ABSTRACT
Formerly a commensal organism of the mucosal surfaces of most healthy individuals, Candida albicans is an opportunistic pathogen that causes infections ranging from superficial to the more life-threatening disseminated infections, especially in the ever-growing population of vulnerable patients in the hospital setting. In these situations, the fungus takes advantage of its host following a disturbance in the host defense system and/or the mucosal microbiota. Overwhelming evidence suggests that the gastrointestinal tract is the main source of disseminated C. albicans infections. Major risk factors for disseminated candidiasis include damage to the mucosal intestinal barrier, immune dysfunction, and dysbiosis of the resident microbiota. A better understanding of C. albicans’ interaction with the intestinal epithelial barrier will be useful for designing future therapies to avoid systemic candidiasis. In this review, we provide an overview of the current knowledge regarding the mechanisms of pathogenicity that allow the fungus to reach and translocate the gut barrier.
Graphical abstract
Invasion of C. albicans through the intestinal epithelial barrier.
Introduction
The importance of the intestinal mucosa in the interaction of candida sp. with its host
Formerly described as a harmless commensal of the human mucosal surfaces, C. albicans is also an opportunistic fungus responsible for candidiasis that can range from superficial to invasive and life-threatening infections in debilitated patients, mostly occurring in the hospital setting. Thus, systemic candidiasis is associated with a high crude mortality ranging from 20 to 49%,1–Citation4 and almost all of the organs can be secondarily infected following the hematogenous dissemination of the fungus. From a pathophysiological point of view, invasive candidiasis has various stages where C. albicans has to: (i) enter the bloodstream, (ii) survive in the blood reservoir and (iii) escape from the bloodstream in order to establish deep-seated infections.Citation5 In the first stages, a clear association between the presence of C. albicans on mucosal surfaces such as the mouth or the vagina and its translocation into the blood has not been established yet. However, various observations strongly suggest that invasive candidiasis are mostly of endogenous origin with the gastrointestinal (GI) tract being the main portal of entry into the bloodstream.Citation5,Citation6 Indeed, molecular typing studies have pointed to gut mycobiota as the main origin of disseminating C. albicans isolates in the blood.Citation7–Citation11 On the host side, immune dysfunction such as neutropenia, damage to the mucosal barrier and dysbiosis of the bacterial microbiota have been identified as major risk factors for invasive candidiasis.Citation9–Citation13 With in vivo investigations, Koh demonstrated a strong association between (i) dysbiosis of the bacterial microbiota, (ii) mechanical alteration of the gut barrier function, (iii) a decrease in the function of neutrophils (PMNs) and (iv) major risks for systemic candidiasis.Citation14
In addition to the role of C. albicans as a commensal or a pathogen and with the advance of Next Generation Sequencing (NGS) for microbiome analysis, recent reports suggest that changes in the fungal microbiota of the gut are linked to the pathogenesis of multiple gastrointestinal (e.g. inflammatory bowel diseases (IBD))Citation15,Citation16 or extra-intestinal disorders (e.g. allergic airway diseases, neurologic disorders, cancer),Citation17 in which, an increase of colonization by fungal species, including C. albicans, was reported compared to healthy subjects.Citation15,Citation18,Citation19
All in all, C. albicans is a versatile yeast able to interact with the gut content of its human host both as a commensal, a pathogen or a major colonizer of the GI tract. This transition from commensalism to pathogenicity involves both host-related factors (e.g. environment of the digestive tract, the gut mucosa, genetics and health status), the microbiota (i.e. dysbiosis), and C. albicans itself. In this context, the purpose of this review is to discuss the interaction between C. albicans and the intestinal epithelial barrier.
Interaction of microorganisms with the intestinal mucosa
Intestinal factors influencing the interaction of micro-organisms with the gut mucosa
Human intestinal microbiota and mycobiota
The human intestinal microbiota is a complex ecosystem, largely dominated by bacteria in terms of number (i.e. around Citation1014 bacterial cells) and species diversity (500 to 1000 species, mainly anaerobes). However, the global gut microbiota also comprises archaea, viruses and eukariota such as parasites and fungi.Citation20 Whereas the precise contribution of gut-associated archaea, viruses and parasites in human physiology remains to be specified, research over the last few decades has given us a more comprehensive view of the contribution of bacteria and fungi to gut-associated microbiota (GM) and the potential impact on human health.Citation21,Citation22 In healthy adults, the composition of the GM is unique to each individual.Citation23 It differs along the digestive tractCitation24 and varies according to environmental and lifestyle stimuli.Citation25
Although specialized in the digestion of nutrients, the GM contributes to the host’s gut homeostasis by (i) modulating the host’s energy metabolism,Citation26 (ii) sustaining gut integrity,Citation26 (iii) preventing gut colonization by pathogensCitation22,Citation26 and (iv) promoting the development and maturation of the gut-associated immune system.Citation26,Citation27 Consequently, an imbalanced GM composition can result in the dysregulation of host homeostasis contributing to the onset or progression of many diseases. Indeed, dysbiosis of the GM is associated with the pathogenesis of intestinal (e.g. IBD, irritable bowel syndrome, coeliac disease) but also extra-intestinal diseases (e.g. allergy, asthma, disruption of the immune system, metabolic diseases, cardiovascular diseases, and neurologic diseases).Citation28–Citation32
Among microbiota components, fungi account for about a quarter of the species diversity but for less than 0.1% of the GM in quantity.Citation7 The human mycobiota comprises 158 genera and 390 species among which only 221 are found in the digestive tract.Citation7 However, despite growing interest in the gut mycobiota, several issues remain to be addressed regarding its precise composition from one individual to another. The most commonly-detected fungi in the human GM (in order of frequency) are the genera Candida, Malassezia, Aspergillus, Debaryomyces, Penicillium, Cladosporium, Trichosporon, Galactomyces, Saccharomyces and Cryptococcus (i.e. the percentage of positive samples reported in gut mycobiome studies being 80, 25, 24, 21, 20, 18, 9, 9, 6 and 4% respectively).Citation33
However, the mycobiota appears less stable than the bacterial microbiota and highly subject to changes brought on by environmental factors, particularly diet (e.g. animal-based versus plant-based diet).Citation33
Although, the role of fungi in the human GM remains poorly understood, recent studies have highlighted the role of diverse fungal communities in host physiology. Symbiotic relationships between fungi inhabiting the human GI tract and the host have indeed been reported to be essential for digestion functions.Citation34 Additionally, other findings have underscored the relationship between the gut mycobiota and human diseases. Associations between the mycobiome biodiversity in IBD or in obesity, and gut environment modifications have been reported.Citation15,Citation35 For example, Mar Rodriguez et al. observed differences in the biodiversity of the mycobiome of obese compared to healthy individuals, suggesting that this fungal dysbiosis contributes to changes in the lipid and glucose metabolisms observed in diabetic patients.Citation35 Finally, antifungal or antibiotic treatments can also lead to microbial dysbiosis and so to a disruption in the balance between bacterial and fungal communities, highlighting the interdependence of fungi and bacteria in the gut.Citation17
Intestinal Epithelial Cells (IECs)
The intestinal epithelium (IE) consists of a monolayer of cells covering a surface of ~400 m2, organized into crypts and villi. Pluripotent intestinal epithelial stem cells residing in the bottom of the crypts ensure a continuous renewal of the IE, where the local environment drives their proliferation and functional differentiation. IE encompass differentiated cell types grouped under the term intestinal epithelial cells (IECs) (i.e. enterocytes, enteroendocrine cells, goblet cells, Paneth cells, Microfold (M) cells and Tuft cells) with specialized functions (i.e. absorption, hormone secretion, mucus secretion, antimicrobial peptide (AMPs) production, antigen sampling and taste-chemosensory responses respectively) in addition to Cup cells whose function remains to be specified.Citation36 Additionally, IECs exert immunoregulatory functions that are critical for the development, maturation and homeostasis of the immune system all along the gut mucosa. Finally, thanks to this complex system of cells and functions, IECs form a physical and biochemical barrier capable of segregating microorganisms from the host with the ability to discriminate commensals from pathogenic microorganisms. IECs respond differentially to commensal or pathogenic microbial signals that consequently reinforce or weaken their barrier function. In parallel, the mucosal biochemical and immune systems orchestrate appropriate immune responses to these microbial signals that can range from the tolerance of commensal microorganisms to anti-pathogenic responses.Citation37
Physical barrier function
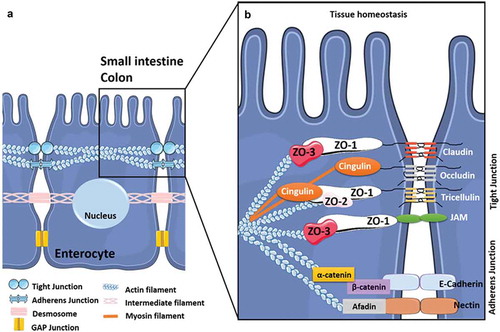
IECs are mostly composed of enterocytes (over 80%) that form a monolayer of differentiated and polarized epithelial cells with an apical side that displays microvilli which form the brush border exposed to the digestive lumen.Citation38 Beyond their role in nutrient and fluid absorption, enterocytes also contribute to the integrity and impermeability of the IE by establishing the intestinal border. They form a physical barrier between the content of the gut lumen and the underlying tissues, limiting the translocation of microbes and their related products.Citation39,Citation40 Thus, the cohesion of the IE relies particularly on junctional intercellular complexes including tight-junctions (TJs) and weak junctions (i.e. adherens-, gap-junctions and desmosomes) (). The composition and functions of these junctional complexes vary by IEC type, especially for TJs,Citation41 but their structural organization has been extensively studied in enterocytes (). Between enterocytes, adherens junctions (AJs) contribute to cell-cell adhesion and intracellular signaling whereas gap-junctions and desmosomes participate in cell-cell communication and adhesion, respectively. Together with AJ, TJs form a seal (i.e. the apical junctional complex) that ensures the architectural cohesion and integrity of the IE. This complex is closely linked to the cytoskeleton that is crucial in maintaining the cohesion and structure of the IECs by extending throughout the cytosol ().Citation42–Citation44 Finally, TJs, the most apically located intercellular junctions, are crucial for the IE’s role of physical barrier since they are targeted by numerous physical and chemical factors,Citation44-Citation51including cytokines, proteases, hormones, neurotransmitters, dietary components, bacterial and fungal toxins, and xenobiotics that can alter TJs and consequently modulate the permeability and integrity of the IE. Commensal bacteria can modulate impermeability and integrity in a homeostatic manner by targeting TJs,Citation47,Citation52–Citation54 whereas enteric pathogens including bacteria, viruses, parasitesCitation55-Citation57 and possibly fungiCitation5 have developed strategies aimed at exploiting TJs to promote their invasiveness into IECs.
Figure 1. The intercellular junctions between enterocytes at the digestive barrier.
(A) Composition and organization of the enterocyte-enterocyte junctions in the intestinal epithelium. Tight junctions (or TJs) are the first intercellular junctions present at the apico-lateral region of enterocytes followed by the adherens junctions (AJs), the desmosomes and finally the GAP junctions at the baso-lateral region. (B) Composition and organization of the TJs and AJs. The TJs and the AJs form circumferential junctions composed by transmembranous proteins, including Claudins, TJ associated Marvel domain containing Occludin, Tricellulin and Marvel D3, and JAMs for TJs and E-cadherin and Nectin for AJs, all connected to the cytoskeleton through various proteins (i.e. Zonula Occludens (ZO1-3) proteins, Cinguline, Catenin and Afadin).
Secretory functions of IECs
IECs also express secretory functions reinforcing the physical barrier role of the epithelial layer and ensuring biochemical functions.
First, enterocytes secrete the glycocalix that mainly contains transmembrane mucin glycoproteins covering the apical dense microvilli surface, which locally protects the enterocyte cell membranes.Citation58 Moreover, the IE is lined with a viscous layer of mucus that is synthesized and extruded at the cell surface by specialized secretory IECs, i.e. the goblet cells,Citation59 that are dispersed throughout the gastro-intestinal tractCitation60 (). In addition to other proteins and lipids, the mucus is predominantly composed of secreted mucins (gel- and non-gel-forming mucins). This class of high molecular weight glycoproteins, strongly O-glycosylated, confer the viscous, gel-like quality to the mucus.Citation58,Citation61,Citation62 MUC2 is the largest gel-forming mucin expressed in the intestinal tract.Citation63 Though its composition and amount vary along the gastrointestinal tract (), this continuous viscous layer lubricates the IE, regulates the transport of molecules from the gut lumen to IECs and forms a physical barrier that (i) protects the IE against acidic and proteolytic environments encountered in the gut lumen and (ii) limits the access of most microorganisms and deleterious substances at the surface of the IE. However, enteric pathogens including eukariota have developed strategies to overcome the mucus layer, enabling them to reach and colonize the epithelial barrier to further cause infectionCitation60 (reviewed inCitation50,Citation64). These include (i) the production of lytic enzymes to disrupt the mucus layerCitation65-Citation70 and/or (ii) the downregulation of mucus production by goblet cells or changes in its composition.Citation71 Additionally, components of the mucus can trigger specific transcriptional responses in enteric pathogens that facilitate their interaction with the epithelial layer and/or promote their virulence.Citation72,Citation73
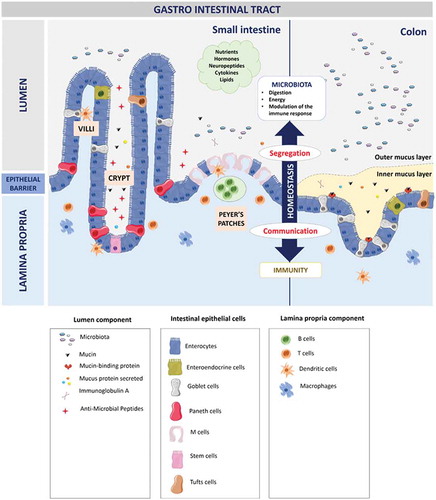
Figure 2. Organization of the gastro-intestinal tract.
The gastro-intestinal tract forms a physical and biochemical barrier capable of segregating microorganisms from the host with the ability to discriminate commensals from pathogenic microorganisms. The physical barrier consists of a monolayer of cells, which includes various intestinal epithelial cell types (IECs) differently organized from the small intestine to the colon. The biochemical barrier consists of a mucus layer whose composition, structure and also properties differ between the small intestine and the colon. In the small intestine, the mucus is composed of a highly dynamic monolayer, not anchored to the surface of the epithelial cells. This monolayer of mucus is permeable to the bacteria. However, the distal peristaltic movements keep the microorganisms away from the surface of the epithelial cells. In the colon, the mucus is organized in two layers: the inner layer and the outer layer. The inner layer is in perpetual renewal (approximately every 1 to 2 hours) in order to remain totally germ-free. This layer is firmly anchored to the epithelial barrier through the interaction between mucins in the mucus and the mucins-binding protein located at the surface of the epithelial cells. Due to its size-exclusion filter function (i.e. exclusion of any element of more than 0.5 μm), the inner layer of mucus is impermeable to microorganisms. Finally, the outer layer is the normal habitat of intestinal commensal microbiota. In addition to the secretion of bioactive molecules such as nutrients, hormones, neuropeptides, cytokines and lipids in the gut lumen, the gut microbiota takes an active part in this permanent remodeling as highlighted in germ-free animals in which a thinner layer of mucus is observed as the result of a decreased number of goblet cells compared with conventional animals.Citation61–Citation63 The outer-most layer of mucus is a reservoir of dense populations of commensal microorganisms whose composition is linked to the existing luminal populations.Citation64 Consequently, normal or altered GM and mycobiota will influence goblet cell function as well as the composition and volume of the mucus layer by mechanisms probably involving both the direct effect of locally released microbial factors and/or the indirect effect of bioactive or immune factors resulting from the host-response to intestinal microbes. Moreover, the mucus layer is also a biochemical barrier thanks to the presence of various secreted antimicrobial factors mostly secreted by the cytoplasmic granule-rich Paneth cells (PCs). These PCs are located at the base of the small intestinal crypt in healthy individuals.Citation65 Various factors, including cholinergic agonists, bacteria and bacterial products (such as lipopolysaccharides and lipoteichoic acid),Citation66,Citation67 lead to the discharge of PC granules from the crypt into the mucus layer, forming a biochemical barrier that is crucial for establishing baseline homeostasis for mucosal and systemic inflammatory response. The PCs mainly secrete a panel of antimicrobial peptides (AMPs), which are released in intestinal mucus layer in the small intestine in humans. The composition of the mucus changes along the GI tract, contributing to the increase of the amount of microorganisms in the digestive microbiota between the small intestine and the colon.
In addition to the being a physical barrier, the mucus layer harbors a biochemical function. It serves as a reservoir for bioactive factors mainly secreted by IECs in order to reinforce the barrier function by maintaining GM-Host homeostasis as well as influencing the composition of the GM. Among these components, antimicrobial factors are essentially secreted by PMNs residing in the lamina propria and by specialized IECs named Paneth Cells (PCs) that reside exclusively at the base of the crypts in the small intestine of healthy individualsCitation74 (). These specialized cells contain granules whose secretion is modulated, among others, by microbes and microbial productsCitation75,Citation76 through the stimulation of specific Pattern Recognition Receptors (PRRs) located at the surface of IECs.Citation27 These granules are mostly enriched in antimicrobial peptides (AMPs) including α- and β-defensins,Citation77,Citation78 lysozyme C,Citation79 secretory group IIA phospholipase A2 (sPLA2),Citation80 Regenerating Islet-Derived 3 Gamma (REG3γ),Citation81 α-1 antitrypsin,Citation82 Angiogenin 4Citation83 and Cathelicidin.Citation84 PCs also release cytokines (IL-17A, TNF-α, IFN-β and IL-1), proteases (Metalloprotease 7), proteins and scaffolding molecules as well as non-granule molecules including Wnt proteins, Epidermal Growth Factor and Notch ligands.Citation82 These non-granule secretions create a specific microenvironment called the “stem cell niche”, that surround the stem cells and is essential for the regeneration and differentiation of specific IECs in the small intestine.Citation82,Citation85
Additional secreted antimicrobial factors are found in the intestinal mucus layers, including secretory Immunoglobulin A (sIgA). sIgA are secreted by IgA+ B cells moving from the germinal center of Peyer’s patches to the lamina propria in intestinal villi.Citation86 They provide an adaptive immune component which is crucial for maintaining gut homeostasis by (i) inhibiting microbial motility and improving the mucus’ ability to trap microorganisms,Citation87 (ii) clearing the microorganisms through peristalsis or mucocilary exclusion,Citation88 (iii) shaping and maintaining the GM compositionCitation89 and (iv) neutralizing the bacterial toxins.Citation90,Citation91
Furthermore, IECs constitutively produce a panel of cytokines, including TGF-α, IL-10, IL-15, and IL-18, that are crucial for the basal recruitment of immune cells, the regulation of IEC growth and so gut homeostasis.Citation92–Citation95 These cytokines also up-regulate the production of other cytokines (i.e. IL-1α or β, IL-6, IL-7, IL-8, TNF-α and TGF- β) and/or chemokines by immune cells during microbial infection.Citation92,Citation96–Citation98
Finally, bacterially derived antimicrobial factors, among them the peptides called bacteriocins, are secreted by gram-positive and gram-negative bacteria in the intestinal mucus layer. These bacteriocins have a restrained antimicrobial activity spectrum that affects the composition of the microbiota (interdependency within bacterial communities).Citation74 Moreover, the gut microbiota produce several metabolites (from anaerobic fermentation of exogenous undigested dietary components) such as short-chain fatty acids (SCFAs), aryl-hydrocarbon receptor ligands or polyamines, which interact with host cells and thus influence immune response.Citation99 Altogether, the small intestine is constantly exposed to bacteria and bacterial products, which explains the constant baseline level of secreted antimicrobial factors in the mucus layer.Citation100,Citation101
Gut mucosal immunity
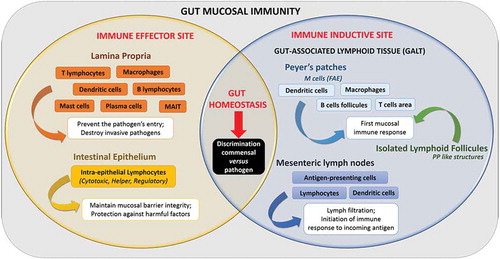
IECs play a central role in the intestinal immune response, acting as frontline sensors between GM and the immunity of the gut mucosa.Citation96 A close relationship exists indeed between the digestive microbiota and the intestinal mucosa, mediated by a complex intestinal immune system. This mucosal immune system (or MIS) is divided into three physically distinct parts: (i) the intestinal epithelial barrier, (ii) the lamina propria (LP) and (iii) the gut-associated lymphoid tissue or GALT, comprising Peyers’ patches (or PPs), isolated lymphoid follicles (or ILF) and mesenteric lymph nodes (or MLNs). Functionally, the intestinal MIS comprises two immune sites, (i) the effector and (ii) the inductive siteCitation102 (). They are challenged with discriminating entero-pathogenic microorganisms from the resident commensal flora, and adapting the immune response as needed, from tolerance to inflammation.Citation103 These aspects have been extensively addressed elsewhereCitation27 and will not be developed in this review. However, the specialized cells of the immune system are of particular interest in the context of this review because they contribute to the translocation of micro-organisms through the gut barrier.
In this context, an important effector site of gut mucosal immunity is the lamina propria layer, which has the key functions of (i) preventing entry/spread of pathogens across gut mucosa and (ii) destroying invasive pathogens.Citation102 Indeed, almost all of the intestinal immune cells are found in the LP layer; this includes innate immune cells (including dendritic cells (DCs), macrophages (Mφ), Mucosal-associated invariant T cells (MAIT), mast cells and innate lymphoid cells), and adaptive immune cells (Lymphoid T cells and plasma cells).Citation104 These immune cells have a crucial function in the interaction with the GM. Among them, the γδ intraepithelial T lymphocytes (that represents the major T cell population within the intestine) respond directly to microbiota signals to promote intestinal homeostasis which (i) helps preserve the integrity of damaged epithelial surfacesCitation105 and (ii) produces innate antimicrobial factors and pro-inflammatory cytokines and chemokines in response to resident bacteria that penetrate the intestinal epithelium and to intestinal injury.Citation106,Citation107 Another important line of immune cells for maintaining intestinal homeostasis are the CD4 regulatory T cells (Tregs). Indeed, Tregs are important response modulators to gut microbiota, notably through the secretion of anti-inflammatory cytokines such as IL-10, which trigger an immunosuppressive response and gut homeostasis.Citation108 In parallel, the innate lymphoid cells (ILCs) contribute to host defense and immune homeostasis within GALT.Citation113 Thus, ILCs are able to rapidly respond to gut microbiota contributing to GALT formation and function (Reviewed in Citation112).
Among GALT, PPs consist of dome-like structures or sub-epithelial domes (SED) containing large B cells follicles and smaller T cells areas, in addition to migratory dendritic cells (DCs) and macrophages (Mφ). These SED are separated from the gut lumen by a single epithelial layer called the Follicle-Associated Epithelium or FAE. The FAE contains epithelial cells, including microfold (M) cells. When compared to the adjacent enterocytes, these M cells display reduced irregular microvilli (short fold-like invagination or microfold) on the apical surface.Citation27 Moreover, M cells express various apical receptors (e.g. integrin β-1, GP2, C5a and poliovirus receptors) that serve as a portal of entry for luminal components.Citation110 At the basolateral membrane, M cells present a large pocket-like invagination that contains B lymphocytes (LB) and T lymphocytes (LT) in equal proportions, Mφ and DCs.Citation111 This particular structure in the M cells act as antigen-presenting cells, sampling gut luminal antigens, macromolecules and microorganisms present in the intestinal lumen in different ways (i.e. transcytosis, phagocytosis, microvesicle shedding) without any degradation.Citation27,Citation112 Consequently, the translocation of antigens through M cells is pivotal for linking the contents of the gut to the immune system, with the aime to trigger an appropriate immune response to the harmless or harmful stimuli in the gut lumen. In a healthy host, this process largely contributes to the tolerance toward antigens derived from diet or the commensal flora through signaling pathways involving an Ets (E-twenty-six-specific sequence) transcription factor, SpiB, which acts as a major regulator of the structural and functional maturation of M cells.Citation113,Citation114
On the contrary, many pathogenic microorganisms including reoviruses, polioviruses,Citation115 and enteric bacteria (e.g. Salmonella typhimurium, Shigella flexneri, Yersina enterocolitica)Citation116–Citation119 have developed strategies to exploit (i) weaknesses in the cell-cell junctions bordering M cells and (ii) the absence of mucus covering these specific cells. Consequently, M cells form a possible “gateway” that allow pathogens to enter and invade the host, triggering deleterious immune responses.Citation120
Candida albicans’ interaction with the intestinal barrier
C. albicans is an opportunistic pathogen that interacts dynamically with its human host. Indeed, when colonizing diverse mucosal surfaces of the human host as a commensal, C. albicans’ extensive phenotypic plasticity makes it capable of causing life-threatening systemic infections.Citation121 It is a polymorphic fungus that exhibits in vivo yeast or hyphal (and possibly chlamydospore) forms, and the ability to transition from one to another is considered a virulence trait of the fungus.Citation122,Citation123 It also expresses metabolic flexibility that largely contributes to its adaptability and virulence to diverse host niches. This phenotypic plasticity is best exemplified by the white and opaque phenotypes that correspond to distinct morphological states of the same isolate able to undergo heritable and reversible epigenetic transitions.Citation124 In addition, this phenotypic and metabolic flexibility allow the fungus to adapt to specific fluctuating external stresses, including nutrient conditions, temperature and pH.Citation125 These stresses drive the switch of C. albicans to the white or opaque state, each displaying specific biological features affecting, among others, mating competency, immunogenicity, virulence and niche specificity.Citation130–Citation135 While white cells are the most clinically relevant form of C. albicans and considered as an all-around phenotype fitting various environmental conditions, opaque cells correspond to a more metabolically specialized form of the fungus.Citation129
The gut environment forms a specific niche characterized by a number of environmental factors including pH, temperature, hypoxia, and the presence of serum or carbon sources. There are also a wide range of microbial and nutrient metabolites that can influence morphological changes in C. albicans and the phenotype of the fungus.Citation136 Interestingly, Pande K et al. observed a morphological switch to a “dark” phenotype in C. albicans cells exposed to the mammalian gut environment.Citation137 Indeed, when the Wor1 transcription factor is necessary and sufficient to drive the switch from the white to the opaque state,Citation138–Citation140 WOR1 overexpression triggered the switch to a Gastrointestinally-IndUced Transition (GUT) morphotype optimized to the gut living in a commensal state.Citation137 Finally, the gut niche appears to be a complex environment capable of driving alternative phenotype switches that promote commensalism or pathogenicity in C. albicans as the result of specific combinations of environmental stresses. However, how and which variations of the gut environment favor the commensal or virulent form of C. albicans remain to be specified. Thus, the passage of C. albicans through the gut mucosal barrier is a complex process depending on host characteristics that are both extrinsic and intrinsic, associating phenotypic switches that promote C. albicans commensalism or pathogenesis.
Notably, the pathophysiology of candidiasis is intimately linked to the ability of C. albicans to adhere to and invade host cell barriers. During these processes, C. albicans has to face different components of the gut mucosal barrier, including the antimicrobial peptide armory and the mucus layer, to successfully reach and then invade the epithelial layer lining the gut mucosa, possibly resulting in blood dissemination.
Interaction of C. albicans with the gut microbiota
In the digestive tract, 221 fungal species are part of the microbiota, mostly yeast and filamentous fungi belonging to the Ascomycota, Basidiomycota and Zygomycota phyla. In healthy humans, this mycobiota encompasses almost 66 genera of fungi, mainly including Saccharomyces and Candida species.Citation7
The equilibrium of the healthy microbiome is under the control of transkingdom interactions between the different inhabitants of the microbiota. Among them, bacterial-fungal interactions are particularly important for the homeostatic state, as highlighted by Iliev et al. who reported the existence of mixed fungal-bacterial biofilms in the GI tract of mice.Citation141 Fungal-bacterial interactions create a complex balance between synergism (e.g. C. albicans/Streptococcus sp.) and/or antagonism (e.g. C. albicans/Lactobacillus sp.) pathways, leading to a state of harmony within the microbiota (reviewed in Wang et al., 2014).Citation142–Citation144
In this context, the GI microbiota exerts anti-Candidal properties through different mechanisms. Indeed, the bacterial microbiota can modulate C. albicans colonization through the production of bacterial metabolites that limits fungal proliferation and virulence.Citation145 Such bacterial metabolites exert antifungal activities in vitro through molecular mechanisms involving the C. albicans’ TOR pathway, a central signaling pathway that controls cell growth in response to environmental nutrient signals in eukariota.Citation146 For instance, Lactobacillus sp. limits C. albicans’ growth and virulence, affecting the germ-tube formationCitation145 in the GI tract through the production of H2O2, organic acidsCitation147 and short chain fatty acids (SCFAs). Thus, bacterial metabolites inhibit C. albicans’ hyphal growth and invasion of human enterocytes by repressing the yeast-to-hyphae transition as well as the inducibility of hyphae specific transcripts.Citation146 In addition, other metabolic products, including dietary products such as tryptophan, exert antifungal activities by enhancing the mucosal reactivity to C. albicans. Indeed, tryptophan promotes the local transcription of a bacterial gene that triggers IL-22 productive cells and the proliferation of IL-22-producing innate lymphoid cells in the GI tract.Citation148 As a result of its pivotal role in innate antifungal resistance,Citation149,Citation150 the resulting increased production of IL-22 promotes resistance to colonization by C. albicans by attenuating inflammation in the IECs and damage caused by C. albicans.Citation149
As a consequence, disturbances in the bacterial community (or dysbiosis) are associated with enhanced C. albicans colonization,Citation151,Citation152 as reported early in the 1970’s.Citation153,Citation154 These observations were underscored in germ-free mice that experienced an increase in colonization as the result of the lack of a bacterial microbiota.Citation155 Various iatrogenic factors including antimicrobial and immunosuppressive therapies have been reported for their capacity to disrupt the GI microbiota, thus modulating C. albicans colonization.Citation144,Citation156,Citation157 Fan et al. showed that antibiotics specifically targeting anaerobic bacteria, Bacteroidetes and Firmicutes, favored colonization of C. albicans in the gut of mice.Citation158 These authors observed that anaerobic bacteria from mature adult microbiota were critical for maintaining resistance to C. albicans since Bacteroides thetaiotamicron was capable of inhibiting colonization through the activation of the Hypoxia Inducible Factor (HIF)-1α.Citation158 This transcription factor is an essential regulator of mammalian innate defense, that, once activated, induces the production of the Cathelicidin-AMPs LL37, a critical immune effector for limiting C. albicans colonization.Citation158 Conversely, the use of antifungal therapies can contribute to fungal dysbiosis in which some species are reduced while others expand. This was recently reported by Wheeler et al. who observed worsening colitis and allergic airway disease in experimental mice models. The administration of oral antifungal therapies correlated with an increase in Aspergillus sp., Wallemia sp. and Epicoccum sp. Colonization, whereas Candida sp. colonization was decreased.Citation17
Notwithstanding iatrogenic factors, many metabolic diseases, including GI disorders, have been linked to disturbances in C. albicans colonization.Citation15,Citation18 Over-colonization observed during bacterial dysbiosis may aggravate the severity of ulcers and inhibit healing associated with IBD.Citation159 Additionally, a reduced diversity in both the fungal and bacterial gut flora, with a specific increase in the Candida genus, was reported in IBD patients.Citation19 Recently, Sokol et al., highlighted a higher proportion of C. albicans in stools of IBD patients compared with healthy subjects, which points to a disease-specific fungal mycobiota dysbiosis.Citation15 Collectively, these data strongly suggest the involvement of C. albicans in the pathogenesis of IBD.
It is now clear that overgrowth of C. albicans on GI mucosal surfaces contributes to disseminated candidiasis, especially in weakened patients in the hospital setting who are exposed to iatrogenic factors that favor C. albicans’ colonization, such as antimicrobial therapies and/or chemotherapies.Citation144,Citation156,Citation157 However, when C. albicans colonization is associated with a metabolic disorder, including GI disorders, it is not clear whether the increased colonization results from or contributes to the pathogenesis of the disorder. Further studies are expected to specify (i) the inter-communication between C. albicans and bacteria at the species or community level in the gut, with emphasis to mixed biofilms and their role in the gut homeostasis and GI disorders; (ii) the influence of C. albicans’ over- or sub-colonization on the host metabolism and local or systemic immune response and (iii) the biological features of C. albicans associated with various digestive disorders.
Interaction of C. albicans with the gut mucosal barrier
Tackling the AMPs armory and the mucus barrier
As seen above, bacteria are essential for an efficient innate immune response against C. albicans, notably through the induction of AMP production, including LL37, histatin 5 and β-defensins.Citation158,Citation160 These AMPs exert anti-Candidal activities by (i) direct induction of cell death through mechanisms promoting pore formation, membrane depolarization and/or osmotic dysregulation in yeasts, (ii) immune modulation (through the promotion of PMNs/monocyte recruitment, TNF-α expression, chemoattraction of immature DCs and T cells) restraining C. albicans proliferation, and (iii) the repression of C. albicans virulence traits (e.g. α-defensin-6 prevents adherence to and invasion into enterocytes as well as biofilm formationCitation161) (Reviewed in Swidergall et al., 2014).Citation160 During commensalism, the continuous but weak Pathogen Associated Molecular Patterns (PAMPs)/PRRs interactions lead to a low level of Nuclear Factor kappa B (NF-kB) pathway activation and so to a basal level of production of AMPs.Citation162,Citation163 Upon increased colonization and infection, a strong up-regulation of the ERK and JNK MAP kinase pathways occurs, boosting inflammatory and damage responses of the host cells that will secondarily increase the production of AMPs by epithelial cells.Citation164
C. albicans has developed a three-phase strategy to avoid the candidacidal activities of AMPs. First, C. albicans secretes peptide effectors which protect against a broad range ofCitation165-Citation167AMPs. This is exemplified by the Msb2 membrane glycoprotein whose extracellular domain, i.e. Msb2*, is released after cleavage in large amount in the extracellular environment, inactivating a large panel of AMPs.Citation167–Citation172 Interestingly, some data suggest the involvement of secreted aspartyl proteases (Saps) in the cleavage and release of Msb2*, while other observations corroborate the putative involvement of Saps as direct inhibitors of AMPs. The proteolytic activities of Saps (Sap1-4, Sap8 and Sap9) were reported to reduce the antifungal activity of the AMP LL-37.Citation173 Second, in response to AMPs, C. albicans is able to express the drug efflux pump gene Flu1, allowing the AMPs to be outsourced from the intracellular compartments of the fungus.Citation172 Third, the fungus is able to regulate signaling pathways, such as the HOG (High-osmolarity glycerol) pathway, which are key elements in survival to various AMPs.Citation174,Citation175 Knowing that the HOG pathway is involved in both the regulation of the ROS production and the production of ATP by the mitochondria,Citation174 and that ROS production and ATP efflux is induced by various AMPs,Citation160 this HOG pathway may exert a key function in the survival of C. albicans to AMPs by downregulating ROS production and ATP efflux.
It is assumed that the AMPs release in the intestinal environment, especially in the mucus layer, is conditioned by the composition of the gut microbiota that interacts with AMP-secreting cells (mainly PCs and PMNs). Consequently, dysbiotic states modify the panel and concentration of AMPs secreted in the gut lumen.Citation176 However, less is known about the direct regulation of the secretion of AMPs by C. albicans and the gut mycobiota. Interestingly, in this context, microarray-based transcriptomic analyses of intestinal epithelial cells interacting with C. albicans did not report overexpression of AMPs-related genes following infection,Citation177 whereas a strong expression of AMPs (especially DEFB4) was observed in oral cells challenged with the fungus.Citation177,Citation178 Once more, these observations corroborate the view that molecular and cellular features characterizing C. albicans interactions depend on the type of host cell. However, no in-depth study has focused on the regulation of AMP secretion by C. albicans in gut-associated PCs and PMNs.
In addition to evading AMPs, C. albicans has to cross the mucus layer to access and adhere to the layer of IECs. The mucins that mostly make up this protective layer modulate the morphology and physiology of C. albicans,Citation179 as exemplified by the mucin polymer Muc5AC expressed in the lung and the stomach that downregulate the expression of a range of genes related to adherence, filamentation and biofilm formation in C. albicans.Citation180 Moreover, the mucin Muc7 has been reported to exert a direct antimicrobial effect upon C. albicans in the oral cavity.Citation171 Nevertheless, the effect of mucins on colonization and dissemination of C. albicans in the gut remains to be specified.Citation181
C. albicans has however developed strategies to circumvent the mucus protective layer. C. albicans is able to adhere to mucins in a pH-independent process involving hydrophobic interactions that allow the non-specific binding of C. albicans to the 66-kDa cleavage product of the 118kDa C-terminal glycoprotein backbone of mucins.Citation182 Whereas the involvement of specific C. albicans’ ligands remains to be specified, Böhm et al. recently identified transcription regulators in C. albicans, including ZCF8, ZFU2 and TRY4, that favor (i) the yeast form of C. albicans more prone to colonize the gut in mice and (ii) adherence to mucins and mucus-producing IECs.Citation183 After adhering to mucins, which is critical in initiating successful colonization to host mucosal surfaces, C. albicans degrades the GI mucus layer by secreting mucinolytic enzymes. Among them, Secreted Aspartyl Protease 2 protein (Sap2p) facilitates mucus penetration by promoting pore formation in the mucus layerCitation184 using a strategy similar to that employed by enteropathogenic bacteria.Citation74,Citation182,Citation185–Citation188 Finally, C. albicans produces other Saps that degrade the sIgA scattered in the mucus layer, thus escaping the host innate immune response in part.Citation189
Interaction of C. albicans with enterocytes
In a susceptible host, once it has evaded the AMPs armory and the mucus layer, C. albicans adhere to IECs, cross the epithelial barrier and finally induce cellular damages leading to the loss of gut epithelium integrity.Citation179
C. albicans adherence to enterocytes
Candida sp. adherence to IEC is a key step in the physiopathological process that contributes both to the colonization and pathogenesis of disseminated candidiasis that mainly originate from the digestive mucosa.Citation189 Heterogeneities have been reported in the adherence properties of different Candida species, C. albicans displays a greater capacity to adhere to epithelial cells in vitro, including to intestinal epithelial cells.Citation190,Citation191 Thus, C. albicans’ adherence to epithelial cells is a complex, dynamic and multifactorial process that requires a close relationship between the fungus and host proteins exposed at cell surfaces.Citation192
The capacity of C. albicans to adhere to host tissues depends on the type of epithelial cell. Indeed, C. albicans yeast forms display decreased adherence to differentiated enterocytes in vitro as compared with oral epithelial cells,Citation10 suggesting that (i) oral cells may express specific molecules enhancing C. albicans attachment or (ii) the brush border lining the enterocyte’s layer displays unique features limiting the fungus’ ability to adhere. Following the first contact, most of the adherent yeast cells switch to the hyphal form,Citation10 probably as a result of the physical contact with the epithelial surface.Citation10,Citation193 This is an important step since the expression of hyphal-associated genes will (i) strengthen adherence through enhanced expression of adhesinsCitation194,Citation195 as well as (ii) favor the expression of hyphal invasins, enabling the fungus to enter the epithelial cells (see below). Another interesting observation from the early stages of interaction is the intimate entrapment of C. albicans hyphal tips within the microvilli that cover the surface of the enterocytes, a phenomenon that probably reinforces the attachment to the epithelial surface.Citation10,Citation196,Citation197 Finally, hyphal forms of the fungus conglomerated to form aggregates that adhered to the surface of enterocytes, a phenomenon that was not observed with oral cells.Citation10 It is possible that aggregation of Candida cells indirectly promote attachment of the fungus to the surface of enterocytes.Citation198 Collectively, these observations strongly suggest that enterocytes express specific cues that influence the attachment of C. albicans. This view is supported by observations from Sohn et al. who investigated the transcriptional response of C. albicans adhering to different substrates, including vaginal and intestinal cells, and abiotic surfaces, for different periods of time.Citation197 These authors reported subsets of genes that were specifically expressed in adherent conditions as compared to suspension cultures. Furthermore, the transcriptional response of the fungus differed significantly depending on the adhesive surface (including enterocytes). The response was globally close early on, but differences became more pronounced as the interaction progressed. These observations suggest the ability of C. albicans to sense and to adapt its transcriptional program to subtle variations in the adherent surface.Citation197
In addition to passive van der Waals forces and hydrophobic interactions that probably contribute to the initial contact of C. albicans on epithelial surfaces,Citation199 expression of surface molecules (adhesins) is a key biological feature of the fungus, allowing its attachment to host tissues (both as a commensal or a pathogen),Citation200 to abiotic surfaces, and to other microorganisms or Candida cellsCitation201 (reviewed inCitation190,Citation202,Citation203). The major C. albicans adhesins are GPI-anchored glycoproteins that are expressed by yeast or hyphal forms of the fungus or both, including the multifunctional agglutinin-like sequence (Alsp) proteins family, the hyphal wall proteins (Hwp) family, the IPF family, the F/Hyphally upregulated protein or Iff/Hyr protein family, and the Epithelial adhesin proteins (Eap) family.Citation190,Citation202 Furthermore, C. albicans expresses numerous other protein adhesins that have not yet been fully characterized in terms of structure and/or capacity to mediate adherence to epithelial cells such as fimbriae, Csh1, Ywp1, Pra1, proteins of the Sap family and others (Reviewed in.Citation190,Citation202–Citation204 Though many of these adhesins have been reported to promote adherence of C. albicans to epithelial cells, few investigations have focused on enterocyte interaction. Sohn et al. observed an upregulation of HWP1 in the early stages of the C. albicans and enterocyte interaction, suggesting its involvement in the attachment of the fungus to the Caco-2 enterocytic cell line.Citation197 Similarly, Hwp2p and Int1p have been reported to contribute to adherence of the fungus to the HT-29 human intestinal epithelial cell line in vitro.Citation196,Citation205 Finally, a C. albicans double mutant for the putative β-glucanase mannoprotein Mp65, defective in hypha and biofilm formation, displayed reduced adherence to plasticCitation206 and epithelial cells including enterocytes.Citation207,Citation208 Given the role of Mp65 in cell wall organization and stability, it is plausible that deletion of the MP65 gene affects the expression of other adhesins at the surface of C. albicans cells, indirectly altering its adherence capabilities.Citation207 However, observations that blocking antibodies specific for Mp65 strongly decrease adherence of wild-type C. albicans to plasticCitation206 and epithelial cellsCitation208 are in line with the direct contribution of Mp65 as an adhesin in C. albicans.
In addition to the protein nature of adhesins and given the high content of the yeast cell wall in carbohydrate components, C. albicans can also adhere to enterocytes through polysaccharidic molecules expressed at the cell wall surface.Citation204,Citation209 Indeed, Timpel et al. reported first that a pmt1∆/∆ deletion mutant, a gene encoding a mannosyltransferase involved in the O-glycosylation pathway, displayed reduced adherence to enterocytic Caco-2 cells, suggesting a role for the O-linked carbohydrate content of the cell wall in the adherence of C. albicans to enterocytes.Citation210 Additional observations reported the contribution of the N-glycosidic part of the cell wall in the adherence of C. albicans to enterocytes. Indeed, in competition experiments based on a panel of carbohydrates, Dalle et al. observed a drastic dose-dependent reduction in the adherence of C. albicans to enterocytic Caco-2 cells with synthetic β-1,2 oligomannosides and to a lesser extent with α-1,2 oligomannosides, two glycans of the N-linked moiety of the cell wall.Citation211 Unlike α-1,2 oligomannosides, β-1, 2 oligomannosides are rare structures in living systems, and their presence has been reported in only few bacterial and yeast species.Citation212,Citation213 However, they are prominently expressed in C. albicans.Citation214 They reside in the yeast wall associated with N-glycans of a high polymer of mannoses bound to a peptide, the phosphopeptidomannan (PPM) and mannoproteins (MPs) as well as to the glycan copula of a glycolipid of the family of mannose-inositol-phosphoceramides, i.e. phospholipomannan (PLM).Citation214,Citation215 They contribute to the virulence of C. albicans thanks to their immunomodulatory and adhesin properties.Citation214 Interestingly, the inhibitory effect obtained with the synthetic β-1, 2 oligomannosides was higher than the other tested carbohydrates including the α-1,2 oligomannosides.Citation211 This strongly suggests that N-glycans specific to the C. albicans cell wall contribute to their attachment to enterocytes. This confirms in vivo observations that reported less C. albicans gastrointestinal colonization in infant mice orally fed with synthetic β-1, 2 oligomannosides, then in these orally fed with α-1,2 oligomannosides.Citation211,Citation216
Altogether, these data support the hypothesis that proteic and carbohydrate adhesins contribute to the attachment of C. albicans to IECs, but further studies are required to better characterize the contribution of each of these adhesins. In addition, soluble factors from the extracellular matrix as well as serum proteins have been suggested as potential mediators of the indirect attachment of C. albicans to mucosal surfaces.Citation204,Citation217 Similarly, specific ligands from host-cell surfaces, including proteins of the integrin and cadherin families, have reportedly facilitated the adherence of C. albicans to epithelial cells.Citation196,Citation218 However, the exact nature of the host factors that influence attachment remains to be specified.
C. albicans invasion into the enterocytes
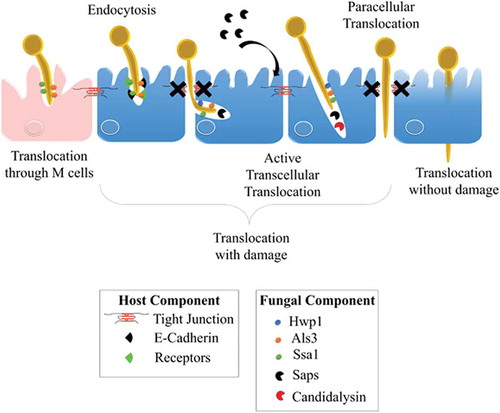
C. albicans has developed several strategies to invade and further damage host epithelial cells. The yeast-to-hyphae transition in C. albicans is pivotal for epithelial invasion,Citation219 contributing to two different well-documented mechanisms of invasion: (i) epithelial-driven endocytosis of hyphal forms,Citation10,Citation220 and (ii) C. albicans-driven active penetration.Citation10,Citation221–Citation223 Endocytosis is considered an early event, seeing as it occurs within the first 4 hours of the interaction,Citation224 whereas invasion at later stages seems mainly driven by active penetration.Citation10,Citation217,Citation225 In fact, these two mechanistically distinct routes of invasion probably occur differentially depending on the epithelial cell type encountered by the fungus, as a result of the temporal and/or spatial availability of the epithelial receptors that are necessary for mediating induced endocytosis.Citation219 For instance, when both mechanisms are present during a C. albicans invasion of oral epithelial cells, active penetration is the only process driving invasion of the enterocytes in vitro at the early stages of the interaction.Citation10 However, recent evidence suggest that endocytosis may occur later in time and that alternate routes may allow the translocation of C. albicans through the gut barrierCitation9 ().
Figure 4. Invasion of C. albicans through the intestinal epithelial barrier.
Schematic representation of the different mechanisms used by C. albicans to translocate through the gut mucosa: (i) the transcellular route, (ii) the paracellular route, (iii) the translocation through M cells, and (iv) the alternate route that may occur. Als3, Agglutinin-Like Sequence 3; Ssa1, Heat shock protein ssa1; Hwp1, Hyphal wall protein 1; Saps, Secreted Aspartyl Proteases; CL, Candidalysin; TJs, Tight Junctions; LDH, Lactate dehydrogenase; Ca2+, Calcium.
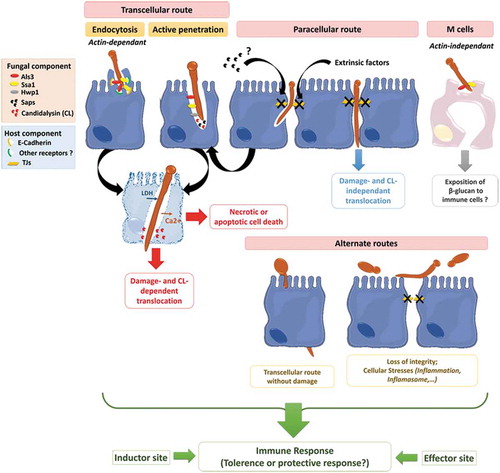
Endocytosis of C. albicans
Endocytosis was first described for C. albicans cells interacting with in vitro models of endothelial or oral epithelial cells. The filamentous forms of the fungus were capable of triggering the formation of pseudopod-like structures at the surface of these host cells,Citation194,Citation220 leading to the partial engulfment of the hyphae and its progressive internalization into host cells.Citation193,Citation194,Citation220 This host-cell mechanism is mainly driven through a Zipper-like actin filament-dependent process as demonstrated by the drastic decrease in the internalization of C. albicans cells into oral and endothelial cells pretreated with the actin polymerization inhibitors Cytochalasin D and Lancuntrulin A.Citation10,Citation194,Citation220 However, additional molecular processes contribute to endocytosis of the fungus, including macropinocytosisCitation10 and the clathrin-dependent endocytic machinery.Citation226 In addition, endocytosis is independent of the viability of the fungus since heat-killed hyphal forms of C. albicans are endocytosed with the same efficiency as live hyphae.Citation10,Citation220 However, endocytosis of dead hyphae was not associated with cellular damage, suggesting that other(s) factor(s) produced by viable forms of the fungus may be necessary to cause cellular damage in both endothelial and oral epithelial cells.Citation220,Citation227
Two fungal invasins were reported as major stimulators of the endocytic machinery.Citation194,Citation228 First, the multifunctional hypha-specific surface protein Als3, a key virulence factor for C. albicans, acts as an invasin by triggering its own endocytosis by endothelial and oral human epithelial cells. N- or E-cadherin expressed at the surface of endothelial and oral epithelial cells, respectively, were identified as putative host cell receptors for this invasin.Citation194,Citation229 Finally, in silico analysis reported molecular similarities between Als3/E-cadherin and E-cadherin/E-cadherin interactions, suggesting the molecular mimicry of Als3 with cadherins.Citation194 Interestingly in the context of C. albicans interaction with intestinal epithelial cells, Goyer et al. reported that Als3 participates in the E-cadherin-independent endocytosis of C. albicans into enterocytes displaying altered tight junctions (TJs) (see below for more details about the role of TJs during C. albicans invasion into enterocytes). Indeed, the specific blockade of E-cadherin by the SHE78-7 neutralizing antibody did not modify internalization of C. albicans into enterocytes displaying altered TJs.Citation9 This corroborated other observations reporting a partial reduction in C. albicans endocytosis into oral epithelial cells when inhibiting E-cadherin function,Citation230 suggesting that other(s) host-cell receptor(s) may contribute to the Als3-driven endocytosis of the fungus. A second invasin, Ssa1, expressed at the surface of C. albicans hyphae and belonging to the Hsp70 family of heat shock proteins, was found to contribute to cadherins-dependent endocytosis of the fungus into endothelial and oral epithelial cells.Citation228 Indeed, C. albicans ssa1∆/∆ exhibited reduced endocytosis into endothelial and oral epithelial cells, that was associated with less cellular damage, a decrease in adherence to endothelial and oral epithelial cells and attenuated virulence in a mouse model of oropharyngeal candidiasis.Citation228 Surprisingly, the C. albicans double mutant ssa1∆/∆als3∆/∆ displayed reduced endocytosis, similar to the single mutant als3∆/∆, suggesting (i) the possible binding of Als3 and Ssa1 to the same host receptor and/or (ii) a cooperative function of both Als3 and Ssa1 acting as a multiprotein complex.Citation228 Finally, other invasins probably contribute directly or indirectly to the endocytosis of C. albicans,Citation217,Citation230 as highlighted by Wächtler et al., who observed reduced endocytosis of killed hyphae of the C. albicans sap1-3∆/∆ and sap4-6∆/∆ deletion mutants into oral cells.Citation217 However, the exact nature of the Saps as well as their direct or indirect involvement in the endocytic mechanism is not yet clear.
Collectively, these observations strongly suggest that endocytosis of C. albicans into epithelial cells is a complex mechanism that involves C. albicans hyphal-invasins including Als3, Ssa1 but possibly other invasins that remain to be specified. The interaction of such invasins with host-receptors, including cadherins but also probably other receptors, is necessary for triggering this process.Citation217 Zhu et al. highlighted that the involvement of Epithelial Growth Factor Receptor (EGFR) and HER2 as cadherin-independent host receptors during the endocytosis of C. albicans into oral epithelial cells.Citation230 However, the exact nature and function of such invasins and host receptors in the endocytosis of C. albicans into enterocytes displaying altered TJs remains to be specified.
Active penetration
Active penetration is a fungal-driven process and requires viable forms of the fungus. Similar to endocytosis, active penetration exploits the yeast-to-hyphae transition of C. albicans, which germinate and produce hyphae that progressively elongate and penetrate the epithelial cells.Citation10,Citation219 This invasion mechanism is of prime interest since it is the only one that allows C. albicans to gain entry to enterocytes with intact intercellular junctions in vitro.Citation9–Citation11 Scanning Electron Microscopy (SEM) observations of Candida-infected enterocytes did not reveal pseudopod-like structures surrounding C. albicans hyphae, which is a typical feature of endocytosis. By contrast, a depression in the cell surface was observed around the C. albicans entry site, probably as the result of an active process of the extending hyphae forcing entry either directly into the apical surface or at intercellular junctions between adjacent enterocytes. These observations were supported by the fact that pretreatment with endocytosis inhibitors does not inhibit the uptake of the fungus into enterocytic Caco-2 cells.Citation10
The contribution of fungal hydrolases to this process has been investigated but remains unclear.Citation231 Notably in the gut barrier, Saps contribute to the active penetration of C. albicans. The C. albicans deletion mutants sap1-3∆/∆ or sap4-6∆/∆Citation217 displayed moderate but significant reduction in their ability to invade enterocytes. In addition, treating the fungus with the aspartic protease inhibitor pepstatin A reduced enterocyte invasion in a dose-dependent manner.Citation10 The role of other hydrolases, including lipases and phospholipases B1, did not obviously contributing to active penetration of C. albicans into enterocytes.Citation10 Altogether these results suggest that active penetration of C. albicans into intestinal cells is likely to occur through molecular mechanisms combining the mechanical pressure exerted by elongating hyphae and the lytic activities of Saps.
Interestingly, Als3, a major invasin that contributes to induced endocytosis, reportedly facilitates the active penetration of C. albicans into oral epithelial cells,Citation194 but probably through an indirect mechanism that favors the binding of the two structures.Citation219 Als3 also seems to play a role in invasion since a C. albicans deletion mutant als3Δ/Δ, which is not defective in hyphal formation, was less able to invade intact enterocytes,Citation9 which are not able to internalize the fungus through endocytosis.Citation10
Interestingly, in an attempt to decipher the molecular basis of the interaction of C. albicans to epithelial cells, Wächtler et al. investigated the abilities to adhere to, invade and cause damage to oral and intestinal epithelial cells, of a set of 26 mutants deleted for genes predicted to be and/or previously reported as major contributors to host-C. albicans interaction. Whereas EFG1 and its upstream regulators appeared essential for all stages of the infection process, other genes displayed discrete but significant functions in specific stages of the interaction. Nine genes were identified as “damage-associated” since the corresponding deletion mutants were still able to invade oral epithelial cells but without damaging them. Surprisingly, six of these mutants (i.e. gpd2∆, gpp1∆, cka2∆, bcr1∆/∆, hwp1∆, bud2∆ and rsr1∆) displayed a strong and significant decrease in invasion into enterocytes.Citation219 This suggests that the genes involved in glycerol accumulation (GPP1 and GPD2) and directed hyphal growth (BUD2 and RSR1) are necessary for the active penetration of C. albicans into enterocytes but not into oral cells.Citation219
Together, these observations strongly support the view that C. albicans requires some specific genes to invade enterocytes but not oral epithelial cells,Citation219 reflecting the ability to adapt to specific cell environments by expressing biological factors that can contribute differentially to specific stages of the infection process.
The paracellular route
Several studies have suggested a paracellular route involving the proteolytic cleavage of the intercellular adherens junction (AJ) protein E-cadherin in both oral epithelial cellsCitation225,Citation232 and IECs.Citation233 AJs are located immediately below the TJs, and are mainly composed of proteins of the cadherin family including E-cadherin. AJs together with TJs form a junction belt that ensures the architectural cohesion of the intestinal epithelium.Citation234,Citation235
While active penetration has been described as the predominant mechanism of Candida invasion into IECs, in 2016 Goyer et al. observed that endocytosis of C. albicans can also occur during interaction with IECs in vitro. They demonstrated that TJs play a protective role in the early steps of the interaction by limiting endocytosis-mediated C. albicans invasion into enterocytes. While intact IECs are mainly invaded through active penetration, pharmacologically altered TJs are invaded by active penetration and induced endocytosis, through molecular mechanisms involving, at least in part, the hyphal-associated invasin Als3. So, in addition to demonstrating the role of TJs in limiting the invasion of IECs by C. albicans, this study highlights the putative harmful effects of extrinsic factors that alter the integrity of TJs, thereby allowing C. albicans to invade through the gut barrier.Citation9 This corroborates reports of clinical situations where therapies or medical practices may alter the permeability of the digestive tract, consequently promoting the translocation and hematogenous dissemination of common residents of the gut microbiota, including C. albicans.Citation236,Citation237 Interestingly, recent findings indicate that C. albicans itself can alter the integrity of TJs. Using an in vitro model of the intestinal barrier, Böhringer et al. observed a barrier breakdown after 8 h of infection, as illustrated by the drastic decrease in the Trans-Epithelial Electrical Resistance (TEER) of C2BBe1 (a clone of Caco-2 cells) intestinal epithelial cells interacting with C. albicans. This breakdown was not related to cell death since C2BBe1 cells were not damaged at this point in time. In addition, the drop in TEER required the fungal yeast-to-hyphae transition since the efg1∆/cph1∆ deletion mutant, unable to produce hyphae, did not induce barrier breakdown or cell death. Along with barrier breakdown, a strong decrease in the level of TJs proteins, including occludin, JAM-A, Claudin −1, −3 and −4, was observed by Western-blotting on whole infected-cell extracts, suggesting that the barrier breakdown results from the loss of these proteins. This was confirmed by immunofluorescence testing which found a progressive extinction of Occludin, Claudin-1 and 4 and JAM-A staining after 10 h of infection. All in all, this study showed that a barrier breakdown occurs during infection by C. albicans and this is related to a decrease in TJs protein levels and their concomitant disappearance from cell-cell borders.Citation177 Recently, Allert et al. observed the ability of the ece1∆/∆ mutant to compromise epithelium integrity and to translocate across the intestinal epithelial barrier through damage-independent factors and without damaging the cells.Citation179 The authors hypothesized that these factors probably alter the cell-cell connections (i.e. TJs or adherens junctions), facilitating paracellular translocation. Focusing on the effect of Saps on the promotion of the degradation of the E-cadherin protein during C. albicans infection, they studied mutants lacking various SAP genes (i.e. sap1-3∆/∆, sap5∆/∆, sap4-6∆/∆ and sap9/10∆/∆). Unfortunately, no translocation defect was observed with the mutants. However, the reduced ability of the sap1-3∆/∆ mutant to translocate through the blank transwell insert without cells cannot rule out the possible minor role of SAPs in the paracellular route of translocation.Citation179
Collectively, these are important observations that point to the paracellular route as a possible mechanism used by C. albicans to translocate through the intestinal barrier. This mechanism may favor the passage of C. albicans cells between enterocytes without invading or damaging intestinal cells.Citation10,Citation193 However, it is clear now that easier access to the basolateral site of intestinal cells promotes fungal invasion through both active penetration and induced endocytosis.Citation9 Finally, C. albicans itself is capable of altering the integrity of TJs, secondarily promoting its own translocation.Citation177 However, the extent to which the paracellular route promotes intercellular transmigration of C. albicans (associated or not to epithelial invasion and damage) remains to be specified.
C. albicans-induced epithelial cell death and mucosal damage
Late stages of the infection of intestinal cells are characterized by a loss of barrier integrity associated with the destruction of IECs invaded by C. albicans ().Citation203 It is possible that the barrier breakdown observed in the late stages, as a consequence of alterations in intercellular junctions,Citation177,Citation233 triggers host-cell signaling pathways that promote inflammation, the activation of inflammasomes and apoptotic events, ultimately leading to necrosis of the enterocytes.Citation43 Invasion by C. albicans also actively contributes to enterocyte damage and cell death.Citation203 Until recently, it was generally believed that Candida invasion resulted in the death of infected epithelial cells possibly through mechanisms involving necrosis and/or apoptosis.Citation231 However, the invasion of epithelial cells per se is not always clearly associated with cell damage.Citation179 In fact, initial invasion of oral, vaginal or intestinal epithelial layers causes little measurable cell death.Citation10,Citation193 Conversely, the later phases of C. albicans invasion into IECs are associated with epithelium damage mainly due to the mechanical elongation of the hyphae.Citation219 However, the identification of a specific virulence factor inducing cell damage has remained elusive. A cytolytic toxin produced by C. albicans, named Candidalysin (CL), was recently identified; it is thought to be the missing link between hyphae formation and damage to host cells.Citation179,Citation238 CL is a 31-amino-acid peptide generated from the pre-protein Ece1 (Extent of cell elongation 1) (271 amino acids), which is encoded by the ECE1 gene belonging to the eight core filamentation genes of C. albicans and highly expressed during hyphae formation.Citation239 Previous studies had shown that the seven lysine-arginine dibasic amino acid motifs present in the Ece1 pre-protein can be recognized and cleaved by a subtilisin-like serine protease, Kex2, in the yeast Golgi apparatus.Citation240 CL is one of the cleaved peptides generated from Ece1, and its α-helical structure allows it to intercalate and permeabilize the host cell membranes and subsequent cell lysis when secreted in sufficient amount. In oral epithelial cells, CL triggers a danger-response signalling pathway and activates epithelial immunity.Citation232 Moreover, the permeability of the cell membrane is enhanced by a positive charge at the carboxy terminus of the peptide which triggers an inward current concomitant with calcium influx.Citation232 Recent observations have confirmed the importance of CL during the translocation of C. albicans through the intestinal barrier in vitro. Allert et al. screened more than 2000 C. albicans gene deletion mutants to identify fungal factors involved in C. albicans gut translocation, leading to the identification of several genes associated with cellular damage including PRN4, orf19.2797, NRP2, AAF1, HMA1, TEA1, orf19.3335, PEP12 and ECE1.Citation179 Focusing on these nine corresponding mutants, eight of them, including ece1∆/∆, showed a significant decrease in their ability to induce damage at 24 h post-infection. Given the critical role of CL for oral and vaginal epithelial cell damage, the authors investigated the role of CL during C. albicans gut translocation using a C. albicans mutant that lacked only the CL-encoding region within ECE1 (ece1 ∆/∆ + ECE1∆184–279 mutant). They observed that both ece1 ∆/∆ and ece1 ∆/∆ + ECE1∆184–279 mutants were unable to induce damage and that their ability to induce damage was partially restored with the administration of synthetic CL. This demonstrated that a combination of both (i) hyphae formation and (ii) CL secretion were required for optimal damage induction.Citation179 Necrotic cell death seems to be the major mechanism involved in damage observed during C. albicans translocation through enterocytes. Indeed, 24 h post-infection, no induced apoptosis (i.e. Annexin V staining) was observed while 40% of IECs were necrotic.Citation179 In addition, damage was associated with a loss of epithelial integrity during the translocation of C. albicans through the intestinal epithelial layer.Citation179 These studies highlight the key role of CL in Candida pathogenesis and provide a molecular explanation for the destructive activity of hyphae on the mucosal surface.Citation239
Interaction of C. albicans with M cells
As mentioned above, several bacterial pathogens exploit M cells to invade mucosal tissues and cross the digestive epithelial barrier to reach the bloodstream.Citation241 This suggests that entry through M cells is an essential step in the pathophysiology of many infectious diseases. In a recent in vitro study, Albac et al. demonstrated that C. albicans co-localizes with and preferentially invades M cells in a model of the enterocytic Caco-2 cells co-cultured with M-like cells, providing novel evidence that the fungus can use M cells as a portal of entry through the intestinal barrier. In addition to active penetration, F-actin-dependent endocytosis contributed to internalization of the fungus into M cells through a mechanism involving hypha-associated invasins such as Ssa1 and Als3.Citation11 Moreover, fungal β-glucans can be endocytosed by M cells which, by virtue of their APC function, expose β-glucans to macrophages and dendritic cells associated with Peyer’s patches, thereby modulating the systemic immune response to C. albicans.Citation236 Altogether these recent findings strongly support the view that understanding the contribution of M cells in sensing C. albicans as a commensal or a pathogen should provide new insights to the commensal role of C. albicans as well as to the pathophysiology and initiation of life-threatening systemic candidiasis. The exact molecular mechanisms by which C. albicans targets and translocates through M cells as well as the relevance of these interactions in vivo remain to be elucidated.
Models to study interaction of C. albicans with the intestinal mucosa
For now, several models have been already used to study the interaction between C. albicans and the epithelial intestinal barrier. Surprisingly, when the gut barrier forms a complex environment involving several cell types, each with specialized functions, the in vitro models already described are mostly based on the use of only one cell type: enterocytes. Only one model investigated the interaction of C. albicans with two co-cultured cell types: enterocytes and M-like cells. However, new models are aimed at specifying the interaction of the fungus with AMPs, mucus and immune cells in the gut environment, as well as with other specialized cells including PCs, and Goblets cells. All of the existing models (i.e. in vivo and in vitro) are summarized in and .
Table 1. In vitro models developed to study the interactions between Candida albicans and the epithelial intestinal barrier.
Table 2. In vivo models described to study the interaction between Candida albicans and the epithelial intestinal barrier Murine models of intravenous disseminated candidiasis.
Conclusions
Formerly described as a commensal that exists on the mucosal surfaces of most healthy individuals, Candida albicans is an opportunistic pathogen that causes infections ranging from superficial to life-threatening disseminated infections, especially in the ever-growing population of vulnerable patients in hospital settings. In these situations, the fungus takes advantage of its host thanks to disturbances in the host-defense system and the mucosal microbiota. Overwhelming evidence suggests that the gastrointestinal tract is the main source of disseminated C. albicans infections. Some of the major risk factors for disseminated candidiasis include damage to the mucosal intestinal barrier, immune dysfunction, and antibiotic-induced dysbiosis of the resident microbiota.
Clearly, translocation of C. albicans through the intestinal barrier into the bloodstream is a critical step in the initiation of invasive candidiasis, which makes it important to understand the mechanisms by which C. albicans interacts with the gut barrier. Given the specific cues shared by intestinal cells and the specific environment of the gut lumen, the bulk of knowledge acquired when studying interaction of C. albicans with other mucosa, including the oral mucosa, is not fully transferable to the behavior of the fungus in the gut context.
A more precise view has emerged recently on how C. albicans translocates through the gut epithelial layer. However, the nature of the fungal and host factors contributing to the translocation remain to be specified. In addition, many questions are still unsolved and remain to be explored. For instance, a lack of knowledge exists with regard to the specific molecular mechanisms allowing IECs to prevent fungal invasion and how C. albicans can hijack them. The mechanisms by which C. albicans interacts with specialized IECs such as PCs and goblet cells and their effects on gut homeostasis and/or the pathogenesis of candidiasis is still unknown. Similarly, the extent to which the maintenance of inter-cellular junctions is crucial to restraining C. albicans invasion and how the fungus alters these junctions is still ongoing. In addition, little is known about how the biochemical components of the gut environment modulate the fungal factors and how C. albicans resists these biochemical factors. Finally, from an immune point of view, the specific mechanisms and pathways involved during host response associated to commensalism or pathogenicity of C. albicans need to be precise in the gut reservoir. For instance, though the contribution of the M cells in the translocation of C. albicans is now obvious, the consequence of this mechanism on the priming of tolerant or protective immune response is still unknown. A better overall understanding of the interaction between C. albicans and the intestinal epithelial barrier will help with the development of future therapies to avoid systemic candidiasis.
Importantly, with the advance of molecular tools to study microbiota components, new insights have emerged with regard to the association of GM dysbiosis with gastrointestinal or extra-gastrointestinal disorders where C. albicans colonization is disturbed. When highlighting the intricacy of bacterial-fungal relationships in the GM,Citation254 the consequences of these complex interactions upon the host and C. albicans virulence needs to be clarified.Citation158 Such information will help with the creation of new strategies to combat candidiasis, taking advantage of the existing interactions between bacteria and fungi in the gut microbiota. Co-infection models that closely mimic the interactions of the digestive microbiota and IECs are needed if we are to fully understand the pathophysiological mechanisms of candidiasis.
Abbreviation
| Adenosin Triphosphate | = | ATP |
| Adherens Junctions | = | AJs |
| Agglutinin-Like Sequence | = | ALS |
| Anti-Microbial Proteins | = | AMPs |
| B Lymphocytes | = | LB |
| Candidalysin | = | CL |
| c-Jun N-terminal Kinase | = | JNK |
| Dendritic Cells | = | DCs |
| Epithelial Adhesin Protein | = | EAP |
| Epithelial Growth Factor Receptor | = | EGFR |
| E-twenty-six-specific sequence | = | Ets |
| Extent of cell elongation 1 | = | Ece1 |
| Extracellular-signal-Regulated Kinase | = | ERK |
| Follicle-Associated Epithelium | = | FAE |
| GastroIntestinal | = | GI |
| Gastrointestinally-IndUced Transition | = | GUT |
| Glyco-Protein 2 | = | GP2 |
| Gut-Associated Lymphoid Tissue | = | GALT |
| Gut-associated Microbiota | = | GM |
| Heat Shock Protein | = | HSP |
| High-osmolarity glycerol | = | HOG |
| Hyphal wall protein | = | Hwp |
| Hypoxia Inducible Factor | = | HIF |
| Inflammatory Bowel Disease | = | IBD |
| Intestinal Epithelial Cells | = | IECs |
| Intestinal Epithelium | = | IE |
| Isolated Lymphoid Follicles | = | ILF |
| Macrophages | = | Mφ |
| Mannoproteins | = | MPs |
| Mesenteric Lymph Nodes | = | MLNs |
| Microfold cells | = | M cells |
| Mitogen-Activated Protein Kinases | = | MAPK |
| Mucosal Immune System | = | MIS |
| Neutrophil cells | = | PMNs |
| Next Generation Sequencing | = | NGS |
| Nuclear Factor kappa B | = | NFkB |
| Paneth Cells | = | PCs |
| Pathogen Associated Molecular Patterns | = | PAMPs |
| Pattern Recognition Receptors | = | PRRs |
| Peyers’ Patches | = | PPs |
| PhosphoLipoMannan | = | PLM |
| PhosphoPeptidoMannan | = | PPM |
| Reactive Oxygen Species | = | ROS |
| Regenerating Islet-Derived 3 Gamma | = | REG3γ |
| Scanning Electron Microscopy | = | SEM |
| Secreted Aspartyl Proteases | = | SAPs |
| Secretory group IIA PhosphoLipase A2 | = | sPLA2 |
| Secretory Immunoglobulin A | = | sIgA |
| Short-Chain Fatty Acids | = | SCFAs |
| Sub-Epithelial Domes | = | SED |
| T Lymphocytes | = | LT |
| Tight Junctions | = | TJs |
| Trans Epithelial Electrical Resistance | = | TEER |
| Zonula Occludens | = | ZO |
Disclosure of interest
The authors report no conflict of interest
Additional information
Funding
References
- Tadec L, Talarmin J-P, Gastinne T, Bretonnière C, Miegeville M, Le Pape P, Morio F. Epidemiology, risk factor, species distribution, antifungal resistance and outcome of Candidemia at a single French hospital: a 7-year study. Mycoses. 2016;59(5):296–303. doi:10.1111/myc.2016.59.issue-5.
- Horn DL, Neofytos D, Anaissie EJ, Fishman JA, Steinbach WJ, Olyaei AJ, et al. Epidemiology and outcomes of candidemia in 2019 patients: data from the prospective antifungal therapy alliance registry. Clin Infect Dis. 2009;48(12):1695–1703. doi:10.1086/597588.
- Arendrup MC, Sulim S, Holm A, Nielsen L, Nielsen SD, Knudsen JD, Drenck NE, Christensen JJ, Johansen HK. Diagnostic issues, clinical characteristics and outcome for patients with fungaemia. J Clin Microbiol. 2011;49(9):3300–3308.
- Gudlaugsson O, Gillespie S, Lee K, Berg JV, Hu J, Messer S, Herwaldt L, Pfaller M, Diekema D. Attributable mortality of nosocomial candidemia, revisited. Clin Infect Dis. 2003;37(9):1172–1177. doi:10.1086/375875.
- Sheppard DC, Filler SG. Host cell invasion by medically important fungi. Cold Spring Harb Perspect Med. 2014 Nov 3;5(1):a019687. doi:10.1101/cshperspect.a019687.
- Zhu W, Filler SG. Interactions of Candida albicans with epithelial cells. Cell Microbiol. 2010 Mar;12(3):273–282. doi:10.1111/j.1462-5822.2009.01412.x.
- Gouba N, Drancourt M. Digestive tract mycobiota: a source of infection. Med Mal Infect. 2015;45(1–2):9–16. doi:10.1016/j.medmal.2015.01.007.
- Saiman L, Ludington E, Pfaller M, Rangel-Frausto S, Wiblin RT, Dawson J, Blumberg HM, Patterson JE, Rinaldi M, Edwards JE, et al. Risk factors for candidemia in neonatal intensive care unit patients. Pediatr Infect Dis J. 2000;19(4):319–324. doi:10.1097/00006454-200004000-00011.
- Goyer M, Loiselet A, Bon F, L’Ollivier C, Laue M, Holland G, Bonnin A, Dalle F. Intestinal Cell Tight Junctions Limit Invasion of Candida albicans through Active Penetration and Endocytosis in the Early Stages of the Interaction of the Fungus with the Intestinal Barrier. PLoS One. 2016;11(3):e0149159. doi:10.1371/journal.pone.0149159.
- Dalle F, Wächtler B, L’Ollivier C, Holland G, Bannert N, Wilson D Labruère C, Bonnin A, Hube B. Cellular interactions of Candida albicans with human oral epithelial cells and enterocytes. Cell Microbiol. 2010 Feb 1;12(2):248–271. doi:10.1111/j.1462-5822.2009.01394.x.
- Albac S, Schmitz A, Lopez-Alayon C, d’Enfert C, Sautour M, Ducreux A, Labruère-Chazal C, Laue M, Holland G, Bonnin A, et al. Candida albicans is able to use M cells as a portal of entry across the intestinal barrier in vitro. Cell Microbiol. 2016 Feb;18(2):195–210. doi:10.1111/cmi.12570.
- Perlroth J, Choi B, Spellberg B. Nosocomial fungal infections: epidemiology, diagnosis, and treatment. Med Mycol. 2007 Jun;45(4):321–346. doi:10.1080/13693780701218689.
- Zaborin A, Smith D, Garfield K, Quensen J, Shakhsheer B, Kade M Tirrell M, Tiedje J, Gilbert JA, Zaborina O, et al. Membership and behavior of ultra-low-diversity pathogen communities present in the gut of humans during prolonged critical illness. mBio. 2014 Sep 23;5(5):e01361–01314. doi:10.1128/mBio.01361-14.
- Koh AY. Murine models of Candida gastrointestinal colonization and dissemination. Eukaryot Cell. 2013 Nov;12(11):1416–1422. doi:10.1128/EC.00196-13.
- Sokol H, Leducq V, Aschard H, Pham H-P, Jegou S, Landman C, Cohen D, Liguori G, Bourrier A, Nion-Larmurier I, et al. Fungal microbiota dysbiosis in IBD. Gut. 2017;66(6):1039–1048. doi:10.1136/gutjnl-2015-310746.
- Botschuijver S, Roeselers G, Levin E, Jonkers DM, Welting O, Heinsbroek SEM, de Weerd HH, Boekhout T, Fornai M, Masclee AA, et al. Intestinal Fungal Dysbiosis Is Associated With Visceral Hypersensitivity in Patients With Irritable Bowel Syndrome and Rats. Gastroenterology. 2017;153(4):1026–1039. doi:10.1053/j.gastro.2017.06.004.
- Wheeler ML, Limon JJ, Bar AS, Leal CA, Gargus M, Tang J, Brown J, Funari VA, Wang HL, Crother TR, et al. Immunological Consequences of Intestinal Fungal Dysbiosis. Cell Host Microbe. 2016 Jun 8;19(6):865–873. doi:10.1016/j.chom.2016.05.003.
- Standaert-Vitse A, Sendid B, Joossens M, François N, Vandewalle-El Khoury P, Branche J, Van Kruiningen H, Jouault T, Rutgeerts P, Gower-Rousseau C, et al. Candida albicans colonization and ASCA in familial Crohn’s disease. Am J Gastroenterol. 2009 Jul;104(7):1745–1753. doi:10.1038/ajg.2009.225.
- Chehoud C, Albenberg LG, Judge C, Hoffmann C, Grunberg S, Bittinger K, Baldassano RN, Lewis JD, Bushman FD, Wu GD. Fungal Signature in the Gut Microbiota of Pediatric Patients With Inflammatory Bowel Disease. Inflamm Bowel Dis. 2015 Aug;21(8):1948–1956. doi:10.1097/MIB.0000000000000454.
- Sommer F, Bäckhed F. The gut microbiota—masters of host development and physiology. Nat Rev Microbiol. 2013;11(4):227. doi:10.1038/nrmicro2974.
- Limon JJ, Skalski JH, Underhill DM. Commensal Fungi in Health and Disease. Cell Host Microbe. 2017 Aug 9;22(2):156–165. doi:10.1016/j.chom.2017.07.002.
- Mohajeri MH, Brummer RJM, Rastall RA, Weersma RK, Harmsen HJM, Faas M, Eggersdorfer M. The role of the microbiome for human health: from basic science to clinical applications. Eur J Nutr. 2018 May 10. doi:10.1007/s00394-018-1703-4.
- Faith JJ, Guruge JL, Charbonneau M, Subramanian S, Seedorf H, Goodman AL, Clemente JC, Knight R, Heath AC, Leibel RL, et al. The long-term stability of the human gut microbiota. Science. 2013;341(6141):1237439. doi:10.1126/science.1237439.
- Sekirov I, Russell SL, Antunes LCM, Finlay BB. Gut microbiota in health and disease. Physiol Rev. 2010;90(3):859–904. doi:10.1152/physrev.00045.2009.
- David LA, Maurice CF, Carmody RN, Gootenberg DB, Button JE, Wolfe BE, Ling AV, Devlin AS, Varma Y, Fischbach MA, et al. Diet rapidly and reproducibly alters the human gut microbiome. Nature. 2014;505(7484):559. doi:10.1038/nature12820.
- Rolhion N, Chassaing B. When pathogenic bacteria meet the intestinal microbiota. Philos Trans R Soc Lond B Biol Sci. 2016 Nov 5;371:1707. doi:10.1098/rstb.2015.0504.
- Allaire JM, Crowley SM, Law HT, Chang S-Y, Ko H-J, Vallance BA. The Intestinal Epithelium: central Coordinator of Mucosal Immunity. Trends Immunol. 2018;39(9):677–696.
- Carding S, Verbeke K, Vipond DT, Corfe BM, Owen LJ. Dysbiosis of the gut microbiota in disease. Microb Ecol Health Dis. 2015 Jan 1;26(s2):26191.
- Cox LM, Weiner HL. Microbiota Signaling Pathways that Influence Neurologic Disease. Neurotherapeutics. 2018 Jan 1;15(1):135–145. doi:10.1007/s13311-017-0598-8.
- Geuking MB, Köller Y, Rupp S, McCoy KD. The interplay between the gut microbiota and the immune system. Gut Microbes. 2014;5(3):411–418. doi:10.4161/gmic.29330.
- Rescigno M. Intestinal microbiota and its effects on the immune system. Cell Microbiol. 2014;16(7):1004–1013. doi:10.1111/cmi.2014.16.issue-7.
- Sittipo P, Lobionda S, Lee YK, Maynard CL. Intestinal microbiota and the immune system in metabolic diseases. J Microbiol. 2018;56(3):154–162. doi:10.1007/s12275-018-7548-y.
- Hallen-Adams HE, Suhr MJ. Fungi in the healthy human gastrointestinal tract. Virulence. 2017 03;8(3):352–358. doi:10.1080/21505594.2016.1247140.
- Iliev ID, Leonardi I. Fungal dysbiosis: immunity and interactions at mucosal barriers. Nat Rev Immunol. 2017 Oct;17(10):635–646. doi:10.1038/nri.2017.55.
- Mar Rodríguez M, Pérez D, Javier Chaves F, Esteve E, Marin-Garcia P, Xifra G, Vendrell J, Jovè M, Pamplona R, Ricart W, et al. Obesity changes the human gut mycobiome. Sci Rep. 2015 Oct;12(5):14600. doi:10.1038/srep14600.
- Gerbe F, Legraverend C, Jay P. The intestinal epithelium tuft cells: specification and function. Cell Mol Life Sci. 2012;69(17):2907–2917. doi:10.1007/s00018-012-0984-7.
- Mowat AM. To respond or not to respond — a personal perspective of intestinal tolerance. Nat Rev Immunol. 2018 Jun;18(6):405–415. doi:10.1038/s41577-018-0002-x.
- Holmes R, Lobley RW. Intestinal brush border revisited. Gut. 1989;30(12):1667. doi:10.1136/gut.30.12.1667.
- Suzuki T. Regulation of intestinal epithelial permeability by tight junctions. Cell Mol Life Sci. 2013 Feb 1;70(4):631–659. doi:10.1007/s00018-012-1070-x.
- Tsukita S, Furuse M, Itoh M. Multifunctional strands in tight junctions. Nat Rev Mol Cell Biol. 2001 Apr;2(4):285–293. doi:10.1038/35067088.
- Pearce SC, Al-Jawadi A, Kishida K, Yu S, Hu M, Fritzky LF, Edelblum KL, Gao N, Ferraris RP. Marked differences in tight junction composition and macromolecular permeability among different intestinal cell types. BMC Biol. 2018;16(1):19. doi:10.1186/s12915-018-0481-z.
- Hartsock A, Nelson WJ. Adherens and tight junctions: structure, function and connections to the actin cytoskeleton. Biochim Biophys Acta BBA-Biomembr. 2008;1778(3):660–669. doi:10.1016/j.bbamem.2007.07.012.
- Assimakopoulos SF, Papageorgiou I, Charonis A. Enterocytes’ tight junctions: from molecules to diseases. World J Gastrointest Pathophysiol. 2011;2(6):123. doi:10.4291/wjgp.v2.i6.123.
- Zihni C, Mills C, Matter K, Balda MS. Tight junctions: from simple barriers to multifunctional molecular gates. Nat Rev Mol Cell Biol. 2016;17(9):564–580. doi:10.1038/nrm.2016.80.
- Bouhet S, Oswald IP. The effects of mycotoxins, fungal food contaminants, on the intestinal epithelial cell-derived innate immune response. Vet Immunol Immunopathol. 2005;108(1–2):199–209. doi:10.1016/j.vetimm.2005.08.010.
- Lewis SA, Berg JR, Kleine TJ. Modulation of epithelial permeability by extracellular macromolecules. Physiol Rev. 1995 Jul;75(3):561–589. doi:10.1152/physrev.1995.75.3.561.
- Ulluwishewa D, Anderson RC, McNabb WC, Moughan PJ, Wells JM, Roy NC. Regulation of tight junction permeability by intestinal bacteria and dietary components. J Nutr. 2011 May;141(5):769–776. doi:10.3945/jn.110.135657.
- Wang W, Uzzau S, Goldblum SE, Fasano A. Human zonulin, a potential modulator of intestinal tight junctions. J Cell Sci. 2000;113:4435–4440.
- Fasano A. Zonulin, regulation of tight junctions, and autoimmune diseases. Ann N Y Acad Sci. 2012;1258(1):25–33. doi:10.1111/j.1749-6632.2012.06538.x.
- Martens EC, Neumann M, Desai MS. Interactions of commensal and pathogenic microorganisms with the intestinal mucosal barrier. Nat Rev Microbiol. 2018;16(8):457–470.
- Tapia R, Kralicek S, Hecht G. EPEC Perturbation of Crb3 and Pals1 Localization Precedes Tight Junction Disruption. Faseb J. 2017;31: 1043–10.
- Anderson RC, Cookson AL, McNabb WC, Park Z, McCann MJ, Kelly WJ, Roy NC. Lactobacillus plantarum MB452 enhances the function of the intestinal barrier by increasing the expression levels of genes involved in tight junction formation. BMC Microbiol. 2010 Dec 9;10:316. doi:10.1186/1471-2180-10-316.
- El Asmar R, Panigrahi P, Bamford P, Berti I, Not T, Coppa GV, Catassi C, Fasano A. Host-dependent activation of the zonulin system is involved in the impairment of the gut barrier function following bacterial colonization. Gastroenterology. 2002;123(5):1607–1615. doi:10.1053/gast.2002.36578.
- Alam A, Neish A. Role of gut microbiota in intestinal wound healing and barrier function. Tissue Barriers. 2018 Jul 3;6(3):1539595. doi:10.1080/21688370.2018.1539595.
- Awad WA, Hess C, Hess M. Enteric Pathogens and Their Toxin-Induced Disruption of the Intestinal Barrier through Alteration of Tight Junctions in Chickens. Toxins. 2017 Feb 10;9(2):pii: E60.
- Torres-Flores JM, Arias CF. Tight Junctions Go Viral!. Viruses. 2015 Sep 23;7(9):5145–5154. doi:10.3390/v7092865.
- Di Genova BM, Tonelli RR. Infection Strategies of Intestinal Parasite Pathogens and Host Cell Responses. Front Microbiol. 2016;7:256. doi:10.3389/fmicb.2016.00256.
- Hattrup CL, Gendler SJ. Structure and function of the cell surface (tethered) mucins. Annu Rev Physiol. 2008;70:431–457. doi:10.1146/annurev.physiol.70.113006.100659.
- Deplancke B, Gaskins HR. Microbial modulation of innate defense: goblet cells and the intestinal mucus layer. Am J Clin Nutr. 2001;73(6):1131S–1141S. doi:10.1093/ajcn/73.6.1131S.
- Naughton J, Duggan G, Bourke B, Clyne M. Interaction of microbes with mucus and mucins: recent developments. Gut Microbes. 2014;5(1):48–52. doi:10.4161/gmic.26680.
- Petersson J, Schreiber O, Hansson GC, Gendler SJ, Velcich A, Lundberg JO et al. Importance and regulation of the colonic mucus barrier in a mouse model of colitis. Am J Physiol-Gastrointest Liver Physiol. 2010 Nov 25;300(2):G327–33. doi:10.1152/ajpgi.00422.2010.
- Kandori H, Hirayama K, Takeda M, Doi K. Histochemical, lectin-histochemical and morphometrical characteristics of intestinal goblet cells of germfree and conventional mice. Exp Anim. 1996;45:155–160.
- Ishikawa K, Satoh Y, Oomori Y, Yamano M, Matsuda M, Ono K. Influence of conventionalization on cecal wall structure of germ-free Wistar rats: quantitative light and qualitative electron microscopic observations. Anat Embryol (Berl). 1989;180(2):191–198. doi:10.1007/BF00309771.
- Li H, Limenitakis JP, Fuhrer T, Geuking MB, Lawson MA, Wyss M, Brugiroux S, Keller I, Macpherson JA, Rupp S, et al. The outer mucus layer hosts a distinct intestinal microbial niche. Nat Commun. 2015;6:8292. doi:10.1038/ncomms9292.
- Ouellette AJ, Darmoul D, Tran D, Huttner KM, Yuan J, Selsted ME. Peptide localization and gene structure of cryptdin 4, a differentially expressed mouse paneth cell alpha-defensin. Infect Immun. 1999 Dec;67(12):6643–6651.
- Porter EM, Liu L, Oren A, Anton PA, Ganz T. Localization of human intestinal defensin 5 in Paneth cell granules. Infect Immun. 1997 Jun;65(6):2389–2395.
- Ghoos Y, Vantrappen G. The cytochemical localization of lysozyme in Paneth cell granules. Histochem J. 1971 May;3(3):175–178. doi:10.1007/BF01002560.
- Longman RJ, Douthwaite J, Sylvester PA, Poulsom R, Corfield AP, Thomas MG, Wright NA. Coordinated localisation of mucins and trefoil peptides in the ulcer associated cell lineage and the gastrointestinal mucosa. Gut. 2000;47(6):792–800.
- Corfield AP. Mucins: a biologically relevant glycan barrier in mucosal protection. Biochim Biophys Acta BBA-Gen Subj. 2015;1850(1):236–252. doi:10.1016/j.bbagen.2014.05.003.
- Johansson MEV, Hansson GC. Immunological aspects of intestinal mucus and mucins. Nat Rev Immunol. 2016 Oct;16(10):639–649. doi:10.1038/nri.2016.88.
- McGuckin MA, Lindén SK, Sutton P, Florin TH. Mucin dynamics and enteric pathogens. Nat Rev Microbiol. 2011 Apr;9(4):265–278. doi:10.1038/nrmicro2538.
- Corfield AP, Wagner SA, Clamp JR, Kriaris MS, Hoskins LC. Mucin degradation in the human colon: production of sialidase, sialate O-acetylesterase, N-acetylneuraminate lyase, arylesterase, and glycosulfatase activities by strains of fecal bacteria. Infect Immun. 1992;60:3971–3978.
- Corfield AP, Wagner SA, O’Donnell LJ, Durdey P, Mountford RA, Clamp JR. The roles of enteric bacterial sialidase, sialateO-acetyl esterase and glycosulfatase in the degradation of human colonic mucin. Glycoconj J. 1993;10(1):72–81. doi:10.1007/BF00731190.
- Haider K, Hossain A, Wanke C, Qadri F, Ali S, Nahar S. Production of mucinase and neuraminidase and binding of Shigella to intestinal mucin. J Diarrhoeal Dis Res. 1993;11(2):88–92.
- Homer KA, Whiley RA, Beighton D. Production of specific glycosidase activities by Streptococcus intermedius strain UNS35 grown in the presence of mucin. J Med Microbiol. 1994;41(3):184–190. doi:10.1099/00222615-41-3-184.
- Moncada DM, Kammanadiminti SJ, Chadee K. Mucin and Toll-like receptors in host defense against intestinal parasites. Trends Parasitol. 2003;19(7):305–311. doi:10.1016/S1471-4922(03)00122-3.
- Lidell ME, Moncada DM, Chadee K, Hansson GC. Entamoeba histolytica cysteine proteases cleave the MUC2 mucin in its C-terminal domain and dissolve the protective colonic mucus gel. Proc Natl Acad Sci. 2006;103(24):9298–9303. doi:10.1073/pnas.0600623103.
- Cooke CL, An HJ, Kim J, Canfield DR, Torres J, Lebrilla CB, Solnick JV. Modification of gastric mucin oligosaccharide expression in rhesus macaques after infection with Helicobacter pylori. Gastroenterology. 2009;137(3):1061–1071. doi:10.1053/j.gastro.2009.04.014.
- Skoog EC, Sjöling A, Navabi N, Holgersson J, Lundin SB, Lindén SK. Human gastric mucins differently regulate Helicobacter pylori proliferation, gene expression and interactions with host cells. PloS One. 2012;7:e36378.
- Tu QV, McGuckin MA, Mendz GL. Campylobacter jejuni response to human mucin MUC2: modulation of colonization and pathogenicity determinants. J Med Microbiol. 2008;57(7):795–802. doi:10.1099/jmm.0.47752-0.
- Bevins CL, Salzman NH. Paneth cells, antimicrobial peptides and maintenance of intestinal homeostasis. Nat Rev Microbiol. 2011 May;9(5):356–368. doi:10.1038/nrmicro2546.
- Ayabe T, Satchell DP, Wilson CL, Parks WC, Selsted ME, Ouellette AJ. Secretion of microbicidal α-defensins by intestinal Paneth cells in response to bacteria. Nat Immunol. 2000 Aug;1(2):113–118. doi:10.1038/77783.
- Satoh Y, Habara Y, Ono K, Kanno T. Carbamylcholine- and catecholamine-induced intracellular calcium dynamics of epithelial cells in mouse ileal crypts. Gastroenterology. 1995 May;108(5):1345–1356. doi:10.1016/0016-5085(95)90681-9.
- Kiyohara H, Egami H, Shibata Y, Murata K, Ohshima S, Ogawa M. Light microscopic immunohistochemical analysis of the distribution of group II phospholipase A2 in human digestive organs. J Histochem Cytochem Off J Histochem Soc. 1992 Nov;40(11):1659–1664. doi:10.1177/40.11.1431054.
- Lasserre C, Colnot C, Bréchot C, Poirier F. HIP/PAP gene, encoding a C-type lectin overexpressed in primary liver cancer, is expressed in nervous system as well as in intestine and pancreas of the postimplantation mouse embryo. Am J Pathol. 1999 May;154(5):1601–1610. doi:10.1016/S0002-9440(10)65413-2.
- Holly MK, Smith JG. Paneth Cells during Viral Infection and Pathogenesis. Viruses [Internet]. 2018 Apr 26 [cited 2018 Jul 4];10(5). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5977218/.
- Hooper LV, Stappenbeck TS, Hong CV, Gordon JI. Angiogenins: a new class of microbicidal proteins involved in innate immunity. Nat Immunol. 2003 Mar;4(3):269–273. doi:10.1038/ni888.
- Hase K. Cell Differentiation Is a Key Determinant of Cathelicidin LL-37/Human Cationic Antimicrobial Protein 18 Expression by Human Colon Epithelium. Infect Immun. 2002 Feb 1;70(2):953–963. doi:10.1128/IAI.70.2.953-963.2002.
- Nakamura K, Sakuragi N, Takakuwa A, Ayabe T. Paneth cell α-defensins and enteric microbiota in health and disease. Biosci Microbiota Food Health. 2016;35(2):57–67. doi:10.12938/bmfh.16-012.
- Million M, Tomas J, Wagner C, Lelouard H, Raoult D, Gorvel J-P. New insights in gut microbiota and mucosal immunity of the small intestine. Hum Microbiome J. 2018 Apr 1;7–8:23–32. doi:10.1016/j.humic.2018.01.004.
- Mantis NJ, Rol N, Corthésy B. Secretory IgA’s complex roles in immunity and mucosal homeostasis in the gut. Mucosal Immunol. 2011 Nov;4(6):603–611. doi:10.1038/mi.2011.41.
- Stokes CR, Soothill JF, Turner MW. Immune exclusion is a function of IgA. Nature. 1975 Jun 26;255(5511):745–746. doi:10.1038/255745a0.
- Pabst O, Cerovic V, Hornef M. Secretory IgA in the Coordination of Establishment and Maintenance of the Microbiota. Trends Immunol. 2016;37(5):287–296. doi:10.1016/j.it.2016.03.002.
- Forbes SJ, Bumpus T, McCarthy EA, Corthésy B, Mantis NJ. Transient suppression of Shigella flexneri type 3 secretion by a protective O-antigen-specific monoclonal IgA. mBio. 2011;2(3):e00042–00011. doi:10.1128/mBio.00042-11.
- Vaerman JP, Derijck-Langendries A, Rits M, Delacroix D. Neutralization of cholera toxin by rat bile secretory IgA antibodies. Immunology. 1985 Mar;54(3):601–603.
- Peterson LW, Artis D. Intestinal epithelial cells: regulators of barrier function and immune homeostasis. Nat Rev Immunol. 2014 Mar;14(3):141–153. doi:10.1038/nri3608.
- Howe KL, Reardon C, Wang A, Nazli A, McKay DM. Transforming growth factor-β regulation of epithelial tight junction proteins enhances barrier function and blocks enterohemorrhagic Escherichia coli O157: H7-induced increased permeability. Am J Pathol. 2005;167(6):1587–1597. doi:10.1016/S0002-9440(10)61243-6.
- Madsen KL, Lewis SA, Tavernini MM, Hibbard J, Fedorak RN. Interleukin 10 prevents cytokine-induced disruption of T84 monolayer barrier integrity and limits chloride secretion. Gastroenterology. 1997;113(1):151–159. doi:10.1016/S0016-5085(97)70090-8.
- Huttner KM, Bevins CL. Antimicrobial peptides as mediators of epithelial host defense. Pediatr Res. 1999;45(6):785. doi:10.1203/00006450-199906000-00001.
- Watanabe M, Ueno Y, Yajima T, Iwao Y, Tsuchiya M, Ishikawa H, Aiso S, Hibi T, Ishii H. Interleukin 7 is produced by human intestinal epithelial cells and regulates the proliferation of intestinal mucosal lymphocytes. J Clin Invest. 1995;95(6):2945–2953. doi:10.1172/JCI118002.
- Dwinell MB, Eckmann L, Leopard JD, Varki NM, Kagnoff MF. Chemokine receptor expression by human intestinal epithelial cells. Gastroenterology. 1999;117(2):359–367. doi:10.1053/gast.1999.0029900359.
- Jung HC, Eckmann L, Yang S-K, Panja A, Fierer J, Morzycka-Wroblewska E, Kagnoff MF. A distinct array of proinflammatory cytokines is expressed in human colon epithelial cells in response to bacterial invasion. J Clin Invest. 1995;95(1):55–65. doi:10.1172/JCI117676.
- Rooks MG, Garrett WS. Gut microbiota, metabolites and host immunity. Nat Rev Immunol. 2016 Jun;16(6):341–352. doi:10.1038/nri.2016.42.
- Schoenborn AA, Furstenberg RJ von, Valsaraj S, Hussain Fs, Stein M, Shanahan MT, Henning SJ, Gulati AS. The enteric microbiota regulates jejunal Paneth cell number and function without impacting intestinal stem cells. Gut Microbes. 2019;10(1):45–58.
- Schoenborn AA, Furstenberg R von, Valsaraj S, Hussain FS, Stein M, Shanahan M, Gulati AS, Henning SJ. The Enteric Microbiota Regulates Paneth Cell Number and Function Without Affecting Intestinal Stem Cells. Gastroenterology. 2017 Apr 1;152(5):S13.
- Ahluwalia B, Magnusson MK, Öhman L. Mucosal immune system of the gastrointestinal tract: maintaining balance between the good and the bad. Scand J Gastroenterol. 2017 Nov;52(11):1185–1193. doi:10.1080/00365521.2017.1349173.
- Hooper LV, Littman DR, Macpherson AJ. Interactions between the microbiota and the immune system. Science. 2012;336(6086):1268–1273. doi:10.1126/science.1223490.
- Ohno H. Intestinal M cells. J Biochem (Tokyo). 2015;159(2):151–160. doi:10.1093/jb/mvv121.
- Chen Y, Chou K, Fuchs E, Havran WL, Boismenu R. Protection of the intestinal mucosa by intraepithelial γδ T cells. Proc Natl Acad Sci. 2002 Oct 29;99(22):14338–14343. doi:10.1073/pnas.212290499.
- Ismail AS, Severson KM, Vaishnava S, Behrendt CL, Yu X, Benjamin JL, Ruhn KA, Hou B, DeFranco AL, Yarovinsky F, et al. γδ intraepithelial lymphocytes are essential mediators of host–microbial homeostasis at the intestinal mucosal surface. Proc Natl Acad Sci. 2011 May 24;108(21):8743–8748. doi:10.1073/pnas.1019574108.
- Smith P, Garrett W. The Gut Microbiota and Mucosal T Cells. Front Microbiol [Internet]. 2011 [cited 2019 Mar 7];2. https://www.frontiersin.org/articles/10.3389/fmicb.2011.00111/full.
- Romani L, Bistoni F, Puccetti P. Adaptation of Candida albicans to the host environment: the role of morphogenesis in virulence and survival in mammalian hosts. Curr Opin Microbiol. 2003;6:338–343.
- Pearson C, Uhlig HH, Powrie F. Lymphoid microenvironments and innate lymphoid cells in the gut. Trends Immunol. 2012 Jun 1;33(6):289–296. doi:10.1016/j.it.2012.04.004.
- Kurashima Y, Kiyono H. Mucosal Ecological Network of Epithelium and Immune Cells for Gut Homeostasis and Tissue Healing. Annu Rev Immunol. 2017;35(1):119–147. doi:10.1146/annurev-immunol-051116-052424.
- Farstad IN, Halstensen TS, Fausa O, Brandtzaeg P. Heterogeneity of M-cell-associated B and T cells in human Peyer’s patches. Immunology. 1994;83:457.
- Collins JK, Cashman S, Morgan J, O’Sullivan GC. The Gastrointestinal Immune System: recognising Microbes in the Gut.
- Kanaya T, Hase K, Takahashi D, Fukuda S, Hoshino K, Sasaki I, Hemmi H, Knoop KA, Kumar N, Sato M, et al. The Ets transcription factor Spi-B is essential for the differentiation of intestinal microfold cells. Nat Immunol. 2012 Jun 17;13(8):729–736. doi:10.1038/ni.2352.
- Sato S, Kaneto S, Shibata N, Takahashi Y, Okura H, Yuki Y, et al. Transcription factor Spi-B-dependent and -independent pathways for the development of Peyer’s patch M cells. Mucosal Immunol. 2013 Jul;6(4):838–846. doi:10.1038/mi.2012.122.
- Ouzilou L, Caliot E, Pelletier I, Prévost M-C, Pringault E, Colbère-Garapin F. Poliovirus transcytosis through M-like cells. J Gen Virol. 2002;83(9):2177–2182. doi:10.1099/0022-1317-83-9-2177.
- Jones BD, Ghori N, Falkow S. Salmonella typhimurium initiates murine infection by penetrating and destroying the specialized epithelial M cells of the Peyer’s patches. J Exp Med. 1994;180:15–23.
- Schulte R, Kerneis S, Klinke S, Bartels H, Preger S, Kraehenbuhl J-P, Pringault E, Autenrieth IB. Translocation of Yersinia enterocolitica across reconstituted intestinal epithelial monolayers is triggered by Yersinia invasin binding to β1 integrins apically expressed on M-like cells. Cell Microbiol. 2000;2(2):173–185. doi:10.1046/j.1462-5822.2000.00047.x.
- Tahoun A, Mahajan S, Paxton E, Malterer G, Donaldson DS, Wang D, Tan A, Gillespie TL, O’Shea M, Roe AJ, et al. Salmonella transforms follicle-associated epithelial cells into M cells to promote intestinal invasion. Cell Host Microbe. 2012;12(5):645–656. doi:10.1016/j.chom.2012.10.009.
- Grassl GA, Bohn E, Müller Y, Bühler OT, Autenrieth IB. Interaction of Yersinia enterocolitica with epithelial cells: invasin beyond invasion. Int J Med Microbiol. 2003;293(1):41–54. doi:10.1078/1438-4221-00243.
- Clark MA, Jepson MA. Intestinal M cells and their role in bacterial infection. Int J Med Microbiol. 2003;293(1):17–39. doi:10.1078/1438-4221-00242.
- Calderone RA, Clancy CJ. Candida and candidiasis. American Society for Microbiology Press. 2002; 8(8): 876.
- Whiteway M, Bachewich C. Morphogenesis in Candida albicans. Annu Rev Microbiol. 2007;61:529–553. doi:10.1146/annurev.micro.61.080706.093341.
- Navarathna DH, Pathirana RU, Lionakis MS, Nickerson KW, Roberts DD. Candida albicans ISW2 regulates chlamydospore suspensor cell formation and virulence in vivo in a mouse model of disseminated candidiasis. PLoS One. 2016;11(10):e0164449. doi:10.1371/journal.pone.0164449.
- Slutsky B, Staebell M, Anderson J, Risen L, Pfaller MT, Soll DR. “White-opaque transition”: a second high-frequency switching system in Candida albicans. J Bacteriol. 1987;169:189–197.
- Ene IV, Lohse MB, Vladu AV, Morschhäuser J, Johnson AD, Bennett RJ. Phenotypic profiling reveals that Candida albicans opaque cells represent a metabolically specialized cell state compared to default white cells. MBio. 2016;7(6):e01269–16. doi:10.1128/mBio.01269-16.
- Lachke SA, Lockhart SR, Daniels KJ, Soll DR. Skin facilitates Candida albicans mating. Infect Immun. 2003;71(9):4970–4976. doi:10.1128/IAI.71.9.4970-4976.2003.
- Lohse MB, Johnson AD. Differential phagocytosis of white versus opaque Candida albicans by Drosophila and mouse phagocytes. PLoS One. 2008;3(1):e1473. doi:10.1371/journal.pone.0001473.
- Lohse MB, Johnson AD. White–opaque switching in Candida albicans. Curr Opin Microbiol. 2009;12(6):650–654. doi:10.1016/j.mib.2009.09.010.
- Morschhäuser J. Regulation of white-opaque switching in Candida albicans. Med Microbiol Immunol (Berl). 2010;199(3):165–172. doi:10.1007/s00430-010-0147-0.
- Miller MG, Johnson AD. White-opaque switching in Candida albicans is controlled by mating-type locus homeodomain proteins and allows efficient mating. Cell. 2002;110:293–302.
- Mallick EM, Bergeron AC, Jones Jr SK, Newman ZR, Brothers KM, Creton R, Wheeler RT, Bennett RJ. Phenotypic plasticity regulates Candida albicans interactions and virulence in the vertebrate host. Front Microbiol. 2016;7:780. doi:10.3389/fmicb.2016.00780.
- van Burik J-AH, Magee PT. Aspects of fungal pathogenesis in humans. Annu Rev Microbiol. 2001;55(1):743–772. doi:10.1146/annurev.micro.55.1.743.
- Pande K, Chen C, Noble SM. Passage through the mammalian gut triggers a phenotypic switch that promotes Candida albicans commensalism. Nat Genet. 2013 Sep;45(9):1088–1091. doi:10.1038/ng.2710.
- Huang G, Wang H, Chou S, Nie X, Chen J, Liu H. Bistable expression of WOR1, a master regulator of white–opaque switching in Candida albicans. Proc Natl Acad Sci. 2006;103(34):12813–12818. doi:10.1073/pnas.0605270103.
- Srikantha T, Borneman AR, Daniels KJ, Pujol C, Wu W, Seringhaus MR, Gerstein M, Yi S, Snyder M, Soll DR. TOS9 regulates white-opaque switching in Candida albicans. Eukaryot Cell. 2006;5(10):1674–1687. doi:10.1128/EC.00252-06.
- Zordan RE, Galgoczy DJ, Johnson AD. Epigenetic properties of white–opaque switching in Candida albicans are based on a self-sustaining transcriptional feedback loop. Proc Natl Acad Sci. 2006;103(34):12807–12812. doi:10.1073/pnas.0605138103.
- Iliev ID, Funari VA, Taylor KD, Nguyen Q, Reyes CN, Strom SP, Brown J, Becker CA, Fleshner PR, Dubinsky M, et al. Interactions between commensal fungi and the C-type lectin receptor Dectin-1 influence colitis. Science. 2012 Jun 8;336(6086):1314–1317. doi:10.1126/science.1221789.
- Neville BA, d’Enfert C, Bougnoux M-E. Candida albicans commensalism in the gastrointestinal tract. FEMS Yeast Res. 2015;15:7.
- Fox EP, Cowley ES, Nobile CJ, Hartooni N, Newman DK, Johnson AD. Anaerobic Bacteria Grow within Candida albicans Biofilms and Induce Biofilm Formation in Suspension Cultures. Curr Biol. 2014 Oct 20;24(20):2411–2416. doi:10.1016/j.cub.2014.08.057.
- Wang ZK, Yang YS, Stefka AT, Sun G, Peng LH. Review article: fungal microbiota and digestive diseases. Aliment Pharmacol Ther. 2014;39(8):751–766. doi:10.1111/apt.12665.
- Noverr MC, Huffnagle GB. Regulation of Candida albicans Morphogenesis by Fatty Acid Metabolites. Infect Immun. 2004 Nov 1;72(11):6206–6210. doi:10.1128/IAI.72.11.6206-6210.2004.
- García C, Tebbji F, Daigneault M, Liu N-N, Köhler JR, Allen-Vercoe E, Sellam A. The Human Gut Microbial Metabolome Modulates Fungal Growth via the TOR Signaling Pathway. mSphere. 2017 Dec;2(6)pii: e00555-17.
- Fitzsimmons N, Berry DR. Inhibition of Candida albicans by Lactobacillus acidophilus: evidence for the involvement of a peroxidase system. Microbios. 1994;80:125–133.
- Zelante T, Iannitti RG, Cunha C, De Luca A, Giovannini G, Pieraccini G, Zecchi R, D’Angelo C, Massi-Benedetti C, Fallarino F, et al. Tryptophan catabolites from microbiota engage aryl hydrocarbon receptor and balance mucosal reactivity via interleukin-22. Immunity. 2013;39(2):372–385. doi:10.1016/j.immuni.2013.05.019.
- De Luca A, Zelante T, D’angelo C, Zagarella S, Fallarino F, Spreca A, Iannitti RG, Bonifazi P, Renauld JC, Bistoni F. IL-22 defines a novel immune pathway of antifungal resistance. Mucosal Immunol. 2010;3(4):361. doi:10.1038/mi.2009.138.
- Puel A, Picard C, Cypowyj S, Lilic D, Abel L, Casanova J-L. Inborn errors of mucocutaneous immunity to Candida albicans in humans: a role for IL-17 cytokines?. Curr Opin Immunol. 2010 Aug 1;22(4):467–474. doi:10.1016/j.coi.2010.06.009.
- Rosenbach A, Dignard D, Pierce JV, Whiteway M, Kumamoto CA. Adaptations of Candida albicans for growth in the mammalian intestinal tract. Eukaryot Cell. 2010;9(7):1075–1086. doi:10.1128/EC.00011-10.
- White SJ, Rosenbach A, Lephart P, Nguyen D, Benjamin A, Tzipori S, Whiteway M, Mecsas J, Kumamoto CA. Self-regulation of Candida albicans population size during GI colonization. PLoS Pathog. 2007;3(12):e184. doi:10.1371/journal.ppat.0030184.
- Savage DC. Microbial interference between indigenous yeast and lactobacilli in the rodent stomach. J Bacteriol. 1969;98:1278–1283.
- Helstrom PB, Balish E. Effect of oral tetracycline, the microbial flora, and the athymic state on gastrointestinal colonization and infection of BALB/c mice with Candida albicans. Infect Immun. 1979;23:764–774.
- Hummel RP, Oestreicher EJ, Maley MP, Macmillan BG. Inhibition ofCandida albicans by Escherichia coli in vitro and in the germfree mouse. J Surg Res. 1973;15(1):53–58. doi:10.1016/0022-4804(73)90163-7.
- Zwielehner J, Lassl C, Hippe B, Pointner A, Switzeny OJ, Remely M, Kitzweger E, Ruckser R, Haslberger AG. Changes in human fecal microbiota due to chemotherapy analyzed by TaqMan-PCR, 454 sequencing and PCR-DGGE fingerprinting. PLoS One. 2011;6(12):e28654. doi:10.1371/journal.pone.0028654.
- Stringer AM, Gibson RJ, Bowen JM, Keefe DM. Chemotherapy-induced modifications to gastrointestinal microflora: evidence and implications of change. Curr Drug Metab. 2009;10(1):79–83. doi:10.2174/138920009787048419.
- Fan D, Coughlin LA, Neubauer MM, Kim J, Kim MS, Zhan X, Simms-Waldrip TR, Xie Y, Hooper LV, Koh AY. Activation of HIF-1α and LL-37 by commensal bacteria inhibits Candida albicans colonization. Nat Med. 2015;21(7):808. doi:10.1038/nm.3871.
- Li J, Chen D, Yu B, He J, Zheng P, Mao X, Yu J, Luo J, Tian G, Huang Z, et al. Fungi in Gastrointestinal Tracts of Human and Mice: from Community to Functions. Microb Ecol. 2018 May;75(4):821–829. doi:10.1007/s00248-017-1122-8.
- Swidergall M, Ernst JF. Interplay between Candida albicans and the antimicrobial peptide armory. Eukaryot Cell. 2014 Jun 20;EC: 00093-14.
- Chairatana P, Chiang I-L, Nolan EM. Human α-Defensin 6 Self-Assembly Prevents Adhesion and Suppresses Virulence Traits of Candida albicans. Biochemistry. 2017 28;56(8):1033–1041. doi:10.1021/acs.biochem.6b01111.
- Vandamme D, Landuyt B, Luyten W, Schoofs L. A comprehensive summary of LL-37, the factotum human cathelicidin peptide. Cell Immunol. 2012 Nov;280(1):22–35. doi:10.1016/j.cellimm.2012.11.009.
- Selsted ME, Ouellette AJ. Mammalian defensins in the antimicrobial immune response. Nat Immunol. 2005 Jun;6(6):551–557. doi:10.1038/ni1206.
- Moyes DL, Runglall M, Murciano C, Shen C, Nayar D, Thavaraj S, Kohli A, Islam A, Mora-Montes H, Challacombe SJ, et al. A Biphasic Innate Immune MAPK Response Discriminates between the Yeast and Hyphal Forms of Candida albicans in Epithelial Cells. Cell Host Microbe. 2010 Sep 16;8(3):225–235. doi:10.1016/j.chom.2010.08.002.
- Swidergall M, Ernst AM, Ernst JF. Candida albicans mucin Msb2 is a broad-range protectant against antimicrobial peptides. Antimicrob Agents Chemother. 2013 Jun 3;AAC: 00862-13.
- Meiller TF, Hube B, Schild L, Shirtliff ME, Scheper MA, Winkler R, Ton A. A novel immune evasion strategy of Candida albicans: proteolytic cleavage of a salivary antimicrobial peptide. PLoS One. 2009;4(4):e5039. doi:10.1371/journal.pone.0005039.
- Puri S, Kumar R, Chadha S, Tati S, Conti HR, Hube B, Cullen PJ, Edgerton M. Secreted aspartic protease cleavage of Candida albicans Msb2 activates Cek1 MAPK signaling affecting biofilm formation and oropharyngeal candidiasis. PLoS One. 2012;7(11):e46020. doi:10.1371/journal.pone.0046020.
- Szafranski-Schneider E, Swidergall M, Cottier F, Tielker D, Román E, Pla J, Ernst JF. Msb2 Shedding Protects Candida albicans against Antimicrobial Peptides. PLoS Pathog [Internet], 2012 Feb 2 [cited 2019 Jan 10];8(2). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3271078/.
- Argimón S, Fanning S, Blankenship JR, Mitchell AP. Interaction between the Candida albicans High-Osmolarity Glycerol (HOG) Pathway and the Response to Human β-Defensins 2 and 3. Eukaryot Cell. 2011 Feb;10(2):272–275. doi:10.1128/EC.00303-10.
- Jung S-I, Finkel JS, Solis NV, Chaili S, Mitchell AP, Yeaman MR, Filler SG. Bcr1 Functions Downstream of Ssd1 To Mediate Antimicrobial Peptide Resistance in Candida albicans. Eukaryot Cell. 2013 Mar;12(3):411–419. doi:10.1128/EC.00285-12.
- Lis M, Liu TT, Barker KS, Rogers PD, Bobek LA. Antimicrobial peptide MUC7 12-mer activates the calcium/calcineurin pathway in Candida albicans. FEMS Yeast Res. 2010 Aug 1;10(5):579–586.
- Li R, Kumar R, Tati S, Puri S, Edgerton MC. albicans Flu1 mediated efflux of salivary Histatin 5 reduces its cytosolic concentration and fungicidal activity. Antimicrob Agents Chemother. 2013;57(4):1832–1839.
- Rapala-Kozik M, Bochenska O, Zawrotniak M, Wolak N, Trebacz G, Gogol M, Ostrowska D, Aoki W, Ueda M, Kozik A. Inactivation of the Antifungal and Immunomodulatory Properties of Human Cathelicidin LL-37 by Aspartic Proteases Produced by the Pathogenic Yeast Candida albicans. Infect Immun. 2015 Jun;83(6):2518–2530. doi:10.1128/IAI.00023-15.
- Alonso-Monge R, Carvaihlo S, Nombela C, Rial E, Pla J. The Hog1 MAP kinase controls respiratory metabolism in the fungal pathogen Candida albicans. Microbiology. 2009;155(2):413–423. doi:10.1099/mic.0.023309-0.
- Vylkova S, Jang WS, Li W, Nayyar N, Edgerton M. Histatin 5 initiates osmotic stress response in Candida albicans via activation of the Hog1 mitogen-activated protein kinase pathway. Eukaryot Cell. 2007;6(10):1876–1888. doi:10.1128/EC.00039-07.
- Ostaff MJ, Stange EF, Wehkamp J. Antimicrobial peptides and gut microbiota in homeostasis and pathology. EMBO Mol Med. 2013 Oct;5(10):1465–1483. doi:10.1002/emmm.201201876.
- Böhringer M, Pohlers S, Schulze S, Albrecht-Eckardt D, Piegsa J, Weber M, Martin R, Hünniger K, Linde J, Guthke R, et al. Candida albicans infection leads to barrier breakdown and a MAPK/NF-κB mediated stress response in the intestinal epithelial cell line C2BBe1. Cell Microbiol. 2016;18(7):889–904.
- Moyes DL, Shen C, Murciano C, Runglall M, Richardson JP, Arno M Aldecoa-Otalora E, Naglik JR. Protection Against Epithelial Damage During Candida albicans Infection Is Mediated by PI3K/Akt and Mammalian Target of Rapamycin Signaling. J Infect Dis. 2014 Jun 1;209(11):1816–1826. doi:10.1093/infdis/jit541.
- Allert S, Förster TM, Svensson C-M, Richardson JP, Pawlik T, Hebecker B, Rudolphi S, Juraschitz M, Schaller M, Blagojevic M, et al. Candida albicans-Induced Epithelial Damage Mediates Translocation through Intestinal Barriers. mBio. 2018 May 7;9(3):e00915–18. doi:10.1128/mBio.00915-18.
- Kavanaugh NL, Zhang AQ, Nobile CJ, Johnson AD, Ribbeck K. Mucins Suppress Virulence Traits of Candida albicans. mBio. 2014 Dec 31;5(6):e01911–14. doi:10.1128/mBio.01911-14.
- Prieto D, Correia I, Pla J, Román E. Adaptation of Candida albicans to commensalism in the gut. Future Microbiol. 2016 Apr 1;11(4):567–583. doi:10.2217/fmb.16.1.
- Repentigny L, De, Aumont F, Bernard K, Belhumeur P. Characterization of Binding of Candida albicans to Small Intestinal Mucin and Its Role in Adherence to Mucosal Epithelial Cells. Infect Immun. 2000 Jan 6;68(6):3172–3179. doi:10.1128/IAI.68.6.3172-3179.2000.
- Böhm L, Torsin S, Tint SH, Eckstein MT, Ludwig T, Pérez JC. The yeast form of the fungus Candida albicans promotes persistence in the gut of gnotobiotic mice. PLoS Pathog [Internet]. 2017 Oct 25 [cited 2018 Jul 17];13(10). https://www.ncbi.nlm.nih.gov/pmc/articles/PMC5673237/.
- Colina AR, Aumont F, Deslauriers N, Belhumeur P, Repentigny LD. Evidence for degradation of gastrointestinal mucin by Candida albicans secretory aspartyl proteinase. Infect Immun. 1996 Jan 11;64(11):4514–4519.
- Crowther RS, Roomi NW, Fahim RE, Forstner JF. Vibrio cholerae metalloproteinase degrades intestinal mucin and facilitates enterotoxin-induced secretion from rat intestine. Biochim Biophys Acta BBA-Gen Subj. 1987;924(3):393–402. doi:10.1016/0304-4165(87)90153-X.
- Mantle M, Husar SD. Adhesion of Yersinia enterocolitica to purified rabbit and human intestinal mucin. Infect Immun. 1993;61:2340–2346.
- Roberton AM, Stanley RA. In vitro utilization of mucin by Bacteroides fragilis. Appl Environ Microbiol. 1982;43:325–330.
- Slomiany BL, Murty VLN, Piotrowski J, Liau YH, Sundaram P, Slomiany A. Glycosulfatase activity of Helicobacter pylori toward gastric mucin. Biochem Biophys Res Commun. 1992;183:506–513.
- Yang Y-L. Virulence factors of Candida species. J Microbiol Immunol Infect. 2003;36:223–228.
- Modrzewska B, Kurnatowski P. Adherence of Candida sp. to host tissues and cells as one of its pathogenicity features. Ann Parasitol. 2015;61:1.
- Falgier C, Kegley S, Podgorski H, Heisel T, Storey K, Bendel CM, Gale CA. Candida species differ in their interactions with immature human gastrointestinal epithelial cells. Pediatr Res. 2011 May;69(5 Pt 1):384–389. doi:10.1203/PDR.0b013e31821269d5.
- Richardson JP, Ho J, Naglik JR. Candida–epithelial Interactions. J Fungi. 2018;4(1):22. doi:10.3390/jof4010022.
- Zakikhany K, Naglik JR, Schmidt-Westhausen A, Holland G, Schaller M, Hube B. In vivo transcript profiling of Candida albicans identifies a gene essential for interepithelial dissemination. Cell Microbiol. 2007 Dec;9(12):2938–2954. doi:10.1111/j.1462-5822.2007.01009.x.
- Phan QT, Myers CL, Fu Y, Sheppard DC, Yeaman MR, Welch WH, Ibrahim AS, Edwards JE Jr, Filler SG. Als3 is a Candida albicans invasin that binds to cadherins and induces endocytosis by host cells. PLoS Biol. 2007 Mar;5(3):e64. doi:10.1371/journal.pbio.0050064.
- Ponniah G, Rollenhagen C, Bahn Y-S, Staab JF, Sundstrom P. State of differentiation defines buccal epithelial cell affinity for cross-linking to Candida albicans Hwp1. J Oral Pathol Med. 2007;36(8):456–467. doi:10.1111/j.1600-0714.2007.00565.x.
- Wiesner SM, Bendel CM, Hess DJ, Erlandsen SL, Wells CL. Adherence of yeast and filamentous forms of Candida albicans to cultured enterocytes. Crit Care Med. 2002 Mar;30(3):677–683. doi:10.1097/00003246-200203000-00030.
- Sohn K, Senyürek I, Fertey J, Königsdorfer A, Joffroy C, Hauser N, Zelt G, Brunner H, Rupp S. An in vitro assay to study the transcriptional response during adherence of Candida albicans to different human epithelia. FEMS Yeast Res. 2006;6(7):1085–1093. doi:10.1111/j.1567-1364.2006.00130.x.
- Li X, Yan Z, Xu J. Quantitative variation of biofilms among strains in natural populations of Candida albicans. Microbiology. 2003;149(2):353–362. doi:10.1099/mic.0.25932-0.
- Höfs S, Mogavero S, Hube B. Interaction of <Emphasis Type=“Italic”>Candida albicans</Emphasis> with host cells: virulence factors, host defense, escape strategies, and the microbiota. J Microbiol. 2016 Mar 1;54(3):149–169.
- Kojic EM, Darouiche RO. Candida Infections of Medical Devices. Clin Microbiol Rev. 2004 Apr 1;17(2):255–267. doi:10.1128/CMR.17.2.255-267.2004.
- Sundstrom P. Adhesins in Candida albicans. Curr Opin Microbiol. 1999;2(4):353–357. doi:10.1016/S1369-5274(99)80044-7.
- de Groot PWJ, Bader O, de Boer AD, Weig M, Chauhan N. Adhesins in human fungal pathogens: glue with plenty of stick. Eukaryot Cell. 2013 Apr;12(4):470–481. doi:10.1128/EC.00364-12.
- Hebecker B, Naglik JR, Hube B, Jacobsen ID. Pathogenicity mechanisms and host response during oral Candida albicans infections. Expert Rev Anti Infect Ther. 2014;12(7):867–879. doi:10.1586/14787210.2014.916210.
- Chaffin WL. Candida albicans cell wall proteins. Microbiol Mol Biol Rev. 2008;72(3):495–544. doi:10.1128/MMBR.00032-07.
- Younes S, Bahnan W, Dimassi HI, Khalaf RA. The Candida albicans Hwp2 is necessary for proper adhesion, biofilm formation and oxidative stress tolerance. Microbiol Res. 2011;166(5):430–436. doi:10.1016/j.micres.2010.08.004.
- Sandini S, La Valle R, De Bernardis F, Macrì C, Cassone A. The 65 kDa mannoprotein gene of Candida albicans encodes a putative β-glucanase adhesin required for hyphal morphogenesis and experimental pathogenicity. Cell Microbiol. 2007;9(5):1223–1238. doi:10.1111/j.1462-5822.2006.00862.x.
- Sandini S, Stringaro A, Arancia S, Colone M, Mondello F, Murtas S, Girolamo A, Mastrangelo N, Bernardis FD. The MP65 gene is required for cell wall integrity, adherence to epithelial cells and biofilm formation in Candida albicans. BMC Microbiol. 2011;11(1):106. doi:10.1186/1471-2180-11-106.
- De Bernardis F, Liu H, O’Mahony R, La Valle R, Bartollino S, Sandini S, Grant S, Brewis N, Tomlinson I, Basset RC, et al. Human domain antibodies against virulence traits of Candida albicans inhibit fungus adherence to vaginal epithelium and protect against experimental vaginal candidiasis. J Infect Dis. 2007 Jan 1;195(1):149–157. doi:10.1086/509891.
- Gil-Bona A, Amador-García A, Gil C, Monteoliva L. The external face of Candida albicans: A proteomic view of the cell surface and the extracellular environment. J Proteomics. 2018 May;30(180):70–79. doi:10.1016/j.jprot.2017.12.002.
- Timpel C, Strahl-Bolsinger S, Ziegelbauer K, Ernst JF. Multiple Functions of Pmt1p-mediated ProteinO-Mannosylation in the Fungal Pathogen Candida albicans. J Biol Chem. 1998;273(33):20837–20846. doi:10.1074/jbc.273.33.20837.
- Dalle F, Jouault T, Trinel PA, Esnault J, Mallet JM, d’Athis P, Poulain D, Bonnin A. Beta-1,2- and alpha-1,2-linked oligomannosides mediate adherence of Candida albicans blastospores to human enterocytes in vitro. Infect Immun. 2003 Dec;71(12):7061–7068. doi:10.1128/IAI.71.12.7061-7068.2003.
- Lindberg B, Leontein K, Lindquist U, Svenson SB, Wrangsell G, Dell A, Rogers M. Structural studies of the O-antigen polysaccharide of Salmonella thompson, serogroup C1 (6, 7). Carbohydr Res. 1988;174:313–322. doi:10.1016/0008-6215(88)85100-0.
- Oxley D, Wilkinson SG. Structure of a mannan isolated from the lipopolysaccharide of the reference strain (S3255) for a new serogroup of Serratia marcescens. Carbohydr Res. 1991;212:213–217. doi:10.1016/0008-6215(91)84058-M.
- Poulain D, Jouault T. Candida albicans cell wall glycans, host receptors and responses: elements for a decisive crosstalk. Curr Opin Microbiol. 2004;7(4):342–349. doi:10.1016/j.mib.2004.06.011.
- Masuoka J. Surface glycans of Candida albicans and other pathogenic fungi: physiological roles, clinical uses, and experimental challenges. Clin Microbiol Rev. 2004;17(2):281–310. doi:10.1128/CMR.17.2.281-310.2004.
- Dromer F, Chevalier R, Sendid B, Improvisi L, Jouault T, Robert R, Mallet JM, Poulain D. Synthetic analogues of β-1, 2 oligomannosides prevent intestinal colonization by the pathogenic yeast Candida albicans. Antimicrob Agents Chemother. 2002;46(12):3869–3876. doi:10.1128/AAC.46.12.3869-3876.2002.
- Wächtler B, Citiulo F, Jablonowski N, Förster S, Dalle F, Schaller M, Wilson D, Hube B. Candida albicans-Epithelial Interactions: dissecting the Roles of Active Penetration, Induced Endocytosis and Host Factors on the Infection Process. PLoS ONE [Internet], 2012 May 14 [cited 2015 Nov 10];7(5). http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3351431/.
- Santoni G, Spreghini E, Lucciarini R, Amantini C, Piccoli M. Involvement of αvβ3 integrin-like receptor and glycosaminoglycans in Candida albicans germ tube adhesion to vitronectin and to a human endothelial cell line. Microb Pathog. 2001;31(4):159–172. doi:10.1006/mpat.2001.0459.
- Wächtler B, Wilson D, Haedicke K, Dalle F, Hube B. From Attachment to Damage: defined Genes of Candida albicans Mediate Adhesion, Invasion and Damage during Interaction with Oral Epithelial Cells. PLoS ONE [Internet]. 2011 Feb 23 [cited 2015 Nov 10];6(2). http://www.ncbi.nlm.nih.gov/pmc/articles/PMC3044159/.
- Park H, Myers CL, Sheppard DC, Phan QT, Sanchez AA, E Edwards J, Filler SG. Role of the fungal Ras-protein kinase A pathway in governing epithelial cell interactions during oropharyngeal candidiasis. Cell Microbiol. 2005;7(4):499–510.
- Felk A, Kretschmar M, Albrecht A, Schaller M, Beinhauer S, Nichterlein T, Sanglard D, Korting HC, Schäfer W, Hube B. Candida albicans hyphal formation and the expression of the Efg1-regulated proteinases Sap4 to Sap6 are required for the invasion of parenchymal organs. Infect Immun. 2002;70(7):3689–3700. doi:10.1128/IAI.70.7.3689-3700.2002.
- Gow NA, Brown AJ, Odds FC. Fungal morphogenesis and host invasion. Curr Opin Microbiol. 2002;5:366–371.
- Naglik JR, Challacombe SJ, Hube B. Candida albicans secreted aspartyl proteinases in virulence and pathogenesis. Microbiol Mol Biol Rev. 2003;67(3):400–428. doi:10.1128/MMBR.67.3.400-428.2003.
- Villar CC, Zhao XR. Candida albicans induces early apoptosis followed by secondary necrosis in oral epithelial cells. Mol Oral Microbiol. 2010;25(3):215–225. doi:10.1111/j.2041-1014.2010.00577.x.
- Villar CC, Kashleva H, Nobile CJ, Mitchell AP, Dongari-Bagtzoglou A. Mucosal tissue invasion by Candida albicans is associated with E-cadherin degradation, mediated by transcription factor Rim101p and protease Sap5p. Infect Immun. 2007 May;75(5):2126–2135. doi:10.1128/IAI.00076-06.
- Moreno-Ruiz E, Galán-Díez M, Zhu W, Fernández-Ruiz E, d’Enfert C, Filler SG, Cossart P, Veiga E. Candida albicans internalization by host cells is mediated by a clathrin-dependent mechanism. Cell Microbiol. 2009;11(8):1179–1189. doi:10.1111/j.1462-5822.2009.01319.x.
- Filler SG, Swerdloff JN, Hobbs C, Luckett PM. Penetration and damage of endothelial cells by Candida albicans. Infect Immun. 1995;63:976–983.
- Sun JN, Solis NV, Phan QT, Bajwa JS, Kashleva H, Thompson A, Liu Y, Dongari-Bagtzoglou A, Edgerton M, Filler SG. Host cell invasion and virulence mediated by Candida albicans Ssa1. PLoS Pathog. 2010 Nov 11;6(11):e1001181. doi:10.1371/journal.ppat.1000975.
- Phan QT, Fratti RA, Prasadarao NV, Edwards JE, Filler SG. N-cadherin mediates endocytosis of Candida albicans by endothelial cells. J Biol Chem. 2005;280(11):10455–10461. doi:10.1074/jbc.M412592200.
- Zhu W, Phan QT, Boontheung P, Solis NV, Loo JA, Filler SG. EGFR and HER2 receptor kinase signaling mediate epithelial cell invasion by Candida albicans during oropharyngeal infection. Proc Natl Acad Sci U S A. 2012 Aug 28;109(35):14194–14199. doi:10.1073/pnas.1117676109.
- Moyes DL, Richardson JP, Naglik JR. Candida albicans-epithelial interactions and pathogenicity mechanisms: scratching the surface. Virulence. 2015 May 19;6(4):338–346. doi:10.1080/21505594.2015.1010982.
- Rollenhagen C, Wöllert T, Langford GM, Sundstrom P. Stimulation of cell motility and expression of late markers of differentiation in human oral keratinocytes by Candida albicans. Cell Microbiol. 2009 Jun;11(6):946–966. doi:10.1111/j.1462-5822.2009.01303.x.
- Frank CF, Hostetter MK. Cleavage of E-cadherin: a mechanism for disruption of the intestinal epithelial barrier by Candida albicans. Transl Res J Lab Clin Med. 2007 Apr;149(4):211–222. doi:10.1016/j.trsl.2006.11.006.
- Gumbiner BM. Cell Adhesion: the Molecular Basis of Tissue Architecture and Morphogenesis. Cell. 1996 Feb 9;84(3):345–357. doi:10.1016/S0092-8674(00)81279-9.
- Banan A, Choudhary S, Zhang Y, Fields JZ, Keshavarzian A. Ethanol-induced barrier dysfunction and its prevention by growth factors in human intestinal monolayers: evidence for oxidative and cytoskeletal mechanisms. J Pharmacol Exp Ther. 1999;291:1075–1085.
- Muskett H, Shahin J, Eyres G, Harvey S, Rowan K, Harrison D. Risk factors for invasive fungal disease in critically ill adult patients: a systematic review. Crit Care. 2011;15(6):R287. doi:10.1186/cc10324.
- Ortega M, Marco F, Soriano A, Almela M, Martínez JA, López J, Pitart C, Mensa J. Candida species bloodstream infection: epidemiology and outcome in a single institution from 1991 to 2008. J Hosp Infect. 2011;77(2):157–161. doi:10.1016/j.jhin.2010.09.026.
- Moyes DL, Wilson D, Richardson JP, Mogavero S, Tang SX, Wernecke J, Höfs S, Gratacap RL, Robbins J, Runglall M, et al. Candidalysin is a fungal peptide toxin critical for mucosal infection. Nature. 2016;532(7597):64–68. doi:10.1038/nature17625.
- Wilson D, Naglik JR, Hube B. The Missing Link between Candida albicans Hyphal Morphogenesis and Host Cell Damage. PLoS Pathog. 2016 Oct;12(10):e1005867. doi:10.1371/journal.ppat.1005867.
- Casadevall A, Pirofski L. Microbiology: ditch the term pathogen. Nature. 2014 Dec 11;516(7530):165–166. doi:10.1038/516165a.
- Kernéis S, Bogdanova A, Kraehenbuhl J-P, Pringault E. Conversion by Peyer’s patch lymphocytes of human enterocytes into M cells that transport bacteria. Science. 1997;277(5328):949–952. doi:10.1126/science.277.5328.949.
- Gullberg E, Leonard M, Karlsson J, Hopkins AM, Brayden D, Baird AW, Artursson P. Expression of specific markers and particle transport in a new human intestinal M-cell model. Biochem Biophys Res Commun. 2000;279(3):808–813. doi:10.1006/bbrc.2000.4038.
- Des Rieux A, Fievez V, Théate I, Mast J, Préat V, Schneider Y-J. An improved in vitro model of human intestinal follicle-associated epithelium to study nanoparticle transport by M cells. Eur J Pharm Sci. 2007;30(5):380–391. doi:10.1016/j.ejps.2006.12.006.
- Hector RF, Yee E, Collins MS. Use of DBA/2N mice in models of systemic candidiasis and pulmonary and systemic aspergillosis. Infect Immun. 1990 May;58(5):1476–1478.
- Papadimitriou JM, Ashman RB. The pathogenesis of acute systemic candidiasis in a susceptible inbred mouse strain. J Pathol. 1986;150(4):257–265. doi:10.1002/path.1711500408.
- MacCallum Donna M, Odds Frank C. Temporal events in the intravenous challenge model for experimental Candida albicans infections in female mice. Mycoses. 2005 Apr 13;48(3):151–161. doi:10.1111/myc.2005.48.issue-3.
- Spellberg B, Ibrahim AS, Edwards Jr. JE, Filler SG. Mice with Disseminated Candidiasis Die of Progressive Sepsis. J Infect Dis. 2005 Jul 15;192(2):336–343. doi:10.1086/jid.2005.192.issue-2.
- Samonis G, Anaissie EJ, Rosenbaum B, Bodey GP. A model of sustained gastrointestinal colonization by Candida albicans in healthy adult mice. Infect Immun. 1990 Jan 6;58(6):1514–1517.
- Clemons KV, Gonzalez GM, Singh G, Imai J, Espiritu M, Parmar R, Stevens DA. Development of an Orogastrointestinal Mucosal Model of Candidiasis with Dissemination to Visceral Organs. Antimicrob Agents Chemother. 2006 Jan 8;50(8):2650–2657. doi:10.1128/AAC.00530-06.
- Koh AY, Köhler JR, Coggshall KT, Van Rooijen N, Pier GB. Mucosal damage and neutropenia are required for Candida albicans dissemination. PLoS Pathog. 2008 Feb 8;4(2):e35. doi:10.1371/journal.ppat.0040035.
- Biasoli MS, Tosello ME, Luque AG, Magaró HM. Adherence, colonization and dissemination of Candida dubliniensis and other Candida species. Med Mycol. 2010 Mar 1;48(2):291–297. doi:10.3109/13693780903114942.
- de Repentigny L. Animal models in the analysis of Candida host–pathogen interactions. Curr Opin Microbiol. 2004;7(4):324–329. doi:10.1016/j.mib.2004.06.001.
- Oswald IP. Role of intestinal epithelial cells in the innate immune defence of the pig intestine. Vet Res. 2006;37(3):359–368. doi:10.1051/vetres:2006006.
- Förster TM, Mogavero S, Dräger A, Graf K, Polke M, Jacobsen ID, Hube B. Enemies and brothers in arms: candida albicans and gram-positive bacteria. Cell Microbiol. 2016 Dec 1;18(12):1709–1715. doi:10.1111/cmi.12570.