°C for 24 h [Citation29].
MiRNA mimic, siRNA construction and transfection
MiR-706 mimics and small interfering RNA sequences that target to lncRNA Gm47283 were synthesized by Gene Pharma (Gene Pharma, Shanghai, China). HL-1 was washed with serum-free medium and then incubated in 5 mL serum-free medium for 4–6 h for transfection. MiR-706 mimics or control vectors (1 µg/mL) and x-treme GENE siRNA (Invitrogen, Carlsbad, USA) were separately mixed with 300 μl serum-free medium for 5 min. Then, two mixtures were combined and incubated at room temperature for 18 min. Finally, the mixture and x-treme GENE siRNA were added to the HL-1 and incubated at 37°C for 24 h.
Luciferase reporter assays
The Ptgs2ʹUTR and lncRNA Gm47283 full-length sequence containing miR-706 binding sites were amplified by PCR. For luciferase assay, cells were seeded in 48-well plates in triplicate, and 40 ng/well luciferase reporter vector, 10 pmol miR-706 mimic or mimic control were co-transfected into plates using lipofectamine 2000 (Invitrogen, Carlsbad, USA). Cells were lysed at 24 h after transfection, and luciferase activity was determined using the Dual Luciferase Reporter Assay kit (Promega, Madison, USA).
Reactive oxygen species (ROS) production assay
HL-1 cell lines were removed the medium and incubated with diluted DCFH-DA (S0033M, Beyotime, Shanghai, China) at 37°C for 20 min. After incubation, the cell lines were washed again. Fluorescence intensity of intracellular ROS was detected by fluorescence microscope (Leica, Heidelberg, Germany) at excitation wavelength of 488 nm.
Measurement of the MDA and SOD levels
Malondialdehyde (MDA) and superoxide dismutase (SOD) were considered as the oxidative stress injury markers. Therefore, the quantities of MDA and SOD were measured by MDA and SOD kits (MDA: S0131S, SOD: S0101S, Beyotime, Shanghai, China) based on the manufacturer’s instructions.
Western blot
Total protein was extracted from the left ventricular myocardium or HL-1 cell lines and lysed via RIPA buffer. Degenerated protein concentration was measured by bichinchoninic acid (BCA) Protein Assay Kit (Beyotime, Shanghai, China). In each experiment, 20 μg proteins were separated by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, 12% or 10% polyacrylamide gels) and transferred to nitrocellulose filter membrane. The membrane was blocked for 2 h with 5% skim milk in PBS at room temperature and then incubated at 4°C overnight with primary antibodies Gpx4 (1:500 dilution, A11243, Abclonal, Wuhan, China), Alox15 (1:1000 dilution, A6864, Abclonal, Wuhan, China), GAPDH (1:2000 dilution, A19056, Abclonal, Wuhan, China), and Ptgs2 (1:1000 dilution, A1253, Abclonal, Wuhan, China). HRP Goat Anti-Rabbit IgG (H + L) antibody (1:5000 dilution, AS014, Abclonal, Wuhan, China) was used as the secondary antibody for 1 h at room temperature. The images were captured by Odyssey v3.0 software. Protein bands were measured by the Image J software.
Quantitative real-time RT-PCR
Total RNA was extracted from the left ventricles and HL-1 cell lines using Trizol reagent (Invitrogen, Waltham, USA) according to manufacturer’s protocols. cDNA was synthesized by reverse transcription reagent kit (TAKARA, RR037A, Shiga, JAPAN). Gene expression was quantified by SYBR Green PCR Master Mix, and detected using ABI 7300 systems. U6 or GAPDH was served as an internal control for miRNAs and lncRNAs/mRNAs, respectively. 2−ΔΔCt relative quantification method was used to calculate gene expression. Primer sequences are listed in Table S3.
Preparation of stem-cell-membrane vesicles
Stem-cell membranes isolated from bone-marrow-derived mesenchymal stem cells according to the published method [Citation30]. When the cells achieved 80% confluence, the stem cells were detached, centrifuged, and lysed. Then, the stem cell membrane was collected by centrifuging at 15,000 g for 30 min. To prepare stem-cell-membrane vesicles (SVs), the obtained stem-cell membranes were dispersed in siRNAs and extruded using a physical extrusion method.
To prepare DiR-labeled SVs, Briefly, a post-insertion technique was used. DiR dye was dissolved in ethanol (5 mg/mL). A total of 100 μL of the DiR solution was then gradually added to 1 mL of the SVs suspensions and incubated at 37°C to obtain DiR-labeled SVs. The ethanol was then removed by rotary evaporation.
Statistical analysis
Data were presented as the Mean ± SEM and analyzed by GraphPad Prism 8.0 software. One-way ANOVA method was used to compare between experimental groups. P < 0.05 was considered as statistically significant.
Results
Previous studies have found that lncRNAs play crucial roles in the regulatory processes of multiple cardiovascular diseases. Here, we performed a bioinformatics analysis to infer the potential pathogenic lncRNAs and found lncRNA Gm47283 was the risk factor in MI. Furthermore, we also found some crucial genes were also dysregulated in MI. We hypothesized that lncRNA might participate in the regulatory processes by regulating these crucial genes. Thus, we performed the molecular biology experiments to reveal the molecular mechanism of lncRNA Gm47283 in MI.
Identification of differentially expressed transcripts in MI
Firstly, we used the bioinformatics pipeline to identify differently expressed genes and lncRNAs at the threshold of |Fold change| >1.5 or P-value <0.05. Here, because of the annotation rates for the dataset, the down-regulated lncRNAs were not identified at the strict thresholds. Therefore, we focused on mining the regulatory information for the dysregulated lncRNAs and genes. As a result, 10 lncRNAs were extracted and considered as the risk factors in MI, including 9 up-regulated lncRNAs and 1 down-regulated lncRNA (). We also identified 659 up-regulated genes, which were significantly enriched in the pathway of TNF signaling, MAPK signaling, and NF-κB signaling pathway () [Citation31–34]. Interestingly, we then extracted the enriched genes in each significant pathways, and results showed that four genes with known function were identified, including Cxcl10, Il6, Il1β, and Ptgs2 (). In addition, to systematically investigate the up-regulated genes in MI, we input all these up-regulated genes into String database and extracted the protein–protein interaction networks (). In the network, we found that some genes were occupied high node degrees, indicating the crucial functions of which they had.
Figure 1. Identification of differentially expressed genes in MI. (a) The heatmap of top 10 differentially expressed lncRNAs in MI from 20 samples. (b) The volcano plot of differentially expressed genes. Top 10 up-regulated genes and down-regulated genes were labeled in the plot. (c) Pathway enrichment analysis of up-regulated genes. Four common genes between TNF signaling and NF-κB signaling pathways were labeled. (d) The visualization of up-regulated genes-induced PPI network. (e) Degree analysis of the PPI network.
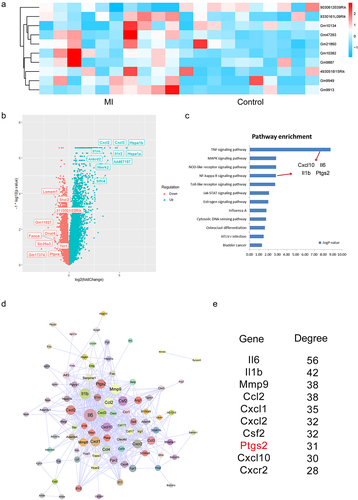
Interestingly, we then used miRanda software to predict the relationship between lncRNAs and miRNAs and used targetScan software to predict the relationship between miRNAs and 4 known genes in . As a result, we found that miR-706 is the potential mediator between lncRNA Gm47283 and Ptgs2 (Supplementary Table S1 and S2).
LncRNA Gm47283 and Ptgs2 were up-regulated in MI
Recently, the ceRNA mechanism of lncRNAs has attracted a lot of research interest. The concept of ceRNAs demonstrates that the RNA molecules containing the binding sites (sequence complementarity) to a particular miRNA can competitively bind to this individual miRNA to reduce its functional availability [Citation35]. LncRNAs can often act as ceRNAs, sponging and sequestering miRNAs, indirectly regulating the target genes of miRNAs, or specifically, releasing the target genes from repression by the targeted miRNA[Citation36]. Next, we prepared the MI models to investigate the regulatory role of lncRNA Gm47283 and Ptgs2 in vivo and vitro. TTC staining showed that significant infarct areas in model groups (), indicating our in vivo models were constructed successfully. Then we used Real-time PCR to detect the expression of lncRNA Gm47283 and Ptgs2 in models, and results showed that the two risk genes were both up-regulated in MI (). Western blots results also validated that Ptgs2 was up-regulated in models ( and Supplementary Figure S1). In addition, we also detected the expression of Alox15 and Gpx4 ( and Supplementary Figure S1). Results showed that Alox15 was up-regulated in MI. Gpx4 was repressed in MI, indicating that ferroptosis activity was activated in MI.
Figure 2. Validation of the expression of lncRNAs and downstream genes in MI. (a) TTC staining of sham hearts and model hearts. (b) Real-time PCR results of lncRNA Gm47283 and Ptgs2. **P < 0.01 vs Sham group. n = 6. (c) Western blot results of Ptgs2, Alox15 and Gpx4 expression. n = 6. (d) ROS staining results of control cardiomyocytes and cardiomyocytes induced by H2O2 and hypoxia. n = 6. (e) Real-time PCR results of lncRNA Gm47283 and Ptgs2 in vitro groups. *P < 0.05 vs Control group. n = 6. (f) Western blot results of Ptgs2, Alox15 and Gpx4 expression. n = 6. Scale bars; 25 μm.
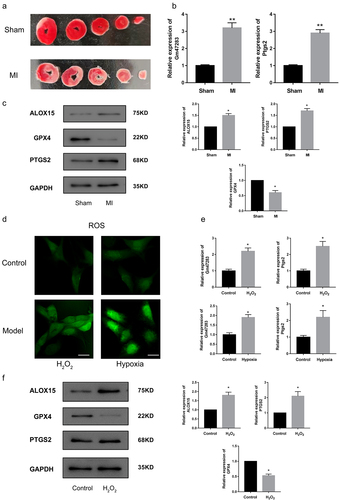
Second, we also used H2O2 and hypoxia to induce in vitro myocardial damage models to detect the expression of lncRNA Gm47283 and Ptgs2. ROS kit results showed that ROS was activated by treating H2O2 and hypoxia (). LncRNA Gm47283 and Ptgs2 were also up-regulated in cellular hypoxia (). Moreover, western blot results showed that Ptgs2 and Alox15 were increased in models but Gpx4 was decreased ( and Supplementary Figure S1), which was in accordant with in vivo model results. We also detected the activity of SOD and MDA. Results showed that SOD was decreased and MDA was increased after H2O2 treatment. Above results validated the predictive factors were up-regulated and ferroptosis was also activated in MI.
LncRNA Gm47283 participated in the pathological processes of MI
The role of lncRNA Gm47283 in MI is unclear. We then investigated the function of lncRNA Gm47283 via loss-of-function and gain-of-function experiments in vitro. First, we constructed a siRNA to inhibit the expression of lncRNA Gm47283. Real-time PCR results validated that lncRNA Gm47283 was efficiently repressed (). Western blot results showed that Ptgs2 was decreased with lncRNA knockdown. Ferroptosis marker Alox15 was up-regulated and Gpx4 was down-regulated ( and Supplementary Figure S1). Furthermore, ROS staining results showed that inhibition of lncRNA Gm47283 could inhibit the ROS activity (). LncRNA Gm47283 knockdown could also increase the quantity of SOD and decrease the quantity of MDA (). These results indicated that inhibition of lncRNA Gm47283 could attenuate cardiomyocyte cellular hypoxiavia repressing ferroptosis. We also overexpressed lncRNA Gm47283 expression via construction of plasmid and Realtime PCR results validated that lncRNA was overexpressed in cardiomyocytes (). We found that over-expression of lncRNA Gm47283 significantly increased the expression of Ptgs2 and Alox15 and repressed the expression of Gpx4 ( and Supplementary Figure S1). In addition, overexpression of lncRNA Gm47283 could increase the level of ROS and MDA and decrease the level of SOD (). Above results indicated that lncRNA Gm47283 played an impaired role in MI via regulating the ferroptosis of cardiomyocyte.
Figure 3. Knockdown of lncRNA Gm47283 alleviates MI. (a) Real-time PCR results of lncRNA Gm47283 knockdown. *P < 0.05 vs Control group. n = 6. (b) Western blot results of Ptgs2, Alox15 and Gpx4 expression in lncRNA knockdown groups. n = 6. (c) ROS staining results of cardiomyocytes in knockdown groups. n = 6. (d) Detection of SOD and MDA levels in lncRNA knockdown groups. *P < 0.05 vs Control group, #P < 0.05 vs H2O2 group. n = 6. (e) Real-time PCR results of lncRNA Gm47283 overexpression. **P < 0.01 vs Control group. n = 6. (f) Western blot results of Ptgs2, Alox15 and Gpx4 expression in lncRNA overexpression groups. n = 6 (g) ROS staining results of cardiomyocytes in lncRNA overexpression groups. n = 6. (h) Detection of SOD and MDA levels in lncRNA overexpression groups. * P < 0.05 vs Control group, #P < 0.05 vs Control+Gm47283 group. n = 6. Scale bars; 25 μm.
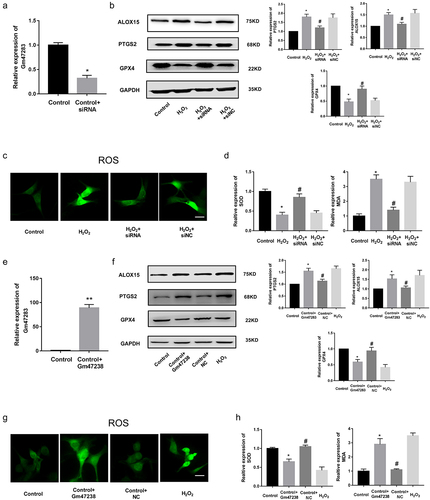
MiR-706 was the regulator of MI
microRNAs negatively regulate gene expression by promoting mRNA degradation or inhibiting mRNA translation [Citation37]. Previous studies also have found that lncRNAs exerted functions by endogenous sponging miRNAs to influence the expression of downstream protein coding genes. Thus, we speculated that lncRNA Gm47283 might regulate MI via ceRNA mechanism. Based on our prediction, we considered that miR-706 was the downstream target of lncRNA Gm47283. MiR-706 was demonstrated as the regulator of oxidative stress in liver fibrogenesis [Citation38]. Then, we performed real-time PCR to detect the expression of miR-706 in MI models. Results showed that miR-706 was down-regulated in MI models (). To investigate the regulatory role of miR-706 in MI, we constructed the miR-706 mimics and AMO-miR-706 to overexpress and inhibit the expression of miR-706. Real-time PCR results showed that miR-706 was induced by mimics and repressed by AMO-miR (). Western blot results showed that miR-706 could inhibit the expression of Ptgs2 and AMO-miR could reverse the repression effects. Ferroptosis marker Alox15 was also repressed by miR-706. Gpx4 expression was increased after mimic treatment ( and Supplementary Figure S1). We also detected the quantity of ROS after miRNA treatment. As a result, ROS staining results showed that overexpression of miR-706 could decrease the ROS activity (). Overexpression of miR-706 could also repress the quantity of MDA and increase the quantity of SOD (). These results indicated that miR-706 was a protective regulator of MI, which participated in MI via regulating cellular ferroptosis level. Moreover, this result also implied that our hypothesis of lncRNA Gm47283/miR-706/Ptgs2 regulatory axis in MI existed.
Figure 4. miR-706 is the regulator of MI. (a) Real-time PCR results of miR-706 expression in MI models. *P < 0.05 vs Control/Sham group. n = 6. (b) Real-time PCR results of miR-706 expression in miRNA interference groups. *P < 0.05 vs Control group. #P < 0.05 vs Control+miR-706 group. n = 6. (c) Western blot results of Ptgs2, Alox15 and Gpx4 expression in miR-706 overexpression or inhibition groups. n = 6. (d) ROS staining results of cardiomyocytes in miR-706 overexpression or inhibition groups. n = 6. (e-f) Detection of SOD and MDA levels in miR-706 overexpression or inhibition groups. *P < 0.05 vs Control group, #P < 0.05 vs H2O2 group, &P < 0.05 vs H2O2+ miR-706 group. n = 6. Scale bars; 25 μm.
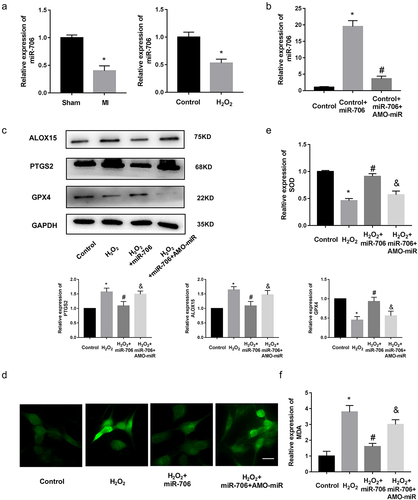
Next, we used luciferase report system to validate the relationship among lncRNA, miRNA, and downstream gene. Briefly, validates the direct binding relationship between miR-706 and Ptgs2. Moreover, we inserted wild-type and mutated-type Ptgs2 into luciferase plasmids. After co-transfection of Ptgs2 wild/mut luciferase plasmids and miR-706 mimic/miRNA NC into cells, results showed that miR-706 mimic sharply repressed the luciferase activity of wide-type Ptgs2 plasmid, but the activity of mutated-type Ptgs2 was not changed with miR-706 mimics (). Then, we established lncRNA Gm47283 plasmid and transfected the plasmid into the luciferase system (). Results showed that lncRNA Gm47283 could rescue the degradation of miR-706 to Ptgs2 (). Furthermore, we also detected the expression of miR-706 expression was regulated after lncRNA Gm47283 inhibition or overexpression ().
Figure 5. Validation of the direct regulatory relationships among lncRNA, miRNA and mRNA. (a) The interaction of miR-706 and Ptgs2. (b) Validation of direct interactions between miR-706 and Ptgs2. *P < 0.05 vs Ptgs2-wt group. n = 3. (c) The interaction of miR-706 and lncRNA Gm47283. (d) Validation of direct interactions among lncRNA Gm47283, miR-706 and Ptgs2. *P < 0.05 vs Ptgs2-wt group, #P < 0.05 vs Ptgs2-wt+miR-706+ Gm47283 group. n = 3. (e) Real-time PCR results of miR-706 expression in lncRNA overexpression and knockdown groups. *P < 0.05 vs Control group. n = 6.
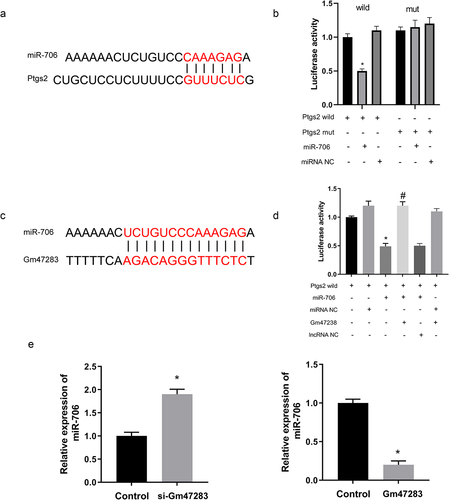
LncRNA Gm47283 regulates MI via miR-706 and Ptgs2
To further investigate the regulatory axis of lncRNA Gm47283/miR-706/Ptgs2 in MI, we performed a co-transfection experiment via co-transfecting lncRNA Gm47283 and miR-706 into cardiomyocyte. Results showed that the enhanced expression of Ptgs2 by lncRNA Gm47283 overexpression was repressed by miR-706 supplement ( and Supplementary Figure S1). We also detected the expression of ferroptosis marker expression via western blot. As a result, we found that Alox15 expression was elevated by lncRNA Gm47283 and then was repressed by miR-706. Gpx4 expression was inhibited by lncRNA overexpression and was rescued by miR-706 ( and Supplementary Figure S1). Additionally, we also detected the quantities of ROS, MDA, and SOD. Results showed that miR-706 could suppress the lncRNA Gm47283 mediated increases of ROS and MDA (). MiR-706 could also rescue the decreased SOD level that was decreased after lncRNA overexpression (). These results indicated that lncRNA Gm47283 could regulate MI via miR-706/Ptgs2/ferroptosis axis.
Figure 6. LncRNA Gm47283 regulates MI via miR-706 and Ptgs2. (a) Western blot results of Ptgs2, Alox15 and Gpx4 expression in co-transfection group. n = 6. (b) ROS staining results of cardiomyocytes in co-transfection groups. n = 6. (c-d) Detection of SOD and MDA levels in co-transfection groups. *P < 0.05 vs Control group, #P < 0.05 vs Gm47283 group. n = 6. Scale bars; 25 μm.
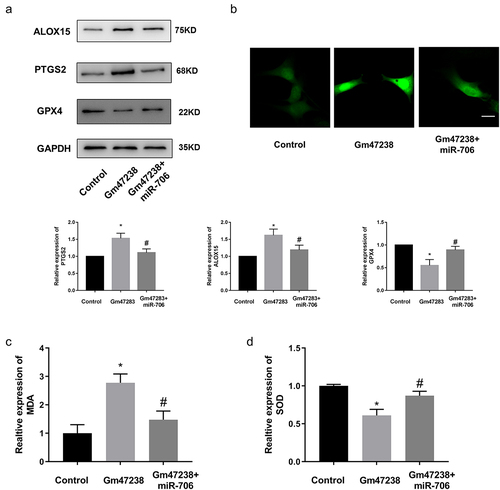
Inhibition of lncRNA Gm47283 attenuated MI in vivo
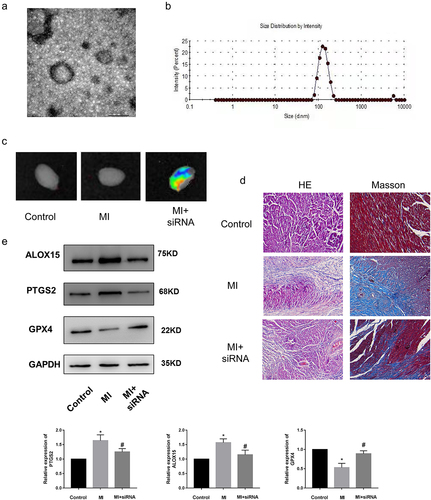
Currently, the regulatory mechanisms of a series of functional ncRNAs have been widely elucidated, but their clinical application has been greatly hindered due to little attention paid to the development of effective in vivo delivery strategies to regulate the biological functions. Moreover, compared to the billions of intracellular ncRNAs that may exert their biological functions, the proportion of ncRNAs currently found to be related to MI is still very low. Therefore, the regulatory mechanism and effective delivery of more functional ncRNAs remain to be explored. We have demonstrated that lncRNA Gm47283 regulated MI via miR-706/Ptgs2/ferroptosis axis. We then performed in vivo inhibition experiments to validate the therapeutic effects of lncRNA Gm47283 in MI models. Here, we used stem cell membrane coated siRNA to target the injury regions. Material characterization experiments showed that the stem cell membrane was successfully coated the siRNA (). More importantly, we also demonstrated that the coated siRNA was aggregated in heart (). After 1 weeks modeling, we found that stem cell coated siRNA could repress the inflammatory infiltration and collagen formation based on HE Staining and Masson staining (). Furthermore, we also detected the expression of Ptgs2 and ferroptosis marker proteins (Alox15 and Gpx4). Results showed that inhibition of lncRNA Gm47283 could decrease the expression of Ptgs2 and Alox15 and rescue the expression of Gpx4 ( and Supplementary Figure S1). These results suggested that heart targeted lncRNA Gm47283 inhibition could be used an effective drug to attenuate MI.
Figure 7. Knockdown of lncRNA Gm47283 via stem cell membrane coated siRNA attenuates MI in vivo. (a) Electron microscopy characterization of the stem cell membrane coated siRNA. (b) Size distribution of the stem cell membrane coated siRNA. (c) In vivo imaging of the hearts in different groups. (d) Masson and HE staining of hearts in different groups. n = 6. Western blot results of Ptgs2, Alox15 and Gpx4 expression in vivo knockdown group. n = 6.
Discussion
MI is the ischemic necrosis of myocardium, on the basis of coronary artery pathological changes, the blood flow of coronary artery is sharply reduced or interrupted, so that the corresponding myocardium appears serious and permanent acute ischemia, eventually bring about the ischemic necrosis of myocardium. It is one of the leading causes of death. In this study, we performed bioinformatics analysis to identify risk lncRNAs in MI and investigated the biological function and mechanism of lncRNA Gm47283 in rat models in vivo and in vitro. As a result, 10 lncRNAs were identified as risk factors from array data. Additionally, we found that TNF and NF-κB pathways were activated in MI, four protein coding genes of which were also extracted, including ferroptosis gene Ptgs2. In vivo and in vitro models also demonstrated that lncRNA Gm47283 and Ptgs2 were activated in MI. Interference of lncRNA Gm47283 expression has the impact on Ptgs2 expression and ferroptosis activity. Mechanically, we found that lncRNA Gm47283 participated in MI through regulating cardiomyocyte ferroptosis activity by targeting miR-706, which could target to the 3ʹUTR of Ptgs2. Moreover, inhibition of lncRNA Gm47283 by stem cell membrane coated siRNA could attenuate MI in vivo ().
Recent studies on miRNAs have renewed our understanding about the regulation of MI. Various studies have provided strong evidence that miRNAs play important roles in MI [Citation39,Citation40]. Notably, the expression of miR-706 was significantly down-regulated in the mice model of MI. In addition, miR-706 in cardiomyocytes was resistant to H2O2-induced damage. Furthermore, more importantly, we found that forced expression of miR-706 attenuated cardiomyocyte ferroptosis. Our study elucidated the anti-ferroptotic effect and mechanisms of miR-706 in MI, and indicated that the exogenous application of miR-706 might be a novel therapeutic strategy for MI. In the present study, enforced expression of miR-706 resulted in a reduction of Ptgs2 in cardiomyocytes. We also verified the interaction of Ptgs2 and miR-706 by luciferase assay. This indicated that the interaction between miR-706 and Ptgs2 3ʹUTR actually existed in vitro. Furthermore, we also investigate the effects of miR-706 negative control on Ptgs2, results showed that the negative control could not affect the expression of Ptgs2 and decrease ROS accumulation (Supplementary Figure S2).
LncRNAs were considered as the crucial regulators in gene post-transcription regulation. In the field of MI, numerous of lncRNAs were demonstrated to participate in the disease regulatory processes via multiple pathways, such as inflammation, apoptosis, and autophagy. Thus, in this study, we wanted to find a novel lncRNA transcript of MI and unveil the regulatory mechanism of it. We firstly performed bioinformatics analysis to call the differentially expressed lncRNAs. Ten up-regulated lncRNAs were identified as risk factors. Real-time PCR results showed that lncRNA Gm47283 was up-regulated in myocardial models in vivo and in vitro. But the regulatory mechanism of lncRNA Gm47283 is unknown. Interestingly, we found that numerous mRNAs were also up-regulated in MI, which were enriched in TNF signaling pathway, MAPK signaling pathway, and NF-κB signaling pathways. Moreover, we found that four up-regulated genes were both enriched in TNF and NF-κB signaling pathways and showed a strong connective activity in the differentially expressed genes-induced PPI network. Whether lncRNA Gm47283 regulated MI via these genes attracted our attention. Based on comprehensive bioinformatics analysis, we found that lncRNA Gm47283 could target to miR-706 to interfere the expression of Ptgs2, which was a known marker of cellular ferroptosis. Thus, we speculated that lncRNA Gm47283 involved in MI via miR-706/Ptgs2/ferroptosis axes.
Ferroptosis is a novel form of regulated cell death identified as iron-dependence, lipid peroxidation collection, and reactive oxygen species (ROS) generation. It is remarkably distinct from other types of classical regulated cell death, such as apoptosis, necrosis, and necroptosis. Ferroptosis participated in various diseases, including cancer, Parkinson’s disease, stroke, and IRI. However, its effect in the progression of AMI remains unclear. Here, we found that Ptgs2 was up-regulated in MI and Ptgs2 was predicted as the downstream target of lncRNA Gm47283/miR-706 axis. Ptgs2 is also known as COX2, which is a well-accepted biomarker for ferroptosis onset. COX2 is induced in cells undergoing ferroptosis [Citation41]. Alox15 was demonstrated as the downstream proteins of COX2 [Citation42,Citation43]. Results showed that Ptgs2 and Alox15 were both up-regulated in MI. In vitro loss and gain function experiments also validated that Ptgs2 participated in MI via serving as the downstream gene of lncRNA Gm47283.
At present, many functional lncRNAs are known, and their regulatory mechanisms have been widely elucidated. However, few efforts have been made to develop effective in vivo regulatory strategies to improve treatment outcomes. RNA interference is the most convenient method because of its strong ability to silence target gene expression. However, due to the polyanion and biomacromolecule characteristics of RNA interference therapies such as siRNA, specific delivery vehicles are required to facilitate siRNA delivery in vivo [Citation44,Citation45]. Nanotechnology has shown great promise for improvement of in vivo siRNA delivery, and several RNA interference nanoparticle platforms have been marketed or entered into early phase clinical trials for the treatment of various diseases. However, effective and safe systemic delivery of siRNAs into heart in vivo remains challenging, due to the complexities of cardiovascular disease.
In this study, we used stem cell membrane coated siRNA to inhibit the expression of lncRNA Gm47283. Stem cell membrane targeted therapy is considered as an advanced tool to deliver the drugs to the injury regions. Thus, here we selected this method and results showed that the stem cell membrane coated siRNA was directly targeted to the injury heart.
Conclusion
Taken together, we identified lncRNA Gm47283 was the top risk factor in MI. Bioinformatics analysis predicted that lncRNA Gm47283 exerted function via targeting miR-706 and Ptgs2. Ptgs2 was also the known regulator of ferroptosis. Inhibition or overexpression of lncRNA Gm47283 could regulate Ptgs2 expression and downstream ferroptosis activity. Overexpression of miR-706 could inhibit the expression of Ptgs2 and the activity of ferroptosis. Our study demonstrated a novel regulatory axis containing lncRNA Gm47283/miR-706/Ptgs2/ferroptosis in MI, which provided a potential mechanism for heart therapeutic treatment in clinic.
Author contributions
Feng Gao designed the overall project. Feng Gao and Yongcheng Zhao carried out the experiments. Other authors participated in some related experiments. Feng Gao drafted the original manuscript and revised the manuscript.
Supplemental Material
Download Zip (1.5 MB)Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Benjamin EJ, Blaha MJ, Chiuve SE, et al. Heart disease and stroke statistics-2017 update: a report from the American Heart Association. Circulation. 2017;135:e146–e603.
- Petrie MC, Jhund PS, Connolly E, et al. High-dose intravenous iron reduces myocardial infarction in patients on haemodialysis. Cardiovasc Res. 2021. DOI:10.1093/cvr/cvab317.
- Qin YY, Huang XR, Zhang J, et al. Neuropeptide Y attenuates cardiac remodeling and deterioration of function following myocardial infarction. Mol Ther. 2021;30(2):881–897.
- Wu X, Wang L, Wang K, et al. ADAR2 increases in exercised heart and protects against myocardial infarction and doxorubicin-induced cardiotoxicity. Mol Ther. 2021;30(1):400–414.
- Sun G, Shen JF, Wei XF, et al. Circular RNA Foxo3 relieves myocardial ischemia/Reperfusion injury by suppressing autophagy via inhibiting HMGB1 by repressing KAT7 in myocardial infarction. J Inflamm Res. 2021;14:6397–6407.
- Janbandhu V, Tallapragada V, Patrick R, et al. Hif-1a suppresses ROS-induced proliferation of cardiac fibroblasts following myocardial infarction . Cell Stem Cell; 2021;29(2):281–297.
- Li Y, Chen X, Jin R, et al. Injectable hydrogel with MSNs/microRNA-21-5p delivery enables both immunomodification and enhanced angiogenesis for myocardial infarction therapy in pigs. Sci Adv. 2021;7(9):eabd6740.
- Sun B, Liu S, Hao R, et al. RGD-PEG-PLA delivers MiR-133 to infarct lesions of acute myocardial infarction model rats for cardiac protection. Pharmaceutics. 2020;12(6):575.
- Zhang Y, Jiao L, Sun L, et al. LncRNA ZFAS1 as a SERCA2a inhibitor to cause intracellular Ca(2+) overload and contractile dysfunction in a mouse model of myocardial infarction. Circ Res. 2018;122:1354–1368.
- Wei L, Sun J, Zhang N, et al. Novel implications of MicroRNAs, long non-coding RNAs and circular RNAs in drug resistance of esophageal cancer. Front Cell Dev Biol. 2021;9:764313.
- Song C, Qi H, Liu Y, et al. Inhibition of lncRNA Gm15834 attenuates autophagy-mediated myocardial hypertrophy via the miR-30b-3p/ULK1 axis in mice. Mol Ther. 2021;29:1120–1137.
- Chen G, Li H, Li X, et al. Loss of long non-coding RNA CRRL promotes cardiomyocyte regeneration and improves cardiac repair by functioning as a competing endogenous RNA. J Mol Cell Cardiol. 2018;122:152–164.
- Wang K, Liu CY, Zhou LY, et al. APF lncRNA regulates autophagy and myocardial infarction by targeting miR-188-3p. Nat Commun. 2015;6:6779.
- Hu H, Wu J, Li D, et al. Knockdown of lncRNA MALAT1 attenuates acute myocardial infarction through miR-320-Pten axis. Biomed Pharmacother. 2018;106:738–746.
- Wu X, Iroegbu CD, Peng J, et al. Cell death and exosomes regulation after myocardial infarction and ischemia-reperfusion. Front Cell Dev Biol. 2021;9:673677.
- Luo MY, Su JH, Gong SX, et al. Ferroptosis: new dawn for overcoming the cardio-cerebrovascular diseases. Front Cell Dev Biol. 2021;9:733908.
- Wang K, Li Z, Sun Y, et al. Dapagliflozin improves cardiac function, remodeling, myocardial apoptosis, and inflammatory cytokines in mice with myocardial infarction. J Cardiovasc Transl Res. 2021. DOI:10.1007/s12265-021-10192-y.
- Zhou L, Sun J, Gu L, et al. Programmed cell death: complex regulatory networks in cardiovascular disease. Front Cell Dev Biol. 2021;9:794879.
- Demuynck R, Efimova I, Naessens F, et al. Immunogenic ferroptosis and where to find it? J Immunother Cancer. 2021;9:e003430.
- Chen Z, Yan Y, Qi C, et al. The role of ferroptosis in cardiovascular disease and its therapeutic significance. Front Cardiovasc Med. 2021;8:733229.
- Liang C, Zhang X, Yang M, et al. Recent progress in ferroptosis inducers for cancer therapy. Adv Mater. 2019;31:e1904197.
- Wang W, Green M, Choi JE, et al. CD8(+) T cells regulate tumour ferroptosis during cancer immunotherapy. Nature. 2019;569:270–274.
- Mahoney-Sanchez L, Bouchaoui H, Ayton S, et al. Ferroptosis and its potential role in the physiopathology of Parkinson’s Disease. Prog Neurobiol. 2021;196:101890.
- Alim I, Caulfield JT, Chen Y, et al. Selenium drives a transcriptional adaptive program to block ferroptosis and treat stroke. Cell. 2019;177:1262–79 e25.
- Szklarczyk D, Gable AL, Nastou KC, et al. The STRING database in 2021: customizable protein-protein networks, and functional characterization of user-uploaded gene/measurement sets. Nucleic Acids Res. 2021;49:D605–D12.
- Agarwal V, Bell GW, Nam JW, et al. Predicting effective microRNA target sites in mammalian mRNAs. Elife. 2015;4:e05005.
- Gao RF, Li X, Xiang HY, et al. The covalent NLRP3-inflammasome inhibitor Oridonin relieves myocardial infarction induced myocardial fibrosis and cardiac remodeling in mice. Int Immunopharmacol. 2021;90:107133.
- Dang Z, Su S, Jin G, et al. Tsantan Sumtang attenuated chronic hypoxia-induced right ventricular structure remodeling and fibrosis by equilibrating local ACE-AngII-AT1R/ACE2-Ang1-7-Mas axis in rat. J Ethnopharmacol. 2020;250:112470.
- Shi P, Li M, Song C, et al. Neutrophil-like cell membrane-coated siRNA of lncRNA AABR07017145.1 therapy for cardiac hypertrophy via inhibiting ferroptosis of CMECs. Mol Ther Nucleic Acids. 2022;27:16–36.
- Wang M, Xin Y, Cao H, et al. Recent advances in mesenchymal stem cell membrane-coated nanoparticles for enhanced drug delivery. Biomater Sci. 2021;9:1088–1103.
- Armstrong AW. Do TNF inhibitors reduce the risk of myocardial infarction in psoriasis patients? JAMA. 2013;309:2043–2044.
- Somasuntharam I, Yehl K, Carroll SL, et al. Knockdown of TNF-alpha by DNAzyme gold nanoparticles as an anti-inflammatory therapy for myocardial infarction. Biomaterials. 2016;83:12–22.
- Ge ZW, Zhu XL, Wang BC, et al. MicroRNA-26b relieves inflammatory response and myocardial remodeling of mice with myocardial infarction by suppression of MAPK pathway through binding to PTGS2. Int J Cardiol. 2019;280:152–159.
- Zhao Z, Qu F, Liu R, et al. Differential expression of miR-142-3p protects cardiomyocytes from myocardial ischemia-reperfusion via TLR4/NFkB axis. J Cell Biochem. 2019. doi:10.1002/jcb.29506.
- Zhang M, Jiang Y, Guo X, et al. Long non-coding RNA cardiac hypertrophy-associated regulator governs cardiac hypertrophy via regulating miR-20b and the downstream PTEN/AKT pathway. J Cell Mol Med. 2019;23:7685–7698.
- Thomson DW, Dinger ME. Endogenous microRNA sponges: evidence and controversy. Nat Rev Genet. 2016;17:272–283.
- Jens M, Rajewsky N. Competition between target sites of regulators shapes post-transcriptional gene regulation. Nat Rev Genet. 2015;16:113–126.
- Yin R, Guo D, Zhang S, et al. miR-706 inhibits the oxidative stress-induced activation of PKCalpha/TAOK1 in liver fibrogenesis. Sci Rep. 2016;6:37509.
- Roy S, Khanna S, Hussain SR, et al. MicroRNA expression in response to murine myocardial infarction: miR-21 regulates fibroblast metalloprotease-2 via phosphatase and tensin homologue. Cardiovasc Res. 2009;82:21–29.
- Dong S, Cheng Y, Yang J, et al. MicroRNA expression signature and the role of microRNA-21 in the early phase of acute myocardial infarction. J Biol Chem. 2009;284:29514–29525.
- Chen C, Chen J, Wang Y, et al. Ferroptosis drives photoreceptor degeneration in mice with defects in all-trans-retinal clearance. J Biol Chem. 2021;296:100187.
- Zhang H, Deng T, Liu R, et al. CAF secreted miR-522 suppresses ferroptosis and promotes acquired chemo-resistance in gastric cancer. Mol Cancer. 2020;19:43.
- Friedmann Angeli JP, Schneider M, Proneth B, et al. Inactivation of the ferroptosis regulator Gpx4 triggers acute renal failure in mice. Nat Cell Biol. 2014;16:1180–1191.
- Whitehead KA, Langer R, Anderson DG. Knocking down barriers: advances in siRNA delivery. Nat Rev Drug Discov. 2009;8:129–138.
- Kanasty R, Dorkin JR, Vegas A, et al. Delivery materials for siRNA therapeutics. Nat Mater. 2013;12:967–977.