ABSTRACT
Long non-coding RNA (lncRNA) cancer susceptibility candidate 7 (CASC7) was reported to be participated in tumor development. This study was carried out to investigate the functions of CASC7 in hepatocellular carcinoma (HCC) progression. The expression of CASC7 and microRNA-30a-5p (miR-30a-5p) in HCC tissues and cells were detected by quantitative Real-time PCR (qRT-PCR). The expression of Krueppel-like factor 10 (KLF10), transforming growth factor-β (TGF-β), and SMAD3 were detected by Western Blot analysis. Transwell assay, flow cytometry, Cell Counting Kit-8 (CCK-8) assay and colony formation assay were performed to evaluate the effects of CASC7, KLF10 and miR-30a-5p on cell function. The relationship among CASC7, KLF10 and miR-30a-5p was evaluated by luciferase reporter assay and bioinformatics analyses. Tumor growth was detected in nude mice. The expression levels of CASC7 were increased and the expression levels of miR-30a-5p were reduced in HCC cells and tissues. Knockdown of CASC7 and overexpression of miR-30a-5p reduced tumor growth as well as HCC cell proliferation, invasion and migration. In HCC tumor tissues, the expression of miR-30a-5p was negatively correlated with the expression of CASC7. Moreover, as a target of miR-30a-5p, KLF10 was regulated by CASC7 and miR-30a-5p, and CASC7 regulated the KLF10/TGF-β/SMAD3 pathway via binding to miR-30a-5p, thereby promoting HCC cell progression.
Graphical Abstract
1. Introduction
HCC has become the sixth most common malignant tumor in the world [Citation1,Citation2], and most of them are diagnosed in late stages with multiple complications and the surgical resection rate is extremely low [Citation3]. At present, the treatment for liver cancer is mainly surgical resection, combined with radiotherapy, chemotherapy and liver transplantation [Citation4]. Most patients often suffer from metastasis and recurrence after surgery [Citation5]. Recent studies have shown that HCC is closely related to abnormal expression of long non-coding RNAs (lncRNAs) and cell signal transductions [Citation6].
LncRNAs are expressed in different cells and tissues and can interact with other macromolecules such as DNAs, miRNAs and proteins [Citation7,Citation8]. With the advance in high-throughput sequencing technologies, various lncRNAs have been identified [Citation9]. Moreover, lncRNAs might serve as the biomarkers for tumor progression and recurrence in HCC patients [Citation10,Citation11]. For example, it has been reported that lncRNA MIAT promotes HCC cell invasion and proliferation via sponging miR-214 [Citation12]. LncRNA cancer susceptibility candidate 7 (CASC7) was revealed to be involved in glioma [Citation13], non-small cell lung cancer [Citation14], and colon cancer [Citation15]. However, the functions of CASC7 in HCC remain unclear.
MicroRNAs (miRNAs) widely exist in many organisms and are involved in the regulation of gene expression [Citation16]. Studies have found that lncRNAs can regulate the expression of miRNAs and tumorigenesis [Citation17]. Moreover, miRNAs are also involved in liver cancer [Citation18] and function as oncogenes or tumor suppressive genes [Citation19]. Meanwhile, miRNAs are related to the clinicopathological characteristics of liver cancer such as pathological type and the degree of malignancy [Citation20]. As a newly discovered miRNA, miR-30a-5p has been reported to be involved in invasion and migration of a variety of cancers [Citation21]. Moreover, studies have found that miR-30a-5p is significantly downregulated and suppresses HCC tumor growth and cell invasion and proliferation in HCC tissues [Citation22]. We therefore speculated that miR-30a-5p may mediate the function of CASC7 in HCC. The TGF-β/SMAD3 signaling participates in HCC migration and metastasis [Citation15,Citation23]. In addition, it has been reported that miRNA-491 is involved in anti-angiogenesis in HCC induced by arsenic trioxide by inhibiting the TGF-β/SMAD3 signaling [Citation14].
Furthermore, the TGF-β/SMAD3 pathway mediated the anti-oncogene of miR-133a/FOSL2 in HCC. These studies suggest that the TGF-β/SMAD3 signaling can be regulated by miRNAs in HCC. We therefore hypothesized that CASC7 might regulate the KLF10/TGF-β/SMAD3 pathway through miR-30a-5p in HCC development. This study was therefore carried out to investigate the role and underlying mechanism of CASC7 in HCC.
2. Methods
2.1. Clinical specimens
The tumor tissues and adjacent non-tumor tissues were obtained from 38 patients who were admitted at the First Affiliated Hospital of Jinzhou Medical University between January 2012 to July 2014. All patients signed the informed consent. Among the 38 patients, 20 patients developed intrahepatic metastases. During 2 years, 18 patients had solitary tumors and no metastasis or recurrence occurred. This study was approved by the Ethics Committee of the aforementioned hospital. The Correlation between clinicopathologic characteristics and the expression of CASC7 was analyzed in HCC ().
Table 1. Correlation between the clinicopathologic characteristics and CASC7 expression in HCC (n = 50)
2.2. Cell lines and cell culture
HepG2, MHCC97H, LO2, HepG2, SK-Hep1, HUH7 and HCCLM3 cell lines were obtained from Tongpai Technology (Shanghai, China). Cells were incubated in DMEM medium with FBS (10%) at 37°C with 5% CO2.
2.3. Cell transfection
ShCASC7, miR-30a-5p inhibitors and mimic and their controls were purchased from Shanghai Gene Pharmaceutical Co., Ltd. The sequences were as follows: shCASC7-1: 5'-TGGAACACATGGTCCA GCACTTTAA-3'; shCASC7-2: 5'-CATCTATT GTCGTGTTTAAGCTT-3'; shNC: 5’-TGGACAC TGGTGACCTCACTAATAA-3'.Lipofectamine 2000 (Invitrogen, Carlsbad, CA) mediated transfection of oligonucleotides into cells was then performed.
2.4. CCK8 assay
Cells (50,000 cells per well) were seeded, and CCK8 solution (100 μL, Liji, Shanghai, China) was added to incubate the cells. Then, the absorbance was measured at 450 nm using a microplate reader (Meigu, Shanghai, China) [Citation24].
2.5. RNA pull-down assay
Cells were transfected with biotin-labeled mutant (Mut)- and the wild-type (WT)-bio-miR-30a-5p (50 nM) for 48 h. After washing with PBS, a specific lysis buffer was added to incubate the cells for 10 min. After pre-coating with BSA, M-280 streptavidin magnetic beads were used to incubate the lysate at 4°C for 3 h. Finally, The binding RNA was purified and its concentration was measured [Citation25].
2.6. Apoptosis assay
Cells (5 x 105 cells/well) were plated and grown to the logarithmic growth phase. Cells were then collected, counted and separated by centrifugation and regenerated by adding 195 μl annexin V-FITC. Propidium iodide (10 μl) and Annexin V-FITC (5 μl) staining solutions were then added. Cells were incubated in darkness for 10–20 min and then placed in an ice bath [Citation26]. Flow cytometry results were then collected (Jiyuan, Guangzhou, China).
2.7. Luciferase reporter assay
Luciferase reporter assay was carried out as previously described [Citation27]. The WT and MUT primers were used to amplify CASC7. pGL3-Bashc luciferase reporter vector carrying the CASC7 fragment was named as CASC7-WT-Luc. The mutant plasmid CASC7-Mut-Luc was constructed by mutagenizing the binding region in CASC7 and miR-30a-5p. pRLTK and miR-30a-5p mimic were co-transfected into cells with CASC7-Mut-Luc and CASC7-WT-Luc. Cells were collected after 6 h of transfection. Luciferase activity was determined using a Luciferase Assay Kit.
2.8. RNA extraction and RT-qPCR
otal RNAs were extracted using TRIzol reagent (Aosentai, Shanghai, China). RT-qPCR was carried out as previously described [Citation27]. The quality of RNA samples was analyzed by NanoDrop 1000. cDNA synthesis and RT-qPCR were carried out using qScript microRNA cDNA synthesis kit and ABI 7,500 real-time PCR system, respectively. The expression levels of CASC7 and miR-30a-5p were calculated using the 2−ΔΔCT method. Gene expression levels were normalized to the internal reference U6 and GADPH. The primer sequences were: U6, 5'-CGCTTCACGAATTTGCGTGTCA-3' (forward) and 5'-GCTTCGGCAGCACATATACTAAAAT-3' (reverse); CASC7, 5'-ATCAACGTCAAGCTGGGA GG-3' (forward) and 5'-CTTGTCCCCCG CTCGTTC-3' (reverse); β-actin, 5'-GACCTCTA TGCCAACACAG-3' (forward) and 5'-AGTACTTGCGCTCAGGAGGA-3’ (reverse); miR-30a-5p,5’-GAGCCAGGTTTCGTGGG CACGTGTGTTAT-3’ (forward) and 5'- ATAACACACGTGCCCACGAAACCTGGCTC-3' (reverse).
2.9. Colony formation assay
Approximately 1,000 cells were plated and cultured in DMEM medium for 7 d. Then, cells were stained with Wright’s stain, followed by staining with Sorensen phosphomolybdate buffer solution and Giemsa dye solution (9:1) for 10 min. Colonies were then counted under a microscope [Citation28].
2.10. Transwell assay
HCC cells were transfected with shCASC7, pcCASC7, miR-30a-5p inhibitor and mimic or NC. Briefly, 3 × 105 HCC cells were treated with 2.5 μg/mL mitomycin C to inhibit cell division for 24 h. After culturing, appropriate amount of trypsin digestion was added, cells were washed with PBS buffer and resuspend in serum-free medium. Then, cells were cultured in the basement with complete medium for 24 h. The superfluous medium was discarded. Cells were then fixed with polyformaldehyde (4%) for 30 min, followed by staining with crystal violet (0.1%) for 10 min. Ten visual fields were randomly selected, cells were observed, photographed, and counted [Citation29].
2.11. Western blot analysis
Total proteins were extracted and separated by SDS-PAGE (10%). Protein samples were transferred onto PVDF membranes, which were blocked with primary antibodies of anti-KLF10 (1:1,000), TGF-b (1:1,000), p-SMAD3 (1:1,000), SMAD3 (1:1,000) and anti-GAPDH antibody (1:1,000) at 4°C overnight. Antibodies were purchased from Shifeng, Shanghai, China. Then anti-rabbit secondary antibody (1:1,000) was added to incubate the membrane [Citation30]. After ECL (Sigma-Aldrich, USA) developed signals, Image J v1.46 software was used.
2.12. IHC staining
Liver cancer tissues were deparaffinized and heated after hydration. The sections were blocked with normal goat serum (10%) and then incubated with anti-Ki-67. The Envision™ ABC kit (Beinuo, Shanghai, China) was used for immunological detection. Finally, the Leica DM4000B/M microscope was used for observation [Citation31].
2.13. Tumor xenograft models in nude mice
Male athymic nude mice were kept under standard conditions. Approximately 5 × 106 HCC cells transfected with CASC7 lentiviral vector or miR-30a-5p inhibitor were separately subcutaneously inoculated into nude mice. All mice were divided to different groups. The tumor size was determined every week for 4 weeks. Mice were sacrificed and tumors were weighted.
2.14. Statistical methods
Data were presented as the mean ± stand deviation (SD). All statistical analyses were performed using the SPSS 13.0 software. Student’s t-test was used for comparisons. p value < 0.05 was considered as significant difference.
3. Results
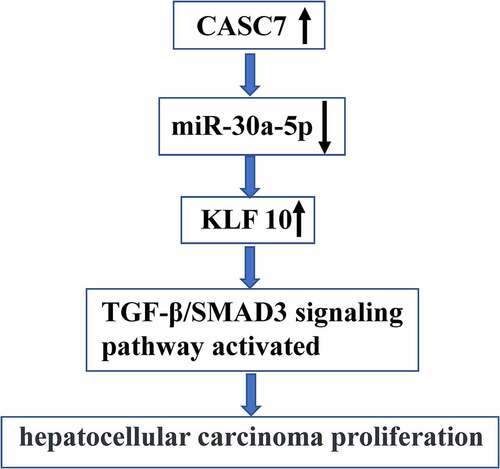
In this study, we aimed to investigate whether and how CASC7 was involved in HCC progression. We found that CASC7 regulated the KLF10/TGF-β/SMAD3 axis via binding to miR-30a-5p, thereby promoting HCC cell progression.
3.1. CASC7 regulated the progression of HCC in vitro
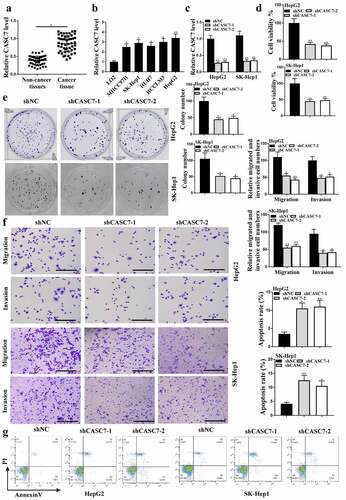
The expression levels of CASC7 in HCC tissues (n = 38) and HUH7, LO2, MHCC97H, SK-hep1, HepG2 cells were increased compared with that in adjacent normal tissues () and HCCLM3 cells (P < 0.05, ). The expression levels of CASC7 in HepG2 cells transfected with shCASC7-1 or shCASC7-2 were significantly reduced, indicating successful transfection (P < 0.05, ). Colony formation () and CCK-8 assay () results showed that shCASC7-1 and shCASC7-2 significantly inhibited HepG2 cell viability and colony formation (P < 0.05). ShCASC7-1 and shCASC7-2 reduced HepG2 cell migration and invasion (, p < 0.05). In addition, shCASC7-1 and shCASC7-2 significantly induced apoptosis of HepG2 cells (P < 0.05, ). Altogether, these results suggested that CASC7 was involved in HCC progression in vitro.
Figure 1. Effects of CASC7 on HCC tumor growth. (a) CASC7 expression in adjacent normal tissues and HCC tissues (n = 38). (b) CASC7 expression in HCC cell lines (c) CASC7 expression in HepG2 cells transfected with sh-NC, shCASC7-1 or shCASC7-2. (d) CCK8 determined Cell viability. (e and f) Colony formation, invasion and migration (100 um). (g) Cell apoptosis rate. ** P < 0.01, * P< 0.05; n= 3.
3.2. MiR-30a-5p regulated HCC progression in vitro
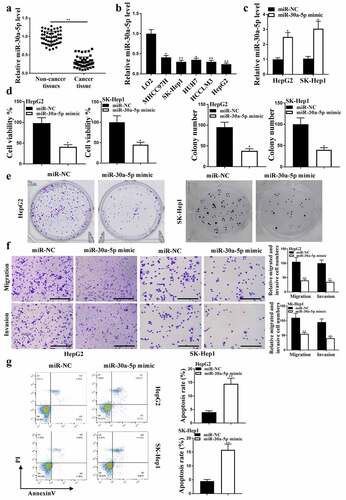
The expression levels of miR-30a-5p were reduced in HCC tissues (, p < 0.01) and HUH7, LO2, MHCC97H, SK-hep1, HepG2 cells (, p < 0.05) compared with that in adjacent normal tissues (n = 38) and HCCLM3 cells. The expression of miR-30a-5p was enhanced after transfection with miR-30a-5p mimic (P < 0.05, ). CCK8 analysis () and colony formation () results showed that miR-30a-5p mimic reduced HepG2 cell viability and colony formation (P < 0.05), as well as HepG2 cell migration and invasion (, p < 0.05), and induced HepG2 cell apoptosis (, p < 0.01). Taken together, these results suggested that miR-30a-5p regulated HCC progression in vitro.
Figure 2. MiR-30a-5p in HCC proliferation. (a) In HCC and adjacent normal tissues (n = 38), miR-30a-5p expression was detected. (b) In HCC cell lines, miR-30a-5p expression was detected. (c) MiR-30a-5p expression in HepG2 and SK-Hep1 cells. (d) CCK8 assay for cell viability. (e and f) Colony formation, migration and invasion (100 um). (g) Cell apoptosis. ** P< 0.01, * P < 0.05; n = 3.
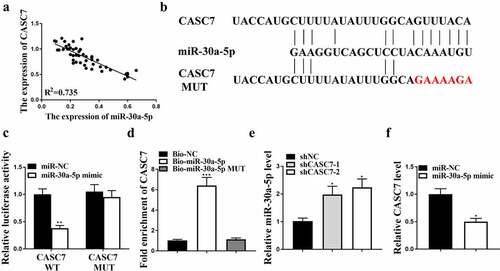
4. MiR-30a-5p was a direct target of CASC7
The expression of CASC7 was inversely correlated with the expression of miR-30a-5p in HCC tissues (R2 = 0.76, ). Starbase v2.0 and Tarbase v7.0 predicted multiple miRNAs that could bind with CASC7, including miR-30a-5p (). Moreover, the luciferase activity was decreased in HepG2 cells co-transfected miR-30a-5p mimic with CASC7-WT (P < 0.05), but not co-transfected CASC7-MUT with miR-30a-5p mimic (). In addition, the expression levels of CASC7 in HepG2 cells treated with miR-30a-5p probe were enhanced compared with HepG2 cells treated with NC probe (P < 0.05, ). In contrast, the expression levels of miR-30a-5p in the shCASC7-1 and shCASC7-2 groups were enhanced (, p < 0.05). The expression levels of CASC7 were reduced after overexpression of miR-30a-5p (P < 0.05, ). Taken together, these results suggested that miR-30a-5p was a direct target of CASC7.
Figure 3. The interaction between miR-30a-5p and CASC7. (a) Correlation between CASC7 and miR-30a-5p. (b) The binding site between miR-30a-5p and CASC7. (c) Relative luciferase activity. (d) The interaction between miR-30a-5p and CASC7 evaluated by RNA pull-down. (e) MiR-30a-5p expression in HepG2 cells. (f) CASC7 expression in HepG2 cells. ** P < 0.01, * P < 0.05, n = 3.
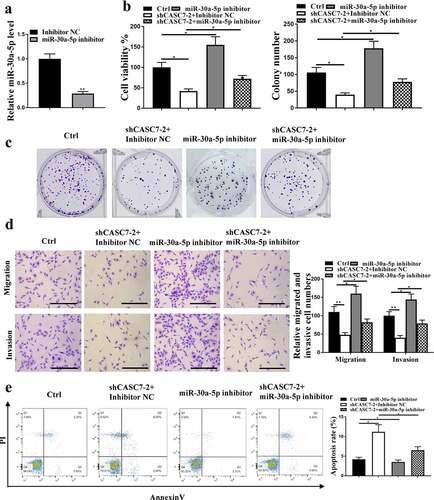
5. MiR-30a-5p mediated the effect of CASC7 on HCC cell proliferation
The expression levels of miR-30a-5p were reduced after inhibition of miR-30a-5p (p < 0.05, ). ShCASC7 reduced HCC cell proliferation (, p < 0.05), which was reversed by miR-30a-5p inhibition. Moreover, shCASC7 significantly inhibited HCC cell invasion and migration, which was also reversed by inhibition of miR-30a-5p (, p < 0.05). Moreover, shCASC7 induced HCC cell apoptosis, which was reversed by miR-30a-5p inhibitor, too (, p < 0.05). Altogether, these results suggest that miR-30a-5p mediated the function of CASC7 in HCC cell proliferation.
Figure 4. The effect of CASC7 on HCC proliferation mediated by miR-30a-5p. (a) miR-30a-5p expression in HepG2 cells. (B, C and D) migration and invasion (100 um), colony formation, cell viability. (e) Cell apoptosis rate. ** P < 0.01, * P < 0.05, n = 3.
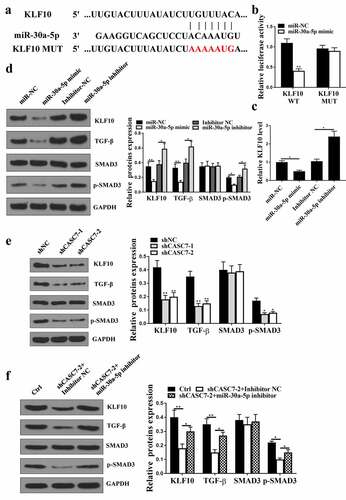
5.1. KLF10 was a direct target of miR-30a-5p
Starbase v2.0 predicted that KLF10 was a potential target of miR-30a-5p (). Co-transfection of KLF10-WT with miR-30a-5p mimic (, p < 0.05) reduced luciferase activity in cells, but not that of the co-transfection of KLF10-MUT with miR-30a-5p mimic (, p > 0.05) in cells. In addition, the expression levels of KLF10 were reduced at both mRNA () and protein () levels in miR-30a-5p mimic group (P < 0.05), but increased in miR-30a-5p inhibitor group (P < 0.05) compared with that in the NC group. The expression levels of p-SMAD3 and TGF-β were decreased in miR-30a-5p mimic group (, p < 0.05), but enhanced in miR-30a-5p inhibitor group (, p < 0.05). Knockdown of CASC7 significantly reduced the expression levels of KLF10, p-SMAD3 and TGF-β (, p < 0.05), which was reversed by miR-30a-5p inhibition (p < 0.05, ). Altogether, these results suggested that KLF10 was a direct target of miR-30a-5p.
Figure 5. KLF10 was predicted. (a) The binding site between miR-30a-5p and KLF10. (b) Relationship between KLF10 and miR-30a-5p evaluated by relative luciferase activity assessment. (c) KLF10 expression in HepG2 cells. (d) KLF10, SMAD3, p-SMAD3, TGF-β and protein expression in HepG2 cells. (e) KLF10, SMAD3, p-SMAD3, TGF-β protein expression in HepG2 cells treated with shCASC7-1 and shCASC7-2. (f) KLF10, SMAD3, p-SMAD3, TGF-β protein expression in HepG2 cells treated with shCASC7 + Inhibitor NC and shCASC7 + miR-30a-5p inhibitor. ** P < 0.01, * P < 0.05, n = 3.
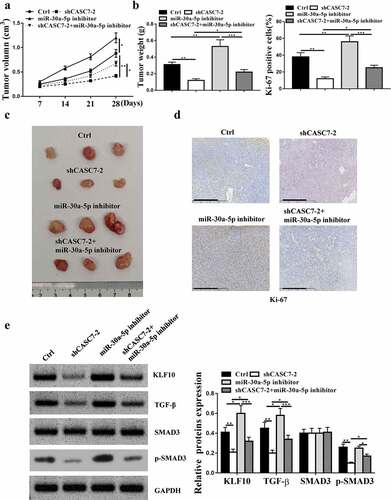
5.2. Down-regulation of CASC7 restrained HCC progression in vivo
It was found that inhibition of miR-30a-5p enhanced the tumor volume and weight, and knockdown of CASC7-2 reduced the tumor volume and weight (, p < 0.05), which was attenuated by inhibition of miR-30a-5p. Immunohistochemical results showed that Ki67 positive cell number was increased in miR-30a-5p inhibitor group and decreased in shCASC7-2 group, which was attenuated by miR-30a-5p inhibitor (). ShCASC7-2 reduced the expression levels of KLF10, TGF-β and p-SMAD3 (P < 0.05), which was attenuated by miR-30a-5p inhibitor (P < 0.05, ). Compared with the Control, inhibitor-NC and sh-NC group, shCASC7-2 significantly reduced the expression levels of KLF10, p-SMAD3 and TGF-β, which was attenuated by miR-30a-5p inhibition (supplementary Figure 1, P < 0.05). Altogether, these results suggested that knockdown of CASC7 inhibited HCC progression in vivo.
6. Discussion
Even with encouraging progress that has been made in the understanding of the molecular mechanisms of liver cancer development, in advanced liver cancer patients, the prognosis remains unfavorable [Citation32]. It is known that altered expression of lncRNAs are closely related to the occurrence and development of liver cancer [Citation33,Citation34]. Here, we showed that the expression levels of CASC7 were increased in HCC tissues and cells. Importantly, CASC7 promoted HCC tumor malignant processes and tumor growth in vivo, suggesting that CASC7 plays an oncogenic role in HCC.
LncRNAs were shown to be involved in pathological processes of human disease [Citation35]. In tumor development process, lncRNAs exert their functions as pro-tumor factors or suppressors in tumors through various mechanisms, such as regulating the expression of neighboring genes or affecting the expression of related genes [Citation36]. Studies have identified many abnormally expressed lncRNAs in HCC, which are involved in HCC prognosis, metastasis and occurrence [Citation37]. For example, the highly upregulated expression of ANRIL is closely related to HCC poor prognosis [Citation38] and induces HCC cell invasion and proliferation [Citation39]. In a variety of malignancies, CASC7 was found to be abnormally expressed. For example, the expression levels of CASC7 in colorectal cancer are significantly decreased, while overexpression of CASC7 can inhibit the survival of colorectal cancer cells [Citation40]. In glioma tissues and cell lines, CASC7 is down-regulated and involved in glioma cell proliferation and apoptosis [Citation13]. Moreover, it was reported that CASC7 suppresses malignant behaviors of breast cancer by regulating the miR-21-5p/FASLG axis [Citation27]. Here, we demonstrated that the expression levels of CASC7 were significantly increased in HCC. Moreover, knockdown of CASC7 inhibited HCC cell invasion, migration, and proliferation and induced HCC cell apoptosis. These suggested that CASC7 acts as an oncogene in the occurrence of HCC. Our findings further enriched our understanding of CASC7 in different types of tumor.
Studies have shown that lncRNAs have sponge adsorption on miRNAs, and thus participate in tumorigenesis [Citation41]. For example, lncRNA DLEU2 sponges miR-30a-5p to accelerate non-small cell lung tumorigenesis and invasion [Citation42]. LncRNA MALAT1 sponges miR-30a-5p to regulate vimentin expression in HCC [Citation43]. Here, miR-30a-5p was found to be a target gene of CASC7. The expression levels of miR-30a-5p were reduced in HCC and reversely correlated with the expression levels of CASC7. In addition, overexpression of miR-30a-5p reduced HCC cell migration, invasion and proliferation and induced apoptosis of HCC cells, and it could reverse the effects of shCASC7 on HCC tumor growth and cell apoptosis, invasion, migration, proliferation. These suggest that miR-29a-3p mediated the function of CASC7 in HCC.
KLF10 is a transcription factor containing a zinc finger protein domain, and TGF-β is an early responsive gene [Citation44]. It has been reported that the TGF-β/SMAD3 axis is closely related to HCC occurrence [Citation45]. Here, KLF10 was a target gene of miR-30a-5p. The expression levels of KLF10, p-SMAD3 and TGF-β were reduced after overexpression of miR-30a-5p. ShCASC71 could reduce the expression levels of KLF10, p-SMAD3 and TGF-β, which was reversed by miR-30a-5p inhibitor. These indicate that CASC7 regulates KLF10/TGF-β/SMAD3 through miR-30a-5p to promote HCC proliferation. In future, we will further investigate the roles of SMAD2 and SMAD4 in HCC proliferation.
Availability of data and materials
The analyzed data sets generated during the study are available from the corresponding author on reasonable request.
Ethics approval and consent to participate
This study was approved by the ethics committee of the First Affiliated Hospital of Jinzhou Medical University. All procedure was followed the guideline of this hospital. Written informed consent was obtained from all individual participants included in this study.
Supplemental Material
Download Zip (1.5 MB)Acknowledgements
The authors would like to express our gratitude for those who have critically reviewed this manuscript and those who give us help during this experiment.
Disclosure statement
No potential conflict of interest was reported by the author(s).
Supplementary material
Supplemental data for this article can be accessed here
Additional information
Funding
References
- Ayuso C, Rimola J, Vilana R, et al. Diagnosis and staging of hepatocellular carcinoma (HCC): current guidelines. Eur J Radiol. 2018;101:72–81.
- Chen L, Guo P, He Y, et al. HCC-derived exosomes elicit HCC progression and recurrence by epithelial-mesenchymal transition through MAPK/ERK signalling pathway. Cell Death Dis. 2018;9(5):1–17.
- Hallager S, Ladelund S, Kjær M, et al. Hepatocellular carcinoma in patients with chronic hepatitis C and cirrhosis in Denmark: a nationwide cohort study. J Viral Hepatitis. 2018;25(1):47–55.
- Balachandrakumar VK, Jabbar NF, White D, et al. Diagnosis and Staging of Hepatocellular Carcinoma (HCC): from mechanisms to management. Cham Spriger. 2019;67–82.
- Kim JM, Joh J-W, N-j Y, et al. Predicting hepatocellular carcinoma recurrence beyond milan criteria after liver resection for solitary hepatocellular carcinoma. J Gastrointest Surg. 2020;24(10):2219–2227.
- He J, Zhao H, Deng D, et al. Screening of significant biomarkers related with prognosis of liver cancer by lncRNA-associated ceRNAs analysis. J Cell Physiol. 2020;235(3):2464–2477.
- Arun G, Spector DL. MALAT1 long non-coding RNA and breast cancer. RNA Biol. 2019;16(6):860–863.
- Dang Y, Wei X, Xue L, et al. Long non-coding RNA in glioma: target miRNA and signaling pathways. Clinical Laboratory. 2018;64(6):887.
- Li J, Li L, Yuan H, et al. Up-regulated lncRNA GAS5 promotes chemosensitivity and apoptosis of triple-negative breast cancer cells. Cell Cycle. 2019;18(16):1965–1975.
- Shang W, Adzika GK, Machuki JO. Molecular mechanisms of circular RNAs, transforming growth factor-β, and long noncoding RNAs in hepatocellular carcinoma. Cancer Med. 2019 2;8(15):6684–6699.
- Yang Z, Chen W. Long non-coding RNA CRNDE promote the progression of tongue squamous cell carcinoma through regulating the PI3K/AKT/mTOR signaling pathway. RSC Adv. 2019;9(37):21381–21390.
- Huang X, Gao Y, Qin J, et al. LncRNA-MIAT promoted proliferation and invasion of HCC cells via sponging miR-214. Am J Physiol Gastrointest Liver Physiol. 2017;314(5):G559.
- Gong X, Liao XY, Huang MY. LncRNA CASC7 inhibits the progression of glioma via regulating Wnt/beta-catenin signaling pathway. Pathol Res Pract. 2019;215(3):564–570.
- Jiang F, Wang X, Liu Q, et al. Inhibition of TGF-beta/SMAD3/NF-kappaB signaling by microRNA-491 is involved in arsenic trioxide-induced anti-angiogenesis in hepatocellular carcinoma cells. Toxicol Lett. 2014;231(1):55–61.
- Liao Z, Chen L, Zhang X, et al. PTPRepsilon acts as a metastatic promoter in hepatocellular carcinoma by facilitating recruitment of SMAD3 to TGF-beta receptor 1. Hepatology. 2020;72(3):997–1012.
- Zhong X, Huang G, Ma Q, et al. Identification of crucial miRNAs and genes in esophageal squamous cell carcinoma by miRNA-mRNA integrated analysis. Medicine (Baltimore). 2019;98(27):e16269.
- Liu Z, Xu S, Dao J, et al. Differential expression of lncRNA/miRNA/mRNA and their related functional networks during the osteogenic/odontogenic differentiation of dental pulp stem cells. J Cell Physiol. 2020;235(4):3350–3361.
- Huang F, Wang B, Zeng J, et al. MicroRNA-374b inhibits liver cancer progression via down regulating programmed cell death-1 expression on cytokine-induced killer cells. Oncol Lett. 2018;15(4):4797–4804.
- Cai Y, Zhan L, Zhang CZ, et al. Effect of Gleditsia sinesis extract on miRNA21 and PTEN gene in liver cancer rat. Zhonghua Gan Zang Bing Za Zhi. 2018;26(2):142–144.
- Chen H, Lan Z, Li Q, et al. Abnormal expression of long noncoding RNA FGD5-AS1 affects the development of periodontitis through regulating miR-142-3p/SOCS6/NF-κB pathway. Artificial Cells. 2019;47(1):2098–2106.
- Li J, L-m Z, Zhang C, et al. The lncRNA FEZF1-AS1 promotes the progression of colorectal cancer through regulating OTX1 and targeting miR-30a-5p. Oncol Res. 2019;28(1):51.
- Zhang SL, Liu Q, Zhang Q, et al. MicroRNA-30a-5p suppresses proliferation, invasion and tumor growth of hepatocellular cancer cells via targeting FOXA1. Oncol Lett. 2017;14(4):5018–5026.
- You Y-K, Luo Q, W-f W, et al. Petchiether A attenuates obstructive nephropathy by suppressing TGF-β/Smad3 and NF-κB signalling. J Cell Mol Med. 2019;23(8):5576–5587.
- Han D, Tong YN, Jia YP, et al. lncRNA EVADR promotes proliferation and migration of human colorectal cancer cell lines. Basic Clini Med. 2018;38:174.
- Ruan X, Li P, Cao H. Identification of transcriptional regulators that bind to long noncoding RNAs by RNA Pull-Down and RNA immunoprecipitation. Methods Mol Biol. 2018;1783:185–191.
- Liu X, Qiu Z, Wang Z, et al. NFBD1/MDC1 participates in the regulation of proliferation and apoptosis in human laryngeal squamous cell carcinoma. Clin Trans Oncology. 2017;20(2):1–8.
- Wang G, Duan P, Liu F, et al. Long non-coding RNA CASC7 suppresses malignant behaviors of breast cancer by regulating miR-21-5p/FASLG axis. Bioengineered. 2021;12(2):11555–11566.
- Zhao W, Geng D, Li S, et al. LncRNA HOTAIR influences cell growth, migration, invasion, and apoptosis via the miR‐20a‐5p/HMGA2 axis in breast cancer. Cancer Med. 2018;7(3):842–855.
- Ridgway PF, Ziprin P, Jones TL, et al. Laparoscopic staging of pancreatic tumors induces increased invasive capacity in vitro. Surg Endosc. 2003;17(2):306–310.
- Tomasello L, Cluts L, Croce CM. Experimental validation of microRNA targets: analysis of microRNA targets through Western blotting. New York NY: Humana Press; 2019.
- Jun L, Tang, Tao T, et al. LncRNA-H19 promotes hepatic lipogenesis by directly regulating miR-130a/PPARγ axis in nonalcoholic fatty liver disease. Biosci Rep. 2019;39(7):BSR20181722.
- Chang W-K, Lee M-Y, Tai Y-H, et al. Does epidural analgesia improve the cancer outcome in hepatocellular carcinoma after resection surgery? A retrospective analysis. J Chin Med Assoc. 2019;82(4):295–299.
- Petruzziello A. Epidemiology of Hepatitis B Virus (HBV) and Hepatitis C Virus (HCV) related hepatocellular carcinoma. Open Virol J. 2018;12(Suppl–1, M3):26–32.
- Marcon PDS, Tovo CV, Kliemann DA, et al. Incidence of hepatocellular carcinoma in patients with chronic liver disease due to hepatitis B or C and coinfected with the human immunodeficiency virus: a retrospective cohort study. World J Gastroenterol. 2018;24(5):613–622.
- Zhou M, Zhao H, Wang X, et al. Analysis of long noncoding RNAs highlights region-specific altered expression patterns and diagnostic roles in Alzheimer’s disease. Brief Bioinform. 2018;20(2):598.
- Li Y, Zhu G, Ma Y, et al. lncRNA CCAT1 contributes to the growth and invasion of gastric cancer via targeting miR-219-1. J Cell Biochem. 2019;120(12):19457–19468.
- Wang Z, Wu Q, Feng S, et al. Identification of four prognostic LncRNAs for survival prediction of patients with hepatocellular carcinoma. Peerj. 2017;5(7):e3575.
- Hua L, Wang C-Y, Yao K-H, et al. High expression of long non-coding RNA ANRIL is associated with poor prognosis in hepatocellular carcinoma. Int J Clin Exp Pathol. 2015;8(3):3076–3082.
- Hsieh AL, Walton ZE, Altman BJ, et al. MYC and metabolism on the path to cancer. Semin Cell Dev Biol. 2015;43:11–21.
- Zhang Z, Fu C, Xu Q, et al. Long non-coding RNA CASC7 inhibits the proliferation and migration of colon cancer cells via inhibiting microRNA-21. Biomedicine & Pharmacotherapy = Biomedecine & Pharmacotherapie. 2017;95:1644–1653.
- Zuo W, Zhang W, Xu F, et al. Long non-coding RNA LINC00485 acts as a microRNA-195 sponge to regulate the chemotherapy sensitivity of lung adenocarcinoma cells to cisplatin by regulating CHEK1. Cancer Cell Int. 2019;19(1):1–16.
- Weiming Wu YZ, Gao E, Yang L, et al. LncRNA DLEU2 accelerates the tumorigenesis and invasion of non-small cell lung cancer by sponging miR-30a-5p. J Cell Mol Med. 2020;24(1):441–450.
- Yujia Pan ST, Cui R, Fan J, et al. Xiaoguang Yu long non-coding MALAT1 functions as a competing endogenous RNA to regulate vimentin expression by sponging miR-30a-5p in hepatocellular carcinoma. Cell Physiol Biochem. 2018;50(1):108–120.
- Zhang Y, Yan J, Pan X. miR-141-3p affects apoptosis and migration of endometrial stromal cells by targeting KLF-12. Pflügers Archiv Euro J Phys. 2019;471(10):1055–1063.
- Cao Q, Liu F, Ji K, et al. MicroRNA-381 inhibits the metastasis of gastric cancer by targeting TMEM16A expression. J Exp Clin Cancer Res. 2017;36(1):29.