Abstract
Physicians and the general public are increasingly using web-based tools to find answers to medical questions. The field of rare diseases is especially challenging and important as shown by the long delay and many mistakes associated with diagnoses. In this paper we review recent initiatives on the use of web search, social media and data mining in data repositories for medical diagnosis. We compare the retrieval accuracy on 56 rare disease cases with known diagnosis for the web search tools google.com, pubmed.gov, omim.org and our own search tool findzebra.com. We give a detailed description of IBM's Watson system and make a rough comparison between findzebra.com and Watson on subsets of the Doctor's dilemma dataset. The recall@10 and recall@20 (fraction of cases where the correct result appears in top 10 and top 20) for the 56 cases are found to be be 29%, 16%, 27% and 59% and 32%, 18%, 34% and 64%, respectively. Thus, FindZebra has a significantly (p < 0.01) higher recall than the other 3 search engines. When tested under the same conditions, Watson and FindZebra showed similar recall@10 accuracy. However, the tests were performed on different subsets of Doctors dilemma questions. Advances in technology and access to high quality data have opened new possibilities for aiding the diagnostic process. Specialized search engines, data mining tools and social media are some of the areas that hold promise.
Introduction
Many diseases are so rare that a general physician is unlikely to see a single case in their whole career.Citation1 Furthermore, the symptoms of rare diseases are often atypical and can point in many different directions. As a result, the correct diagnosis is often delayed for several years.Citation1 As a supplement to existing flowcharts for various rare diseases, a web tool can suggest possible diagnoses to physicians in an easily accessible way, diagnoses which might not be on the flowcharts since no flow chart is able to cover all the 5000+ rare diseases in existence.Citation1 Given a list of possible diagnoses, it is easier to request specific biochemical or genetic tests to confirm or refute the diagnosis than it would be to request the tests based on atypical symptoms alone. In the latter case, the requested tests tend to be common laboratory tests which might not uncover a specific rare disease, thereby contributing to the delay of the correct diagnosis.
The Cause of Diagnostic Errors
Mark Graber and co-workers have in a series of papers identified a number of causes for diagnostic errors.Citation2,3,Citation4 They observed that lack of knowledge is rarely the cause of cognitive errors in medicine but rather involve defective synthesis of the available information. Graber et al classify the 3 most common errors as:
Context errors. The diagnostic possibilities are too restrictive, for example gastrointestinal causes are not considered for chest pain symptoms.
Availability errors. A more likely (common) or a more familiar diagnosis is preferred.
Premature closure. Once a plausible diagnosis is identified alternatives are no longer considered.
Arguably many of these errors occur because the physician has very limited time to consider each case either alone or with peers with complementary expertise.Citation5 Web-based tools thus appear as an obvious candidate to confront these types of errors because they offer fast access to information and potentially fast communication with peers. In the remainder of the paper we will describe recent initiatives within web search, data mining (with special focus on IBM's recent Watson system) and social media methodology for this. We compare the different tools against each other on 2 benchmark data sets with paired query (mainly list of symptoms) and a known diagnosis. The sections on search and social media are kept relatively brief because search has been discussed extensively by us elsewhereCitation6,7 and because the results of social media initiatives are still hard to quantify. We conclude the paper with a discussion of how these tools may affect the different types of errors.
Web Search for Diagnosis
Physicians use the web as an important resource for medical information.Citation8,9 Google search and PubMed are arguably the most popular interfaces to the web for physicians although specialized resources are also widely used. Google indexes (collects, parses and stores) more public web data than anyone else and PubMed hosts the largest database of medical abstracts in the world.
In order to compare these different web search engines, it is necessary define a suitable performance metric. When a search engine is used as an aid for compiling a differential diagnosis, we are not just interested in the most likely disease, but rather in a list of the most likely diseases. A reasonable performance metric could therefore be the probability of having the correct diagnosis in an automatically generated differential diagnosis consisting of e.g. Ten candidate diseases. This number is called recall@10, and is how frequent the correct diagnosis appear among the first 10 search results. In this paper we therefore use recall@10 and recall@20 to compare the performance of the different search engines, both general purpose (Google search), general medical literature search (PubMed) and specialized to rare diseases (FindZebra and omim.org).
Performance of Web Search Tools
In our studyCitation6 from 2013, we showed that the ranking algorithm used in the search engine has a big influence on the quality of the results returned. In that study we queried Google, PubMed and the previous version of FindZebra with lists of symptoms for a collection of 56 rare disease cases and compared the returned results with the known diagnosis. In November 2014 we performed the same test again for the current version of PubMed and FindZebra. Since a majority of the articles indexed by FindZebra are Omim articles, we also performed the test for Omim.org. Since the publication of our previous study Google has indexed both the article (containing all question/answer pairs) and all the case studies used in the test. This means that we would need a new set of test questions in order to obtain a fair estimate of the current recall performance of Google. Instead of creating a new test set we have used the estimated performance for Google from 2013 as a proxy for the current performance. The results of the test can be seen in . Google, PubMed and Omim had a success rate of a one-third or less whereas the current version of FindZebra was able to retrieve the correct diagnosis among the top 20 returned results in about 2-third of the 56 cases. One could argue that the superior performance of FindZebra over Google is be due to the fact that Google has to consider a much larger search space. However, in Google advanced search (found in the Settings menu in Google Search) it is possible to specify the web domains used for indexing, and using the same domains as indexed by FindZebra did not lead to better results in.Citation6 This has led us to the conclusion that specialized search has a definite role to play in domains such as rare disease diagnosis. The 56 questions and the results for each of the search engines can be found here: http://ec2–54–148–37-23.us-west-2.compute.amazonaws.com:8080/QueriesAndResultsFinalForWeb.xls.
Figure 1. Performance of different web based search tools on a query collection consisting of 56 queries from Ref. 6.
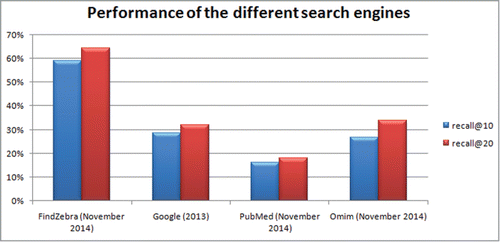
The ranking algorithm used in Google search is highly specialized with more than 200 parameters optimized by continuous A/B-testing on web users. A/B-testing involves dividing a fraction of the web users into a test group and a control group and then perform a statistical analysis in order to select the best possible parameters. This means that Google search will become better and better on average at answering the queries it receives. But there is no guarantee that the queries in specialized domains with little search volume will see an increase in performance. Furthermore, Google has to index huge amounts of data and the results of a search query must be available within milliseconds. This huge volume of data puts a computational constraint on the complexity of the ranking algorithm. A specialized search engine, on the other hand, typically only has to deal with a relatively small document collection (FindZebra, for example, is based on a document collection of only 35.000 documents). Therefore, a specialized search engine typically has the option of using practically every tool available within the fields of machine learning and information retrieval. Until recently, however, the research in FindZebra has been focused on how to include meta-information in the articles rather than finding an optimal document model for ranking. FindZebra currently uses a simple yet highly effective algorithm that finds and ranks documents by comparing word counts in the query string to words counts in an expanded document corpus. Another advantage that a specialized search engine typically has compared to a general search engine is that it can more easily incorporate the use of meta-information. For example, Find-Zebra uses information such as symptoms, symptom equivalence or the fact that 2 articles describe the same disease.This type of information has the potential to substantially improve the search results and has already been used with success in several other search applications. Examples of such applications are the Phenomizer project for hereditary diseases (compbio.charite.de/phenomizer),Citation10,11,Citation12 SimulConsult (simulconsult.com), WebMD (symptoms.webmd.com), POSSUM (www.possum.net.au) and London Medical Databases (www.lmdatabases.com/). Phenomizer is a tool that uses the Human Phenotype Ontology (HPO) to correlate phenotypic abnormalities with genetic disorders. WebMD's symptom checker uses symptom information in order to compile a differential diagnosis. SimulConsult specializes in neurology and genetics, and has a feature where a medical professional can enter symptoms after which the system will give a differential diagnosis based on a probabilistic analysis. London Dysmorphology Database is a database on rare dysmorphic syndromes that provides information on genetic syndromes. It also has a large database of photographic information, and includes the possibility to register undiagnosed or unreported cases. POSSUM is a dysmorphology database that contains textual and photographic information on more than 3000 malformations, metabolic, teratogenic, chromosomal and skeletal syndromes. An overview of these tools are given in . In our performance comparison, summarised in , we have limited ourselves to testing the tools that has a web search interface because this testing requires no additional medical knowledge and can be performed fast.
Table 1. Overview of the different diagnostic tools mentioned in the article
That non-Google ranking algorithms do not necessarily work well is underlined by the poor performance of PubMed. This probably simply reflects that the search functionality in PubMed is designed for retrieving information for the more informed case of knowing for example the author name or keywords. Therefore it is a poor choice of tool for compiling a differential diagnosis. The main data source of FindZebra is omim.org (Online Mendelian Inheritance in Man). Omim is an online database specializing in human genes and genetic phenotypes and contains information on all Mendelian disorders and over 12.000 genes. 22.000 of the the 35.000 documents indexed by FindZebra are based on articles from OMIM. Omim.org offers a Google like interface to their articles (i.e. a simple search field) and focuses exclusively on genetic diseases. Since most rare disease queries are on genetic diseases, we would expect that the performance of omim.org is not much worse than FindZebra. Our previous results, however, show that omim.org has a diffculty ranking the correct results high. Looking in details (see http://ec2–54–148–37–23.us-west-2.compute.amazonaws.com:8080/QueriesAndResultsFinalForWeb.xls), we see that even in the cases where the FindZebra hit is an OMIM article, the same article is ranked lower on omim.org. This suggests that there is potential for optimizing the ranking algorithm in omim.org or that getting the full potential out of omim.org requires refinements of the search strings.
Non-Professional use of Web Tool for Diagnosis
Physicians are not the only ones using the web for diagnosis. A recent studyCitation13 showed that 35% of all American adults have used the internet for diagnosis. Of these “online diagnosers,” 46% went to a physician with their findings and a surprising 41% of these got a confirmation of their diagnosis. In a previous study it was shown that the diagnostic success rate for a medical professional (on a collection of 26 test cases) was approximately 58% (when using Google as the only tool).Citation9 Although the set-up is not same in the 2 studies this is indicative that the success rate might not be that different between professionals and non-professionals. It is especially interesting how the web was used for diagnosis: 77% of the online diagnosers began their exploration using a search engine such as Google, Bing, or Yahoo. Another 13% began at a website specializing in health information, such as WebMD and the remaining 10% started the exploration in other places such as Wikipedia or Facebook. Considering the results summarized in , the success rate of online diagnosers might improve substantially if more specialized tools were utilized instead of a generalized search engine. It should be noted, however, that there are mixed opinionsCitation14,15,Citation16 whether these tools in the hands of non-professionals will provide an overall benefit for diagnosis. One of the major concerns that has been voiced is that the tools might be used by a non-professional as a diagnostic procedure where search results are interpreted as diagnostic conclusions. This might lead to unnecessary high anxiety levels or might affect the user's decision on whether or not to consult a physician. But like it or not, non-professional “diagnosis” is a phenomenon that is likely to grow in the years to come.
Data-Mining for Diagnosis
The use of computer systems as an aid in medical diagnosis is nothing new. The earliest clinical diagnosis decision support systems (CDDSS) date back to the 1970s. All of these early systems used knowledge in a structured form (e.g., manually constructed knowledge bases or relational databases). Two examples of such early (1970s) systems are the Internist-I systemCitation17 and the MYCIN system.Citation18 The Internist-I system used a database of diseases, symptoms and sensitivities (the fraction of patients with a disease, who have the symptom). The MYCIN system was a rule-based expert system designed to diagnose infectious blood diseases based on a long series of yes/no or simple textual questions. During the 1990s systems with more sophisticated probabilistic reasoning, such as e.g. the Iliad system,Citation19 began to emerge. These systems were, however, still based on structured knowledge. One of the major weaknesses of systems relying on structured knowledge alone is that experts are needed in order to update or expand the knowledge bases. This means that such systems can be diffcult to keep up to date, especially in a field like medicine, where the amount of information found in textbooks, case studies, research articles and so forth doubles approximately every 5 y.Citation20 As a consequence of this exponential increase in information, systems for medical diagnosis relying on structured information alone are able to use only an increasingly small fraction of all the available information. This, of course, does not mean that it is impossible to construct useful tools using structured information alone. OMIM, Possum and London Dysmorphology Database are good examples of very useful tools relying almost purely on structured information. Even though these tools are kept up to date within their domain, there is an increasing amount of (unstructured) information that is not used, and perhaps these tools could be improved by exploiting this extra information. Our search engine FindZebra initially started at the other end of the spectrum relying purely on unstructured information, but it has since then evolved to use both structured and unstructured information. It is our clear impression that using both types of information sources at the same time creates a huge synergetic effect.
Since the late 1990s, several attempts have been made at building CDDSS systems based at least partly on data mining instead of handwritten rules.Citation6,21, Citation22 One of the latest attempts was based on an open-domain question answering system called Watson,Citation21 developed by IBM over a 4-year period from 2007 to 2011. The initial purpose of Watson was to see if it was possible to build a computer system able to compete against the best human players in the popular game of Jeopardy!. Jeopardy! is a quiz game where 3 contestants compete against each other at answering a series of questions posed by a quiz master. The first contestant to hit a buzzer after hearing a question gets the right to answer that question. Points are earned by answering correctly and lost by answering incorrectly. The winner of the game is the contestant with the most points at the end. The project became a huge success, and Watson played many games against celebrated Jeopardy! champions and was in fact able to beat the 2 greatest players of all time, Ken Jennings and Brad Rutter in a televised match in February 2011.The success of Watson prompted IBM to put the question-answering technology underlying Watson (called DeepQA) to other uses. Computationally, answering a question in a game of Jeopardy! is very similar to the task of compiling a differential diagnosis: Based on a natural language query (e.g., “Jewish boy age 16 suffering from monthly seizures, sleep deficiency, aggressive and irritable when woken, highly increased sexual appetite and hunger”), the system has to be able to quickly figure out what the question is about, and search a wide variety of heterogeneous, unstructured knowledge sources such as medical textbooks, databases and articles for an answer. Based on this search, the system then has to construct a list of candidate answers and evaluate how likely each of the candidate answers are. The similarity in question type between Jeopardy! and the task of compiling a differential diagnosis meant that IBM was able to turn Watson into a state-of-the-art CDDSSCitation21 relatively fast.
Design of Watson
It took 2-dozen researchers approximately 4 y to develop the DeepQA technology. The technology became extremely complex and consists of more than 500.000 lines of code.Citation23 The overall design is simple, though, and can roughly be broken down into 5 stages:
Question and topic analysis. In this stage, natural language processing is used to discern the nature of the question in order to construct a structured search query.
Hypotheses generation. The search query is used to generate a list of potential hypotheses, e.g. “The disease is Gilbert's syndrome”
Evidence and hypotheses scoring. Each hypothesis is scored along different evidence dimensions (e.g., symptoms, gender, demographics), and the evidence scores are combined in order to score the hypothesis
Ranking. The hypotheses are combined and a ranked list of hypotheses is generated based on the hypothesis scores.
Answer and confidence. In the last stage a ranked list of answers is generated and a level of confidence is calculated for each hypothesis.
Performance of Watson
In order to estimate the recall@10 performance, the developers of medical Watson used a set of 5000 medical questions from the American College of Physicians (ACP). The questions have been used in a Jeopardy!-like competition, called Doctor's Dilemma, that medical interns and residents participate in once a year. 1322 of the questions were used for training and a separate set of 188 questions were used for testing. The results are divided into 4 stages of domain adaption with the following recall@10 performance:
Core is the original Watson Jeopardy! system, 40%.
Core+content is as in 1), but with medical content adaption, 54%.
Core+content+train is as in 2), but trained on Doctors dilemma questions instead of normal Jeopardy! questions, 74%.
Core+content+train+func is as in 3) but with some further tweaks to the Watson engine in order to optimize it for medical questions and answers, 77%. We tried obtain this test set from the IBM Watson team in order to make a comparison between FindZebra and Watson. Unfortunately, we had no luck obtaining these questions. Instead we obtained a set of 3000 Doctors's dilemma questions directly from ACP and performed the same kind of test as the one performed by the developers of Watson (but on a different subset of the Doctor's dilemma questions). First we removed questions not related to diagnosis (e.g., questions in the category ethics) and questions requiring inspection of images. Then we removed all questions where the answer was not a disease (i.e., we removed questions where the answer text did not refer to a disease found on either Wikipedia, Orphanet, Omim, Nord, Gard or Genetics Home Reference). Synonyms of disease names were resolved using the UMLS database. This gave a total of 546 questions that we ran through FindZebra and found a recall©10 of 53%.
The test was performed using a highly conservative computer program: only when a 100% match was found we reported it as a success. For example, in the question syndrome name for hypothalamic hypogonadism and anosmia, the correct answer is Kallmann's syndrome. However, Idiopathic hypogonadotropic hypogonadism with anosmia has been known under the name Kallmanm's syndrome, and 18 OMIM articles about this disease figure among the first 20 search results. Also, Kallmann's syndrome with spastic paraplegia is found as result 3. The answer is nevertheless considered wrong because an exact textual match was not found. This conservative approach thus only gives a lower bound for the recall@10 performance, but it avoids having to deal with subjective opinions of which answers can be considered as correct.
A table showing the recall@20 and recall@10 performance for each of the major categories in the Doctor's dilemma dataset can be found in . Even though it might seem that FindZebra performs better on some question categories than others, there is no statistical evidence to support this claim (we have tested the hypothesis that the recall@n was independent of question category by using a Pearson χ2 test, resulting in a p-value of 0.20 for recall@20 and a p-value of 0.53 for recall@10).
Figure 2. Performance of FindZebra on each of the major disease categories in the Doctor's dilemma dataset.
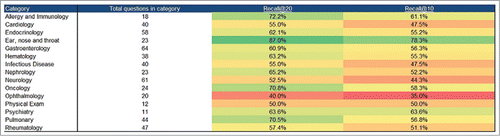
Because we do not have access to the actual test questions used by the Watson team, it is only possible to make a very rough comparison between Watson and FindZebra. When Watson and FindZebra are given the same prerequisites (i.e. no training on the Doctor's dilemma questions), they seem to perform equally well. However, it should be noted that Watson has a much better performance if allowed to train on Doctors dilemma questions. But since search engine queries do not show anything resembling the structure found in a typical Doctors dilemma question, it remains to be seen how much of this performance gain can actually be transferred to a real setting.
Results for each of the 546 questions can be found at http://ec2–54–148–37–23.us-west-2.compute.amazonaws.com:8080/QueriesAndResultsFinalForWeb.xls, and the version of FindZebra used to conduct the tests reported in this paper (including the test on the 56 cases mentioned previously) can be found here: http://ec2-54-148-37-23.us-west-2.compute.amazonaws.com:8080. This version is identical to findzebra.com running on October 2014. Note that the rank found on the web page is often a couple of ranks better than reported in the Excel file due to clustering of documents describing the same disease.
Use of CDDSS System in Practice
Evidence suggests that CDDSS systems have the potential to reduce diagnostic errors and improve quality of care.Citation24,25,Citation26,27 Furthermore, studies have shown that users of the systems are actually satisfied with the results they get from the support tools.Citation28 Despite of this, many physicians are reluctant to use the systems in practice on a day-to-day basis,Citation28 even when the support tools are well integrated into their workflow.Citation29,30, Citation31,32, Citation33 In an older review article on the use of the first generations of CDDSS systems,Citation34 Miller concludes that even though niche systems for specific areas are perceived as very useful, the perceived usefulness of the more general systems is less certain, despite the fact that these systems can often suggest diagnoses that even expert physicians have not considered. One of the key assumptions behind FindZebra is based on these findings, i.e., that a medical professional will typically not use a CDDSS system on a daily basis, but only when facing a diffcult problem. So instead of building a universal question answering system being able to diagnose all kinds of diseases, FindZebra is designed to be a fast, easy to use system, optimized for the types of queries that the system is expected to encounter in practice, namely queries regarding diseases not encountered often.
Social Media for Diagnosis – Collective Intelligence and Authority of Experts
It is hard to come up with quantitative statements about the effect of social media on medical diagnosis because evidence has not so far been collected systematically and it is still early days with the organization and tools taking shape. But it is clear that the potential is hugeCitation35 because the web allows for quick knowledge sharing and peer networks with the required knowledge base can be formed quickly for the case in question. There is already anecdotal evidence of diagnosisCitation36 and a commercial company crowdmed.com has made a business case of combining prediction markets with collective intelligence to increase the accuracy of diagnosis of unsolved cases. Examples of social media dedicated to health care professionals are doximity.com, sermo.com and vis.dk. If these initiatives mature to become an integral part of how physicians work with diagnosis then we can expect to see this affect diagnosis of rare diseases in the coming years.
Conclusion
Diagnostic errors are primarily caused by defective synthesis of the available information. Clinical diagnosis decision support systems (CDDSSs) have the potential of reducing such types of error, but many physicians are nevertheless reluctant to use them on a day to day basis, regardless of how well they are integrated into the workflow. This means that a medical professional is only likely to use a CDDSS when he or she is facing a particularly diffcult problem, such as a rare disease. Our tool FindZebra is an example of a search engine designed to solve exactly this kind of problem.
We have already seen a widespread use of the internet to search for health related information, both among professionals and non-professionals. There are mixed opinions whether these tools in the hands of non-professionals have a positive effect on diagnosis. But non-professional “diagnosis” is a phenomenon that is likely to grow in the years to come, and considering that millions of people (just in the United States) consult their physician based on a self-diagnosis each year, even a modest increase in diagnostic performance can have a huge impact, and save time, money and lives. The overall performance of tools such as Google Search has meant that the diagnostic capabilities of non-professionals have reached a relatively high level. In this and in previous articles we have shown that more specialized tools might have the capabilities to increase this diagnostic performance even further, for professionals and non-professionals alike.
For the busy medical professional speed is clearly important for practical usefulness. This argues for simple user-interfaces that require no training or use of specialized medical vocabulary. Robust machine learning based interpretation of free text queries is therefore a major challenge for the developer of such a system. Combining ideas from free text search and menu/fixed vocabulary based CDDSSs may in the end provide the most accurate and useful systems.
Many initiatives based upon information technology and new means of knowledge sharing are appearing in these years, and the rapid growth of websites such as doximity.com, sermo.com and others indicate that it is an area with a lot of potential. However, the full effect of these social media sites on the diagnostic process, remains to be seen.
Disclosure of Potential Conflicts of Interest
The authors have made FindZebra, but apart from that have no conflicts of interest.
Authors' Contributions
DS, HLJ and OW conceived the study. DS and OW were the main responsible for writing and all authors assessed and approved the final version.
Acknowledgments
The authors wish to thank the Lundbeck Foundation for supporting the FindZebra research project and ACP for use of the Doctors dilemma questions.
References
- The UK Strategy for Rare Diseases. https://www.gov.uk/government/uploads/system/uploads/attachment_data/file/260562/UK_Strategy_for_Rare_Diseases.pdf
- Graber ML, Franklin N, Gordon R. Diagnostic error in internal medicine. Arch Intern Med 2005; 165(13):1493-9; PMID:16009864
- Berner ES, Graber ML. Overconfidence as a cause of diagnostic error in medicine. Am J Med 2008; 121(5):2-23; PMID:18187063
- Graber M, Gordon R, Franklin N. Reducing diagnostic errors in medicine: what's the goal? Acad Med 2002; 77(10):981-92; PMID:12377672
- Singh H, Giardina TD, Meyer AN, Forjuoh SN, Reis MD, Thomas EJ. Types and origins of diagnostic errors in primary care settings. JAMA Intern Med 2013; 173(6):418-25; PMID:23440149
- Dragusin R, Petcu P, Lioma C, Larsen B, Jørgensen HL, Cox IJ, Hansen LK, Ingwersen P, Winther O. Findzebra: A search engine for rare diseases. Int J Med Inform 2013; 82(6):528-38; PMID:23462700
- Radu Dragusin PP.e.a. Specialised tools are needed when searching the web for rare disease diagnoses. Rare Dis 2013; 1(2):528–38.
- Cartright M-A, White RW, Horvitz E. Intentions and attention in exploratory health search. In: Proceedings of the 34th International ACM SIGIR Conference on Research and Development in Information Retrieval, 2011; pp. 65-74. ACM
- Tang H, Ng JHK. Googling for a diagnosis—use of google as a diagnostic aid: internet based study. Bmj 2006; 333(7579):1143-5; PMID:17098763
- Köhler S, Schulz MH, Krawitz P, Bauer S, Dolken S, Ott CE, Mundlos C, Horn D, Mundlos S, Robinson PN. Clinical diagnostics in human genetics with semantic similarity searches in ontologies. Am J Hum Genet 2009; 85(4):457-64.
- Köhler S, Doelken SC, Mungall CJ, Bauer S, Firth HV, Bailleul-Forestier I, Black GC, Brown DL, Brudno M, Campbell J, et al. The human phenotype ontology project: linking molecular biology and disease through phenotype data. Nucleic Acids Res 2014; 42(D1):966-74.
- The Phenomizer Project. http://compbio.charite.de/phenomizer
- Health Online 2013. http://www.pewinternet.org/∼/media/Files/Reports/PIPHealthOnline.pdf
- Jutel A. Self-diagnosis: a discursive systematic review of the medical literature. J Participat Med 2010; 15:8.
- Ruiz ME. Risks of self-medication practices. Curr Drug Saf 2010; 5(4):315-23; PMID:20297863
- White RW, Horvitz E. Experiences with web search on medical concerns and self diagnosis. In: AMIA Annual Symposium Proceedings, 2009; vol. 2009, p. 696. American Medical Informatics Association.
- Myers J. The background of internist i and qmr. In: Proceedings of ACM Conference on History of Medical Informatics, 1987; pp. 195-197. ACM
- Buchanan BG, Shortliffe EH. Rule Based Expert Systems: The Mycin Experiments of the Stanford Heuristic Programming Project (The Addison-Wesley Series in Artificial Intelligence). Addison-Wesley Longman Publishing Co., Inc., Boston 1984.
- HR Warner, P.H.e.a. Iliad as an expert consultant to teach differential diagnosis. Proc Annu Symp Comput Appl Med Care 1988; 371-6.
- Sood A, Ghosh A, et al. Literature search using pubmed: an essential tool for practicing evidence-based medicine. J Assoc Physicians India 2006; 54(R):303
- Ferrucci D, Levas A, Bagchi S, Gondek D, Mueller ET. Watson: Beyond jeopardy! Artificial Intelligence 2013; 199:93-105.
- Ramnarayan P, Tomlinson A, Kulkarni G, Rao A, Britto J, et al. A novel diagnostic aid (isabel): development and preliminary evaluation of clinical performance. Medinfo 2004; 11(2):1091-5.
- Ferrucci D, Brown E, Chu-Carroll J, Fan J, Gondek D, Kalyanpur AA, Lally A, Murdock JW, Nyberg E, Prager J, et al. Building watson: An overview of the deepqa project. AI Magazine 2010; 31(3):59-79.
- Delaney BC. Potential for improving patient safety by computerized decision support systems. Family Practice 2008; 25(3):137-8; PMID:18583355
- Kawamoto K, Houlihan CA, Balas EA, Lobach DF. Improving clinical practice using clinical decision support systems: a systematic review of trials to identify features critical to success. Bmj 2005; 330(7494):765; PMID:15767266
- Sim I, Gorman P, Greenes RA, Haynes RB, Kaplan B, Lehmann H, Tang PC. Clinical decision support systems for the practice of evidence-based medicine. J Am Med Inform Assoc 2001; 8(6):527-34; PMID:11687560
- Classen DC. Clinical decision support systems to improve clinical practice and quality of care. Jama 1998; 280(15):1360-1; PMID:9794318
- Bauer BA, Lee M, Bergstrom L, Wahner-Roedler DL, Bundrick J, Litin S, Hoffer E, Kim RJ, Famiglietti K, Barnett GO, et al. Internal medicine resident satisfaction with a diagnostic decision support system (dxplain) introduced on a teaching hospital service. In: Proceedings of the AMIA Symposium, 2002; p. 31. American Medical Informatics Association.
- Eccles M, McColl E, Steen N, Rousseau N, Grimshaw J, Parkin D, Purves I. Effect of computerised evidence based guidelines on management of asthma and angina in adults in primary care: cluster randomised controlled trial. Bmj 2002; 325(7370):941; PMID:12399345
- Smith WR. Evidence for the effectiveness of techniques to change physician behavior. Chest J 2000; 118(2 suppl):8-17.
- Militello L, Patterson ES, Tripp-Reimer T, Asch SM, Fung CH, Glassman P, Anders S, Doebbeling B. Clinical reminders: why don't they use them? In: Proceedings of the Human Factors and Ergonomics Society Annual Meeting, 2004; vol. 48, pp. 1651-5. SAGE Publications.
- Patterson ES, Doebbeling BN, Fung CH, Militello L, Anders S, Asch SM. Identifying barriers to the effective use of clinical reminders: bootstrapping multiple methods. J Biomed Inform 2005; 38(3):189-99; PMID:15896692
- Berner ES, Maisiak RS, Heudebert GR, Young KR Jr. Clinician performance and prominence of diagnoses displayed by a clinical diagnostic decision support system. In: AMIA Annual Symposium Proceedings, 2003; vol. 2003, p. 76. American Medical Informatics Association.
- Miller RA. Medical diagnostic decision support systems—past, present, and future a threaded bibliography and brief commentary. J Am Med Inform Assoc 1994; 1(1):8-27; PMID:7719792
- Digital Trends. http://www.digitaltrends.com/social-media/the-internet-and-healthcare/#!FeBUb
- The Medical Futurist. http://themedicalfuturist.com/
